Abstract
Background:
Multiple System Atrophy is a rare neurodegenerative disease with alpha-synuclein aggregation in glial cytoplasmic inclusions and either predominant olivopontocerebellar atrophy or striatonigral degeneration, leading to dysautonomia, parkinsonism, and cerebellar ataxia. One prior genome-wide association study in mainly clinically diagnosed patients with Multiple System Atrophy failed to identify genetic variants predisposing for the disease.
Objective:
Since the clinical diagnosis of Multiple System Atrophy yields a high rate of misdiagnosis when compared to the neuropathological gold standard, we studied only autopsy-confirmed cases.
Methods:
We studied common genetic variations in Multiple System Atrophy cases (N = 731) and controls (N = 2898).
Results:
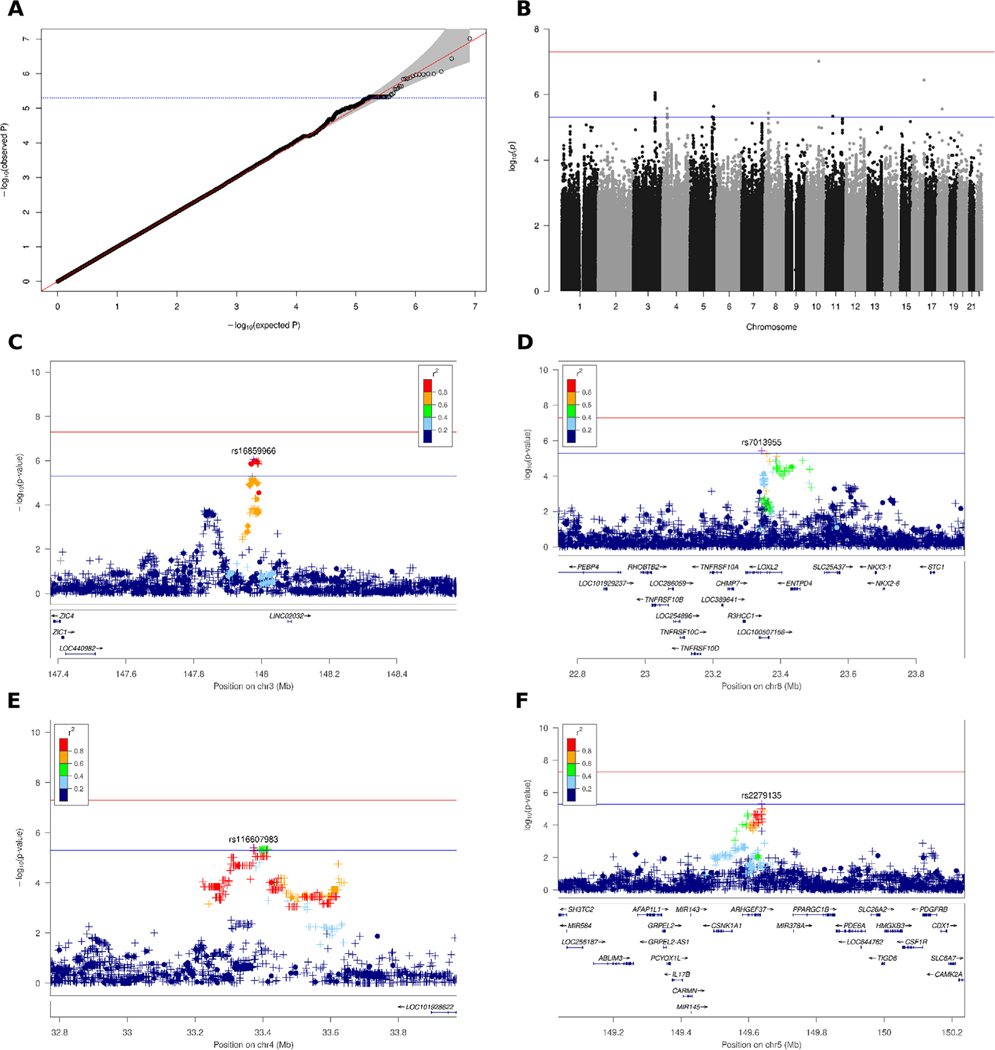
The most strongly disease-associated markers were rs16859966 on chromosome 3, rs7013955 on chromosome 8, and rs116607983 on chromosome 4 with P-values below 5 × 10−6, all of which were supported by at least one additional genotyped and several imputed single nucleotide polymorphisms. The genes closest to the chromosome 3 locus are ZIC1 and ZIC4 encoding the zinc finger proteins of cerebellum 1 and 4 (ZIC1 and ZIC4).
Interpretation:
Since mutations of ZIC1 and ZIC4 and paraneoplastic autoantibodies directed against ZIC4 are associated with severe cerebellar dysfunction, we conducted immunohistochemical analyses in brain tissue of the frontal cortex and the cerebellum from 24 Multiple System Atrophy patients. Strong immunohistochemical expression of ZIC4 was detected in a subset of neurons of the dentate nucleus in all healthy controls and in patients with striatonigral degeneration, whereas ZIC4-immunoreactive neurons were significantly reduced inpatients with olivopontocerebellar atrophy. These findings point to a potential ZIC4-mediated vulnerability of neurons in Multiple System Atrophy.
