Abstract
We have previously described rat insulinoma INS-1-derived cell lines with robust or poor glucose-stimulated insulin secretion (GSIS). In the current study, we have further resolved these lines into three classes: class 1, glucose-unresponsive/glucagon-expressing; class 2, glucose-unresponsive/glucagon-negative; and class 3, glucose-responsive/glucagon-negative. The transcription factor Nkx2.2 was expressed with relative abundance of 3.3, 1.0, and 1.0 in class 1, class 2, and class 3 cells, respectively, whereas Nkx6.1 expression had the opposite trend: 1.0, 2.6, and 6.4 in class 1, class 2, and class 3 cells, respectively. In class 1 cells, overexpressed Nkx6.1 suppressed glucagon expression but did not affect the levels of several other prominent beta cell transcription factors. RNA interference (RNAi)-mediated suppression of Nkx6.1 in class 3 cells resulted in a doubling of glucagon mRNA, with no effect on Pdx1 levels, whereas suppression of Pdx1 in class 3 cells caused a 12-fold increase in glucagon transcript levels, demonstrating independent effects of Nkx6.1 and Pdx1 on glucagon expression in beta cell lines. RNAi-mediated suppression of Nkx6.1 expression in class 3 cells also caused a decrease in GSIS from 13.9- to 3.7-fold, whereas suppression of Pdx1 reduced absolute amounts of insulin secretion without affecting fold response. Finally, RNAi-mediated suppression of Nkx6.1 mRNA in primary rat islets was accompanied by a significant decrease in GSIS relative to control cells. In sum, our studies have revealed roles for Nkx6.1 in suppression of glucagon expression and control of GSIS in islet beta cells.
Type 1 and type 2 diabetes are both diseases of insulin deficiency, but with different etiologies. Type 1 diabetes occurs when pancreatic islet beta cells are destroyed by the host immune system. Type 2 diabetes involves loss of key beta cell functions, such as glucose-stimulated insulin secretion (GSIS), and a gradual loss of beta cell mass by nonautoimmune mechanisms. The recent success of a human islet transplant trial in patients with type 1 diabetes (1) has focused fresh attention on cell-based insulin replacement as a method for treating the disease. However, a major obstacle to broad application of this therapeutic approach is the inadequate supply of fully differentiated human islets. In the case of type 2 diabetes, several classes of drugs have been developed for stimulation of insulin secretion, but complications in their use, including loss of efficacy over time and episodic hypoglycemia, have been noted (2). Thus, a clearer understanding of the factors that control the development of beta cells is critical for ultimate success in creation of “surrogate” cells for insulin replacement therapy in type 1 diabetes and for better understanding of potential drug targets for enhancing beta cell function and mass in type 2 diabetes.
Recently, our laboratory described a set of robustly and poorly glucose-responsive subclones of the rat insulinoma cell line INS-1 (3). These lines were procured by stable transfection of the parental INS-1 cell line (4) with a plasmid containing a neomycin resistance gene. In the current study, we have begun to exploit the robustly and poorly glucose responsive INS-1-derived cell lines for the purpose of defining the genes that control key functions of the differentiated beta cell. As a first step in this process, we have used a “candidate genes” approach, involving assay of the expression level of a set of known genes in several independent robustly versus poorly glucose-responsive lines, resulting in identification of three differentially expressed genes: glucagon, Nkx2.2, and Nkx6.1. Experimental manipulation of Nkx6.1 expression in these cell lines demonstrates roles for this transcription factor in the regulation of glucagon expression and control of GSIS.
Materials and Methods
Preparation and Use of Recombinant Adenoviruses. INS-1-derived cell lines were prepared and cultured as described in refs. 3 and 5. The cDNA encoding hamster Nkx6.1 (6) was used to prepare a recombinant adenovirus (AdCMV-Nkx6.1) by following the methods described in refs. 7 and 8. A virus containing the bacterial β-galactosidase gene (AdCMV-βGAL) was used as a control (9). Purified viruses were used to treat cell lines at multiplicities of infection ranging from 10 to 2,500 for 2 h. Assays and analyses were undertaken 24-48 h later.
