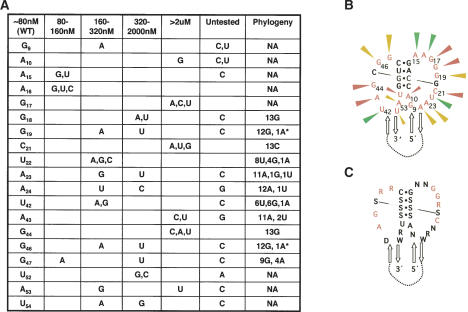
Figure 5.
Mutational analysis of the non-Watson-Crick-paired regions of Δkc2. (A) Indicated nucleotides and substituted mutations for each were assessed for binding to KH2 by filter binding assay. Results are summarized as a comparison to wild-type Δkc2 Kd (80 nM) and grouped by descending Kd. Untested mutations were those in which substitution of the indicated nucleotide at that position resulted in global misfolding of Δkc2 as predicted by mfold 3.1. Phylogeny refers to the number of occurrences of a particular nucleotide out of 13 different sequences (kc1-kc13). NA (not applicable) means those nucleotides are from the fixed regions of the selected clones and are invariant. Asterisks denote the G19-G46 pair that covaries with one A19-A46 base pair. (B) Δkc2 RNA ligand redrawn to focus on loop sequences. Probable non-Watson-Crick-paired nucleotides (tested in A) are shown in red, Watson-Crick interacting nucleotides are shown in black, and 5′ and 3′ stems are shown as anti-parallel arrows. Nucleotides in which mutational analysis showed essentially no tolerance for mutation and phylogenetic variation are indicated by red arrowheads, those with some tolerance for mutation and phylogenetic variation are indicated by yellow arrowheads, and those that are most permissive for mutation and phylogenetic variation are indicated by green arrowheads. (C) Consensus of Δkc2 RNA KH2 ligand based upon experimental data. Red nucleotides are those most likely to be involved in KH2 binding. K is a G or U, R is an A or G, W is an A or U, N can be any nucleotide, and S refers to a Watson-Crick base pair.