Keywords: multiple system atrophy, genome-wide association study, autopsy-confirmed, ZIC1, ZIC4
Multiple system atrophy (MSA) is a rapidly progressive rare neurodegenerative disease presenting with variable combinations of dysautonomia, parkinsonism, and cerebellar ataxia.1 Two forms of MSA can be clinically distinguished, characterized by either predominant parkinsonism or predominant cerebellar symptoms.2 Its estimated prevalence is 3.4–4.9 cases per 100,000 individuals in the general population, and 7.8 cases per 100,000 in persons older than 40 years.3 The mean survival time from disease onset is 6–10 years.4,5 Currently, only limited symptomatic treatments and no disease-modifying therapies are available.6
The typical symptoms of MSA are caused by the progressive degeneration of neurons in different brain regions, particularly in the substantia nigra, striatum, inferior olivary nucleus, pons, and cerebellum, but also other parts of the central nervous systems, emphasizing the multisystem character of MSA.2,7 The histological hallmarks in brains of patients with MSA are glial cytoplasmic inclusions (Papp–Lantos bodies) in oligodendrocytes containing aggregated and misfolded α-synuclein.8 Neuropathologically, two subtypes can be distinguished, one with predominant olivopontocerebellar atrophy (OPCA), the other with mainly striatonigral degeneration (SND).9,10 In addition, a mixed phenotype displaying features of both OPCA and SND is found in the brains of some patients.9,10
The pathogenesis of MSA is unclear. MSA is considered a sporadic disease.11 Epidemiological studies have investigated the influence of environmental factors in MSA, including exposure to farming-related factors (pesticides, solvents, mycotoxins, dust, fuels, oils, fertilizers, animals) and certain lifestyles (consumption of well water, rural living, diet, and physical activity).12–14 Apart from a marginal effect of pesticides, no other environmental factors have been convincingly associated with an increased risk for development of MSA.12−14
Hypothesis-driven candidate gene studies have been inconsistent with respect to variants that might be associated with MSA. Associations of MSA with the genes COQ2, SNCA, MAPT, and PRNP have been discussed.15–20 One prior genome-wide association study (GWAS) did not identify hits of statistical significance at a genome-wide level, despite the analysis of 918 cases and 3864 controls.21 This GWAS had mainly included clinically diagnosed MSA cases. It needs to be stressed that clinical diagnosis is frequently not accurate in MSA. For example, a recent clinicopathological study demonstrated a false-positive diagnosis at autopsy in 38% of patients with clinically diagnosed MSA.22
To avoid inclusion of misdiagnosed patients in the GWAS described in this study, we included only autopsy-confirmed cases and appropriate ethnicity-matched controls.
Subjects and Methods
Patient Recruitment
Ethical approval had been obtained from all responsible ethics committees. All participants had given written consent.
Neuropathologists at each recruitment site (Table 1) based the definite neuropathological diagnosis of MSA on histopathological criteria, taking into account glial cytoplasmic inclusions immunoreactive for α-synuclein in characteristic anatomical distribution as a defining feature of MSA.23 Age, sex, disease history (including disease onset and duration), and neuropathological findings were recorded in a standardized manner for all cases.
TABLE 1.
Recruitment centers and brain bank sources
| City, Country | Source | MSA cases |
|---|---|---|
|
| ||
| Zürich, Switzerland | Institute of Neuropathology, University Hospital of Zurich, Zurich, Switzerland | 1 |
| Göttingen, Germany | University Medical Center Göttingen, Department of Neurology and Paracelsus-Elena-Klinik, 34,128 Kassel, Germany | 2 |
| San Francisco, CA, USA | University of California, San Francisco, CA, USA | 2 |
| Vancouver, BC, Canada | University of British Columbia, Department of Pathology and Laboratory Medicine | 2 |
| New York, NY, USA | Mount Sinai NBTR | 3 |
| Atlanta, GA, USA | Emory University, Department of Neurology & Pathology | 4 |
| Los Angeles, CA, USA | The Human Brain and Spinal Fluid Resource Center | 4 |
| Stockholm, Sweden | Department of Neurology, Karolinska University Hospital, Stockholm, Sweden | 4 |
| Vienna, Austria | Institute of Neurology, Medical University of Vienna | 6 |
| Newcastle upon Tyne, UK | Newcastle Brain Tissue Resource, Newcastle University, Campus for Ageing and Vitality, Newcastle upon Tyne NE4 5PL, UK | 7 |
| Chicago, IL, USA | University of Chicago, Department of Neurology | 8 |
| Indiana, IN, USA | Indiana University School of Medicine | 8 |
| San Diego, CA, USA | San Diego Shiley-Marcos AD Research Center, University of California | 8 |
| Tübingen, Germany | Department of Neuropathology, University Hospital of Tübingen, Tübingen, Germany | 8 |
| Madrid, Spain | Centro de Biología Molecular “Severo Ochoa,” c/Nicolás Cabrera, 1, Universidad Autónoma de Madrid, Cantoblanco, Madrid, Spain | 10 |
| Seattle, WA, USA | Department of Pathology, University of Washington, Seattle, WA, USA | 10 |
| Prague, Czech Republic | Department of Pathology and Molecular Medicine, Thomayer University Hospital, Prague | 12 |
| Sydney, NSW, Australia | Brain and Mind Centre, Sydney Medical School, The University of Sydney, Sydney, NSW, Australia | 12 |
| Arizona, AZ, USA | Banner Sun Health Research Institute | 13 |
| Parkville, VIC, Australia | Australian Brain Bank Network, Howard Florey Laboratories, The Florey Institute of Neuroscience and Mental Health | 13 |
| Dallas, TX, USA | Alzheimer’s Disease Center, University of Texas Southwestern Medical Center, Dallas, Texas, USA | 15 |
| Rosthern, SK, Canada | Saskatoon Health Region/University of Saskatchewan, Rosthern; and Movement Disorders | 17 |
| Paris, France | Raymond Escourolle Neuropathology Department, Groupe Hospitalier Pitié-Salpêtrière, Paris, France | 20 |
| London, UK | Imperial College London | 22 |
| Baltimore, MD, USA | Johns Hopkins Medical Institution Brain Resource Center, MD, USA | 24 |
| London, UK | MRC London Neurodegenerative Diseases Brain Bank, Institute of Psychiatry, Psychology and Neuroscience, King’s College | 26 |
| Munich, Germany | Neurobiobank Munich, Center for Neuropathology and Prion Research, Ludwig-Maximilians University | 29 |
| Boston, MA, USA | Massachusetts General Hospital | 30 |
| Barcelona, Spain | Neurological Tissue Bank of the Biobanc-Hospital Clinic-IDIBAPS | 34 |
| Amsterdam, the Netherlands | Alzheimer Center | 36 |
| Ann Arbor, MI, USA | University of Michigan, Department of Pathology, University of Michigan Medical School, Ann Arbor, MI, USA | 37 |
| Miami, FL, USA | UM Brain Endowment Bank, an NIH NeuroBioBank | 45 |
| Philadelphia, PA, USA | The Penn FTD Center — University of Pennsylvania, USA | 54 |
| Jacksonville, FL, USA | Department of Neuroscience, Mayo Clinic, Jacksonville | 205 |
| Total | 731 | |
MSA, multiple system atrophy.