Small interfering RNA (siRNA) sequences corresponding to rat Nkx6.1 (GAGCACGCTTGGCCTATTC) and rat Pdx1 (GAAAGAGGAAGATAAGAAA) were cloned into vector EH006 and used for construction of Ad-siNkx6.1 and Ad-siPdx1 recombinant adenoviruses by the methods described in refs. 10-12. Adenoviruses containing siRNA sequences corresponding to the Photinus pyralis luciferase gene, GL2 (Ad-siLuc) (11), or a random siRNA sequence (GAGACCCTATCCGTGATTA) (Ad-siRNAcontrol) were used as controls. These viral stocks were used to treat cell lines at an multiplicity of infection of 20 for 18 h. Assays and analyses were undertaken 96 h later.
GSIS: Insulin and Glucagon Content. Cells were grown in culture medium containing 11 mM glucose. Insulin secretion was measured by static incubation as described in ref. 3 after a switch to culture medium containing 5 mM glucose for 12 h. For measurements of insulin and glucagon content, cells were extracted with 1 M acetic acid/0.1% BSA. Media and extract samples were analyzed for insulin or glucagon concentrations with the insulin Coat-a-Count kit (Diagnostic Products, Los Angeles) (13, 14) or a glucagon kit (Linco Research, St. Charles, MO).
Semiquantitative Multiplex PCR and Real-Time PCR Measurements of RNA Levels. Total RNA was isolated and purified by using TRIzol reagent (Invitrogen) or RNeasy (Qiagen, Valencia, CA). Super-Script II (Invitrogen) or iScript (Bio-Rad) was used for first-strand synthesis of cDNA using 0.5-1.0 μg of RNA.
Semiquantitative multiplex PCR was performed as described in ref. 15. Primers were optimized to yield products between 160-280 bp, and reactions were carried out at cycles in the exponential range of product formation (between 16-24 cycles, dependent on target). Standard band/volume analysis using local background correction was used to quantify PCR products. Candidate gene expression profiling in the various cell lines was performed with primers specific for islet hormones (insulin, glucagon, and somatostatin), enzymes and transporters (glucose-6-phosphatase, glucokinase, glucose transporter-2 (GLUT2), hexokinases 1 and 2, and glyceraldehyde dehydrogenase), transcription factors (Brn4, E47, HNF-1α, IB1, NeuroD, Nkx2.2, Nkx6.1, Pax4, and Pdx1) and glycogen targeting subunits (PTG and GL). Other studies involved measurements of α-tubulin, G6PDH, Pax6, HNF3α (FOXO1), ratNkx6.1, hamster Nkx6.1, and total Nkx6.1 (rat plus hamster). Primer sequences for each of these targets are not provided here because of space considerations but can be obtained by contacting the authors. Real-time PCR was performed with 20 ng of cDNA by using the Applied Biosystems Prism 7000 sequence detection system, software, and reagents (16).
EMSA and Chromatin Immunoprecipitation (ChIP) Assay. EMSA analysis was performed as described in ref. 17 by using 5 μg of nuclear extract protein. In reactions for which supershift assays were performed, 1 μl of undiluted anti-Nkx6.1 or anti-Pdx1 antisera was also added. EMSA was performed with an oligonucleotide corresponding to the G1 element of the rat glucagon promoter: 5′-GAACAAAACCCCATTATTTACAGATGAGAA-3′ (top strand shown).
ChIP assays were performed as detailed in refs. 18 and 19. Antisera used in the immunoprecipitations were either anti-Nkx6.1 antiserum or normal rabbit serum. Each ChIP assay was quantitated in triplicate by real-time PCR for recovery of either the rat glucagon or myoD1 promoters. Forward and reverse primers used to amplify the glucagon gene were 5′-GGATCCTTCAGAGAGCTGAATAG and 5′-GGCAAGCTTCACCAGGGTGCTGTG. Forward and reverse primers used to amplify the myoD1 gene were 5′-CCACTTCGTCCTTGGCTCAAC and 5′-GGGATACCAGGCACAGCATAGG.