Controls were ethnically matched to cases and either derived from biobanks KORA-gen24 or popGen25 (Europe sites) or from a North American site (Alzheimer’s Disease Genetics Consortium).26 The Alzheimer’s Disease Genetics Consortium assembled and genotyped DNA from subjects enrolled in the 29 NIA-Alzheimer’s Disease Centers located across the United States. For this study, the Alzheimer’s Disease Genetics Consortium provided a subset of mostly clinical, cognitively normal controls. Patients and controls were of North-Western European and African American ancestry.
DNA Extraction
We isolated DNA from 30 mg frozen cerebellar cortex using QIAamp DNA Mini Kit (Qiagen, Venlo, the Netherlands). DNA extraction was performed at German Center for Neurodegenerative Diseases (DZNE, Munich, Germany). DNA was stored at −80°C until use. DNA concentration was measured using a NanoDrop Spectro-photometer. DNA quality was determined by gel electrophoresis.
Genotyping
All samples were genotyped on Infinium Global Screening Arrays (Illumina, San Diego, CA, USA). The cases were genotyped at the Institute of Clinical Molecular Biology, Kiel University, Germany. The samples were genotyped in one batch on array version 2.0 for cases and version 1.0 for controls. Genotypes were called using Illumina Genome studio according to the manufacturer’s instructions using in-house cluster files.
Quality Control and Imputation
We used PLINK (v. 1.9) [1] and R (v. 3.6.3)27 for all analyses. Only variants successfully genotyped in both the patient and the control populations were included in the subsequent analyses. Variants with multicharacter allele codes, insertions, deletions, duplicated markers, and all A/T and G/C variants were excluded. We excluded all samples discordant between reported and genotypic sex. Missing sex was imputed, and samples with ambiguously imputed sex were discarded. After a first step of filtering out samples and variants with call rate of less than 85%, we excluded variants with an individual call rate of less than 98% in a second filtering step. Next, we removed variants with a minor allele frequency <0.01, a significant deviation from Hardy–Weinberg equilibrium (P < 1 × 10−6) in controls, or informative missingness (P < 1 × 10−5). Subsequently, we excluded individuals with a variant call rate of <98% or an outlying heterozygosity rate (mean ± 3 standard deviations). We used a pruned dataset containing only markers in low linkage-disequilibrium regions (pairwise r2 < 0.2) to test for duplicated individuals and cryptic relatedness (Pihat > 0.125) using pairwise genome-wide estimates of the proportion of identity by descent. For each detected sample pair we excluded the individual with a lower call rate. Ethnical outliers were identified by a principal-component analysis (PCA) together with the publically available 1000 Genomes data with known ethnicities.28 Because the study population has genetically a mainly European ancestry, as ascertained by the PCA, we determined a European center and excluded samples more than 1.5 times the maximal European Euclidean distance away from this center. After a first association analysis of genotyped single nucleotide polymorphisms (SNPs) only, we inspected visually the cluster plots of all variants with a P value <1 × 10−5 and discarded variants without adequate cluster separation. Imputation was carried out on the quality-assured dataset using the TOPMed Imputation Server, which employs Eagle2 for phasing and minimac4 for the imputation of genotypes.29,30 The most likely genotype is used in downstream analyses. Variants were again filtered for minor allele frequency and deviation from Hardy–Weinberg equilibrium in controls with the same thresholds as before. In addition, SNPs with an imputation quality score R2 < 0.7 were excluded, leaving 8,131,900 variants for analyses. As a final step of the quality-control procedure, we used the R package PCAmatchR to ethnically match cases to controls with a 1:4 ratio to overcome possible difficulties with population stratification, leading to 3240 individuals for the analyses.31
Association Analysis
We used logistic regression to test the additive genetic model of each marker for association with disease status. Following scree plot analysis, we incorporated the first two dimensions of the PCA and sex as covariates. We used a genome-wide significance threshold of P < 5 × 10−8 and the threshold of P < 5 × 10−6 for suggestive association. Conditional analyses, including, in turn, each SNP with a suggestive association as additional covariate, were conducted to identify adjacent independent signals. Furthermore, we tested for clumps of correlated SNPs, ie, to assess how many independent loci had been associated, and determined the number of variants supporting the lead SNP at each locus, ie, variants with P values less than the clumping threshold of 5 × 10−5 are in linkage disequilibrium (r2 ≥ 0.4) and not farther than 250 kb away from the respective SNP. Visualization of the results was carried out with R and LocusZoom32 for regional plots. Variant positions in this article are reported on human genome version 38 (GRCh38/hg38).
Immunohistochemistry on MSA Patients’ Brain
Formalin-fixed and paraffin-embedded (FFPE) tissues from patients with MSA and controls without neurological or psychiatric diseases were obtained from the Neurobiobank Munich (Germany). All autopsy cases of the Neurobiobank Munich were collected on the basis of an informed consent according to the guidelines of the ethics commission of the Ludwig-Maximilians-University (Munich, Germany; #345–13). MSA cases had been diagnosed according to established histopathological diagnostic criteria.10,23
For ZIC4 immunohistochemistry, 5-μm-thick sections of FFPE tissues of the frontal cortex and the cerebellar hemisphere, including the dentate nucleus, were prepared. After deparaffinization, heat-induced epitope retrieval was performed in Tris/EDTA, pH 9, at 95°C for 30 minutes. For blocking of endogenous peroxidase and unspecific protein binding, the sections were incubated with 5% H2O2 in methanol for 20 minutes and I-Block reagent (Applied Biosystems, Waltham, MA, USA) for 15 minutes. Subsequently, ZIC4 primary antibody (rabbit, polyclonal; Merck/Sigma-Aldrich, Darmstadt, Germany) was applied overnight at 4°C at a dilution of 1:100. Signal detection was performed using the DCS ChromoLine DAB kit (DCS, Hamburg, Germany) according to the manufacturer’s instructions. Sections were counterstained for 1 minute with Mayer’s hemalum solution (Waldeck, Münster, Germany).
To determine the fractions of ZIC4-positive neurons of all neurons in the dentate nucleus, we scanned stained slides using a slide scanner (Axio Scan. Z1; Zeiss, Oberkochen, Germany) and visualized using the free ZEN lite software (v. 3.3; Zeiss). For statistical evaluation of the data, Student t test was used, and statistical significance was defined as P < 0.05.
Results
Patient Sample
From the initial sample of 731 cases, 13 cases had to be excluded because of insufficient tissue quality. After thorough quality control and filtering, 648 cases and 2592 controls covering 8,131,900 variants were included in the association analysis (Fig. 1). The number of excluded samples and variants in each step of the quality-control procedure is shown in Tables S1 and S2.
FIG. 1.
Flowchart sample quality control. SD, standard deviation. [Color figure can be viewed at wileyonlinelibrary.com]
Association Results
We performed logistic regression incorporating sex and determined the first two dimensions of PCA as covariates using the scree plot method. The genomic inflation factor of λ = 1.01 (unimputed λ = 1.01; Fig. S1) indicates that no significant population stratification was present (Fig. 2A). We did not identify any disease-associated variants with a P value less than the genome-wide significance threshold of P < 5 × 10−8, but suggestive associations with P < 5 × 10−6 at 10 different loci (Fig. 2B) with the leading SNP at each locus shown in Table 2. Conditional analyses, including, in turn, any SNP with P < 5 × 10−6, excluded the presence of multiple independent signals at each locus. All variants with suggestive associations are listed in Table S3. The most noteworthy hits were rs16859966 on chromosome 3 (P = 8.6 × 10−7; odds ratio [OR], 1.58; 95% confidence interval [CI]: 1.32–1.89), rs7013955 on chromosome 8 (P = 3.7 × 10−6; OR, 1.8; 95% CI: 1.40–2.31), and rs116607983 on chromosome 4 (P = 4.0 × 10−6; OR, 2.93; 95% CI: 1.86–4.63), which were supported by at least one additional genotype, as well as several imputed SNPs with P values less than the clumping threshold of 5 × 10−5 as discovered in the clumping analysis (Table 2). The genes closest to the chromosome 3 locus are the Long Intergenic Non-Protein Coding RNA 2032 (LINC02032) approximately 100 kb downstream and the zinc-finger proteins of cerebellum 1 and 4 genes (ZIC1, ZIC4), located roughly 600 kb upstream (Fig. 2E). The top SNP rs7013955 on chromosome 8 maps to the lysyl oxidaselike 2 gene (LOXL2; Fig. 2D). The association signal around SNP rs116607983 on chromosome 4 is located in a region devoid of protein-coding genes approximately 2000 kb to either side (Fig. 2E). A fourth locus on chromosome 5 (rs2279135) was also supported by multiple clumped SNPs, but all SNPs, including the lead SNP, were imputed (Table 2). Several variants clumped at the chromosome 5 locus were located in the ARHGEF37 gene, coding for Rho Guanine Nucleotide Exchange Factor 37 (Fig. 2F). None of the identified SNPs is an expression quantitative trait locus in brain tissues according the Genotype Tissue Expression project.33 At four of the six remaining loci with variants exhibiting suggestive associations, at most two supporting SNPs were present, which were all imputed; in the other two loci, no supporting SNPs could be found in the clumping analysis (Table 2, Fig. S2). We did not investigate these loci further because it is unlikely that they represent valid associations. No significant associations with Bonferroni-adjusted P values were detected with previously reported Parkinson’s disease associations from a meta-analysis of 17 datasets from a Parkinson’s disease GWAS (Table S4).34
FIG. 2.
Association plots for multiple system atrophy (MSA). (A) QQ (quantile-quantile) plot based on 8,109,760 variants after imputation. (B) Manhattan plot showing –log10 P values from logistic regression on imputed variants with sex and two principal components as covariates plotted against their chromosomal position. The red and blue lines indicate the genome-wide significance threshold of 5 × 10–8 and threshold for suggestive associations of 5 × 10–6, respectively. (C) Regional plot for the association between MSA and variants on chromosome 3 in the genomic region from 147.4 to 148.6 Mb. A circle represents a genotyped variant and a plus symbol an imputed variant. The r2 metric displays the pairwise linkage-disequilibrium (LD) between the leading and the respective variant. The bottom part shows gene positions. (D) Regional plot for associations on chromosome 8 in the genomic region from 22.7 to 23.9 Mb. (E) Regional plot for associations on chromosome 4 in the genomic region from 32.8 to 34.0 Mb. (F) Regional plot for associations on chromosome 5 in the genomic region from 149.0 to 150.2 Mb. [Color figure can be viewed at wileyonlinelibrary.com]
TABLE 2.
Top SNPs at each locus with P < 5 × 10−6
| MAF | No. of SNPs in Clump | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| CHR | dbSNP ID | BP | Minor allele | Cases | Controls | OR (95% CI) | P | IM/GT | Total | GT | IM |
|
| |||||||||||
| 10 | rs4933352 | 85,280,795 | G | 0.42 | 0.52 | 0.71 (0.62–0.80) | 9.7E-08 | IM | 2 | 0 | 2 |
| 16 | rs79418449 | 80,515,374 | C | 0.04 | 0.02 | 2.54 (1.77–3.63) | 3.7E-07 | GT | 1 | 1 | 0 |
| 3 | rs16859966 | 147,976,678 | G | 0.17 | 0.12 | 1.58 (1.32–1.89) | 8.6E-07 | IM | 45 | 24 | 21 |
| 5 | rs114019803 | 159,559,041 | T | 0.02 | 0.01 | 3.36 (2.03–5.56) | 2.3E-06 | IM | 3 | 0 | 3 |
| 4 | rs933953 | 31,356,173 | C | 0.25 | 0.32 | 0.71 (0.62–0.82) | 2.6E-06 | IM | 1 | 0 | 1 |
| 18 | rs116914137 | 30,589,500 | A | 0.05 | 0.02 | 2.17 (1.57–3.00) | 2.8E-06 | IM | 3 | 0 | 3 |
| 8 | rs7013955 | 23,343,590 | A | 0.08 | 0.05 | 1.80 (1.40–2.31) | 3.7E-06 | IM | 20 | 1 | 19 |
| 4 | rs116607983 | 33,372,461 | A | 0.03 | 0.01 | 2.93 (1.86–4.63) | 4.0E-06 | IM | 88 | 3 | 85 |
| 11 | rs141819348 | 47,698,235 | T | 0.05 | 0.03 | 2.10 (1.53–2.88) | 4.6E-06 | IM | 3 | 0 | 3 |
| 5 | rs2279135 | 149,637,742 | C | 0.32 | 0.27 | 1.39 (1.21–1.60) | 4.8E-06 | IM | 24 | 0 | 24 |
Results from an association analysis with logistic regression including sex and the first two dimensions of principal-component analysis (PCA) as covariates in 648 cases with MSA and 2898 controls. For an OR < 1, the minor allele has a protective effect, whereas an OR > 1 indicates that the minor allele is associated with an increased risk for development of the disease. Only the leading SNP at each locus with a suggestive association between the disease status and a variant is reported. Table S3 lists all suggestive associations.
SNP, single-nucleotide polymorphism; CHR, chromosome; dbSNP, database of single-nucleotide polymorphism; BP, base-pair coordinates according to human reference genome GRCh38; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval; IM, imputed; GT, genotyped.
ZIC4 Immunohistochemistry on MSA Patients’ Brain
ZIC4 and ZIC1 are known to play a critical role in the embryonal development of the cerebellum. Heterozygous deletions comprising the ZIC1 and ZIC4 locus have been associated with the Dandy–Walker malformation, a rare congenital condition characterized by a hypoplastic cerebellar vermis and an enlarged fourth ventricle.35,36 In mice, deletions of ZIC1 and ZIC4 lead to a striking phenotype similar to the Dandy–Walker malformation with cerebellar hypoplasia and foliation defects.35,36 In addition, paraneoplastic autoantibodies against ZIC4 protein are linked to severe cerebellar dys-function and degeneration.37,38
Because cerebellar degeneration and corresponding symptoms are also a central hallmark of MSA, we decided to follow up on a potential role of ZIC4 in MSA patient brains by performing immunohistochemical stainings. For ZIC1, no primary antibody was appropriately sensitive and specific on human tissue in our hands. Thus, FFPE tissues of the cerebellum and, for comparison, the frontal cortex of patients with MSA (n = 10 SND, n = 14 OPCA/mixed phenotype) and healthy controls (b = 5) were stained with antibodies raised against ZIC4.
Nuclear and cytoplasmic staining of frontal cortex neurons was observed in all brains examined without differences between healthy controls and patients with MSA (Fig. 3A–C). In the cerebellar dentate nucleus, we found strong expression of ZIC4 in a subset of neurons in healthy controls, as well as patients with MSA with predominant SND (Fig. 3D,E,G,H). In contrast, patients with MSA with mixed subtype or OPCA showed reduced numbers of ZIC4-positive neurons, which were furthermore only weakly stained (Fig. 3F,I). Quantification of the proportions of ZIC4-positive neurons among the total number of dentate nucleus neurons depicted relatively constant proportions in healthy controls and patients with MSA-SND (33.2% ± 0.0% vs 32.6% 0.0%), whereas in patients with MSA-OPCA or MSA-mixed phenotype, we found significantly lower percentages of ZIC4-positive neurons (15.5% ± 0.1%) (Fig. 3J).
FIG. 3.

ZIC4 immunohistochemical staining of multiple system atrophy (MSA) patients and control brains. Representative ZIC4 immunohistochemical stainings of different brain regions (antibodies binding specifically to antigens in biological tissues, eg, brain tissue) of a control without neurodegenerative disease (A, D, G) and two MSA patients with striatonigral degeneration (SND) (B, E, H) and mixed subtype (C, F, I), respectively. (A–C) Nuclear and cytoplasmic expression of ZIC4 (brown staining) was detected in a comparable manner in the frontal cortex of healthy controls and patients with MSA. In the cerebellar dentate nucleus (dotted lines in D–I) of healthy controls and patients with SND, a constant subset of neurons stained strongly positive for ZIC4, whereas in patients with olivopontocerebellar atrophy (OPCA) or mixed subtype, only weak staining could be observed, and the number of ZIC4-positive neurons was clearly reduced (D–I, with higher magnification in G–I). (J) Quantification of ZIC4-immunoreactive neurons in relation to the total number of neurons of the dentate nucleus depicted on the entire slide showed significantly reduced fractions of ZIC4-immunoreactive neurons in patients with either mixed subtype (light blue) or OPCA (dark blue) compared with SND or controls without neurodegenerative disease, while no difference was seen between patients with SND and healthy controls. Scale bars: 100 μm (A–C), 200 μm (D–F), 50 μm (G–I). [Color figure can be viewed at wileyonlinelibrary.com]
Discussion
As part of the study, brain banks were contacted worldwide, and all available white MSA brains were included. As in the prior GWAS with 918 predominantly clinically diagnosed MSA patients, our current GWAS of 648 patients with autopsy-confirmed MSA did not identify disease-associated common variants less than the genome-wide significance threshold. Previously, hypothesis-driven candidate gene studies found inconsistent results for genetic variants and genes potentially associated with MSA. An association of MSA with genetic variants in COQ2, SNCA, MAPT, and PRNP had been discussed.16–20,39 However, these genes have not been convincingly confirmed in other candidate gene studies and have not been associated in a previous MSA GWAS.21 This preceding GWAS analyzed 918 mostly clinical cases and 3864 controls. Overall, this GWAS did not identify any genome-wide significant hits. Because our prior GWAS of 219 patients with autopsy-confirmed corticobasal degeneration did identify significant disease-associated common variants, our current findings strongly suggest that the genetic contribution to disease risk is smaller in MSA.40
Nevertheless, our study demonstrates several suggestive associations at different loci, which may provide relevant hypotheses for follow-up investigations into the pathogenesis of MSA.
Specifically, we identified a variant on chromosome 3 (rs16859966; P = 8.6 × 10−7; OR, 1.58; 95% CI: 1.32–1.89) located upstream of ZIC1 and ZIC4. ZIC1 and ZIC4 are located in close genomic proximity to each other and encode transcription factors highly expressed in different brain areas.41,42
Proper function of these proteins is critical for the development of the CNS, particularly the cerebellum.36 Although no effect of rs16859966 on ZIC1 or ZIC4 expression is recorded in the Genotype Tissue Expression database, rare genetic variants or deletions in ZIC1 or ZIC4 result in congenital cerebellar defects.35,36,43 A heterozygous deletion of ZIC1 and ZIC4 causes the Dandy–Walker malformation, a developmental disorder of the cerebellum.35,44 Remarkably, two recent epigenomic analyses in brain tissue of MSA point to ZIC4.45,46 Moreover, paraneoplastic autoantibodies against ZIC4 induce cerebellar degeneration.38 Due to the pronounced cerebellar degeneration in MSA, we followed up on a possible role of ZIC4 In MSA.
Although we could detect a relatively constant proportion of approximately one-third ZIC4-positive neurons among all neurons in the cerebellar dentate nucleus in healthy controls and patients with MSASND, cases with MSA-OPCA or the mixed MSA phenotype showed significantly lower fractions of ZIC4-positive neurons. This finding suggests that ZIC4 may be involved in the neurodegeneration in MSA. The involvement of ZIC4 mutations in the Dandy–Walker cerebellar malformation and the paraneoplastic ZIC4 autoantibody–associated cerebellar degeneration could suggest a pathomechanism in MSA, by which altered ZIC4 expression could increase neuronal vulnerability. Further analyses of a potential functional interaction of α-synuclein and ZIC4 are currently ongoing.
Explorative analysis of PD-related associations identified by GWAS yielded no significant association in MSA when adjusting for multiple testing. However, for unadjusted P values, five SNPs reached a significance threshold of P< 0.05,which might be interesting to study further.
This study has a major limitation. Typically, a GWAS is conceptualized as a two-stage design with a discovery stage and a replication stage and supposedly achieving “genome-wide significance” in the discovery stage. The P values in the replication stage should remain significant after Bonferroni correction. Due to the limited number of autopsy-confirmed MSA cases worldwide, we could not conduct a two-stage procedure, let alone a further independent replication. In view of the aforementioned diagnostic uncertainty in clinical cases, a replication in predominantly clinically diagnosed MSA cases did not seem desirable.
Therefore, we strongly encourage bringing MSA cases to autopsy and conducting a further independent replication study to confirm or refute the hypotheses provided by our study.
Supplementary Material
Acknowledgments:
We thank all those who contributed toward our research, particularly the patients and families who donated brain tissue—without their donation this study would not have been possible. We thank Vanessa Boll and Lena Jaschkowitz for excellent technical assistance. This study received support from the Deutsche Parkinson Gesellschaft; the Else-Kröner-Fresenius-Stiftung; the CurePSP foundation; the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site, Alzheimer’s Disease Genetics Consortium (ADGC) National Institute on Aging (NIA) grant U01AG032984; the National Alzheimer’s Coordinating Center (NACC) NIA grant U01 AG016976; the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy, ID 390857198); the Hannover Cluster RESIST (EXC 2155, ID 390874280) and the Kiel Cluster (EXC2167, Precision Medicine in Chronic Inflammation [PMI]); DFG grant HO2402/18-1; VolkswagenStiftung (Niedersächsisches Vorab); Petermax-Müller Foundation (Etiology and Therapy of Synucleinopathies and Tauopathies); ERARE18-124 (MSA-omics) under the frame of E-Rare-3; and the ERA-Net for Research on Rare Diseases. Data for this study were prepared, archived, and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (NIA grant U24-AG041689). Samples from the KORA biobank have been included in this study. The KORA study was initiated and financed by the Helmholtz Zentrum München - German Research Center for Environmental Health, which is supported by the German Federal Ministry of Education and Research (BMBF) and the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Max-imilians-Universität, as part of LMUinnovativ. Samples from the National Cell Repository for Alzheimer’s Disease, which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the NIA, were used in this study. We thank contributors who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible. A part of the samples was collected at the Mayo Clinic. The Mayo Clinic is an American Parkinson Disease Association (APDA) Mayo Clinic Information and Referral Center, an APDA Center for Advanced Research, and is supported by a Lewy Body Dementia Center Without Walls grant U54NS110435 (to D.D. and O.A.R.). The brain bank was supported, in part, by the Mangurian Foundation Lewy Body Dementia Program at Mayo Clinic. The London Neurodegenerative Diseases Brain Bank, King’s College London was supported by the MRC (Medical Research Council, UK) and the Brains for Dementia Research project (jointly funded by the Alzheimer’s Society and Alzheimer’s Research UK). Data were contributed to this study by the Center on Alpha-synuclein Strains in Alzheimer Disease & Related Dementias at the University of Pennsylvania Perelman School of Medicine (grant U19 AG062418 to J.Q.T., principal investigator); the former Morris K. Udall Center at the University of Pennsylvania Perelman School of Medicine (grant P50 NS053488 to J.Q.T., principal investigator); and the NIA (grants P01-AG066597 and AG072979; formerly AG010124). Parts of the samples were provided by the University of Washington Alzheimer’s Disease Research Center (NIH P50 AG005136 and P30 AG066509) and BioRepository and Integrated Neuropathology Laboratory, with support from the Nancy and Buster Alvord Endowment (C.D. K.). Open Access funding enabled and organized by Projekt DEAL.
Footnotes
Relevant conflicts of interest/financial disclosures: Nothing to report.
Supporting Data
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Fanciulli A, Stankovic I, Krismer F, Seppi K, Levin J, Wenning GK. Multiple system atrophy. Int Rev Neurobiol 2019;149:137–192. [DOI] [PubMed] [Google Scholar]
- 2.Gilman S, Wenning GK, Low PA, et al. Second consensus statement on the diagnosis of multiple system atrophy. Neurology 2008;71(9): 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schrag A, Wenning GK, Quinn N, Ben-Shlomo Y. Survival in multiple system atrophy. Mov Disord 2008;23(2):294–296. [DOI] [PubMed] [Google Scholar]
- 4.Klockgether T, Ludtke R, Kramer B, et al. The natural history of degenerative ataxia: a retrospective study in 466 patients. Brain 1998;121(Pt 4):589–600. [DOI] [PubMed] [Google Scholar]
- 5.Wenning GK, Geser F, Krismer F, et al. The natural history of multiple system atrophy: a prospective European cohort study. Lancet Neurol 2013;12(3):264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohrer G, Hoglinger GU, Levin J. Symptomatic therapy of multiple system atrophy. Auton Neurosci 2018;211:26–30. [DOI] [PubMed] [Google Scholar]
- 7.Walsh RR, Krismer F, Galpern WR, et al. Recommendations of the global multiple system atrophy research roadmap meeting. Neurology 2018;90(2):74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jellinger KA, Lantos PL. Papp-Lantos inclusions and the pathogenesis of multiple system atrophy: an update. Acta Neuropathol 2010; 119(6):657–667. [DOI] [PubMed] [Google Scholar]
- 9.Jellinger KA, Seppi K, Wenning GK. Grading of neuropathology in multiple system atrophy: proposal for a novel scale. Mov Disord 2005;20(Suppl 12):S29–S36. [DOI] [PubMed] [Google Scholar]
- 10.Ozawa T, Paviour D, Quinn NP, et al. The spectrum of pathological involvement of the striatonigral and olivopontocerebellar systems in multiple system atrophy: clinicopathological correlations. Brain 2004;127(Pt 12):2657–2671. [DOI] [PubMed] [Google Scholar]
- 11.Fanciulli A, Wenning GK. Multiple-system atrophy. N Engl J Med 2015;372(3):249–263. [DOI] [PubMed] [Google Scholar]
- 12.Chrysostome V, Tison F, Yekhlef F, Sourgen C, Baldi I, Dartigues JF. Epidemiology of multiple system atrophy: a prevalence and pilot risk factor study in Aquitaine, France. Neuroepidemiology 2004;23(4):201–208. [DOI] [PubMed] [Google Scholar]
- 13.Nee LE, Gomez MR, Dambrosia J, Bale S, Eldridge R, Polinsky RJ. Environmental-occupational risk factors and familial associations in multiple system atrophy: a preliminary investigation. Clin Auton Res 1991;1(1):9–13. [DOI] [PubMed] [Google Scholar]
- 14.Vanacore N, Bonifati V, Fabbrini G, et al. Case-control study of multiple system atrophy. Mov Disord 2005;20(2):158–163. [DOI] [PubMed] [Google Scholar]
- 15.Simon-Sanchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 2009;41(12):1308–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schottlaender LV, Houlden H. Multiple-system atrophy brain Bank C. mutant COQ2 in multiple-system atrophy. N Engl J Med 2014; 371(1):81. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Wei QQ, Ou R, et al. Genetic variants of SNCA are associated with susceptibility to Parkinson’s disease but not amyotrophic lateral sclerosis or multiple system atrophy in a Chinese population. PLoS One 2015;10(7):e0133776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbe C, Heckman MG, Lorenzo-Betancor O, et al. MAPT haplotype diversity in multiple system atrophy. Parkinsonism Relat Disord 2016;30:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labbe C, Ogaki K, Lorenzo-Betancor O, et al. Role for the microtubule-associated protein tau variant p.A152T in risk of alpha-synucleinopathies. Neurology 2015;85(19):1680–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronchi D, Di Biase E, Franco G, et al. Mutational analysis of COQ2 in patients with MSA in Italy. Neurobiol Aging 2016;45:213 e211–213 e212. [DOI] [PubMed] [Google Scholar]
- 21.Sailer A, Scholz SW, Nalls MA, et al. A genome-wide association study in multiple system atrophy. Neurology 2016;87(15):1591–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koga S, Aoki N, Uitti RJ, et al. When DLB, PD, and PSP masquerade as MSA: an autopsy study of 134 patients. Neurology 2015; 85(5):404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trojanowski JQ, Revesz T. Neuropathology working group on MSA. Proposed neuropathological criteria for the post mortem diagnosis of multiple system atrophy. Neuropathol Appl Neurobiol 2007;33(6):615–620. [DOI] [PubMed] [Google Scholar]
- 24.Wichmann HE, Gieger C, Illig T, Group MKS. KORA-gen–resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen 2005;67(Suppl 1):S26–S30. [DOI] [PubMed] [Google Scholar]
- 25.Nothlings U, Krawczak M. PopGen. A population-based biobank with prospective follow-up of a control group. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2012;55(6–7):831–835. [DOI] [PubMed] [Google Scholar]
- 26.Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 2019;51(3): 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Team RC. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 28.Auton A, Abecasis GR, Altshuler DM, et al. A global reference for human genetic variation. Nature 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taliun D, Harris DN, Kessler MD, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. Nature 2021; 590(7845):290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loh PR, Danecek P, Palamara PF, et al. Reference-based phasing using the haplotype reference consortium panel. Nat Genet 2016; 48(11):1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown DW, Myers TA, Machiela MJ. PCAmatchR: a flexible R package for optimal case–control matching using weighted principal components. Bioinformatics 2020;37(8):1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G et al. Genetic effects on gene expression across human tissues. Nature 2017;550(7675):204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol 2019;18(12):1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet 2004; 36(10):1053–1055. [DOI] [PubMed] [Google Scholar]
- 36.Blank MC, Grinberg I, Aryee E, et al. Multiple developmental programs are altered by loss of Zic1 and Zic4 to cause Dandy-Walker malformation cerebellar pathogenesis. Development 2011;138(6): 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bataller L, Wade DF, Fuller GN, Rosenfeld MR, Dalmau J. Cerebellar degeneration and autoimmunity to zinc-finger proteins of the cerebellum. Neurology 2002;59(12):1985–1987. [DOI] [PubMed] [Google Scholar]
- 38.Bataller L, Wade DF, Graus F, Stacey HD, Rosenfeld MR, Dalmau J. Antibodies to Zic4 in paraneoplastic neurologic disorders and small-cell lung cancer. Neurology 2004;62(5):778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scholz SW, Houlden H, Schulte C, et al. SNCA variants are associated with increased risk for multiple system atrophy. Ann Neurol 2009;65(5):610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouri N, Ross OA, Dombroski B, et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun 2015;6:7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347(6220): 1260419. [DOI] [PubMed] [Google Scholar]
- 42.Ali RG, Bellchambers HM, Arkell RM. Zinc fingers of the cerebellum (Zic): transcription factors and co-factors. Int J Biochem Cell Biol 2012;44(11):2065–2068. [DOI] [PubMed] [Google Scholar]
- 43.Ogura H, Aruga J, Mikoshiba K. Behavioral abnormalities of Zic1 and Zic2 mutant mice: implications as models for human neurological disorders. Behav Genet 2001;31(3):317–324. [DOI] [PubMed] [Google Scholar]
- 44.Tohyama J, Kato M, Kawasaki S, et al. Dandy-Walker malformation associated with heterozygous ZIC1 and ZIC4 deletion: report of a new patient. Am J Med Genet A 2011;155A(1):130–133. [DOI] [PubMed] [Google Scholar]
- 45.Bettencourt C, Piras IS, Foti SC, et al. Epigenomics and transcriptomics analyses of multiple system atrophy brain tissue supports a role for inflammatory processes in disease pathogenesis. Acta Neuropathol Commun 2020;8(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rydbirk R, Folke J, Busato F, et al. Epigenetic modulation of AREL1 and increased HLA expression in brains of multiple system atrophy patients. Acta Neuropathol Commun 2020;8(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.