Transient Transfection Assays. A total of 2 μg of plasmid DNA consisting of 1.0 μg of a luciferase reporter plasmid under control of the proximal 450 bp of the rat glucagon promoter (pFoxLucGlucagon), 0.25 μg of pBAT12.Nkx6.1 or pBAT12 lacking a cDNA insert, and the balance consisting of a CMV promoter-driven β-galactosidase reporter plasmid) was mixed with 6 ml of TransFast reagent (Promega) and used to transfect αTC1.6 cells according to the manufacturer's protocol. Additional control experiments used plasmids with luciferase under control of thymidine kinase, CMV, or no promoter in place of 1 μg of pFoxLucGlucagon. Cells were harvested 48 h after transfection, and luciferase activities were measured by using a commercially available assay kit (Promega) and an FB15 luminometer (Zylux, Oak Ridge, TN).
Islet Isolation and Statistical Methods. Islets were isolated from male Wistar rats by pancreatic perfusion as described in ref. 20. GSIS assays were performed as described in ref. 21. Statistical significance was determined by using a two-tailed Student t test.
Results
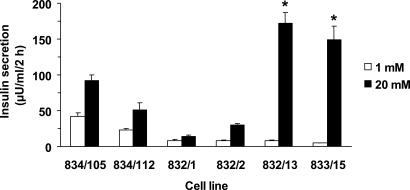
GSIS in INS-1-Derived Cell Lines. GSIS was measured in six independent INS-1-derived cell lines (3, 5). Four of the cell lines were poorly glucose-responsive (lines 834/105, 834/112, 832/1, and 832/2 with average responses of 2.4-, 2.1-, 2.2-, and 3.8-fold, respectively, as glucose was raised from 1 to 20 mM), whereas two cell lines were robustly glucose-responsive (lines 833/15 and 832/13 with average responses of 21- and 30-fold, respectively) (Fig. 1). These six lines were used for subsequent experiments.
Fig. 1.
GSIS from INS-1-derived cell lines. Insulin secretion was measured at 1 or 20 mM glucose in a 2-h static incubation assay in the indicated INS-1-derived cell lines. Data represent the mean ± SEM for three to four independent assays, each performed in triplicate. *, Cell lines 832/13 and 833/15 had larger fold responses to stimulatory glucose than the other four cell lines, with P < 0.0025.
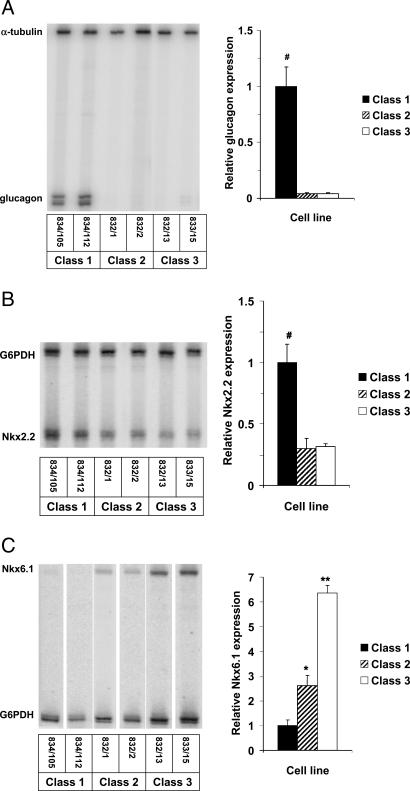
Candidate Gene Screen. The levels of mRNA encoding a panel of 20 transcription factors, metabolic enzymes, or hormones (listed in Materials and Methods) thought to serve as markers or mediators of beta cell differentiation were measured by semiquantitative multiplex PCR; among these, three were differentially expressed in the six INS-1-derived cell lines. The expression pattern for the gene encoding glucagon identified new subclasses of INS-1-derived cell lines (Fig. 2A). Thus, a subset of the poorly glucose-responsive cell lines (834/105 and 834/112) were found to contain substantial quantities of the mRNA encoding glucagon and are hereafter referred to as class 1 cells. Two other poorly glucose-responsive cell lines (832/1 and 832/2) contained little glucagon transcript and are hereafter referred to as class 2 cells. Finally, the two robustly glucose-responsive cell lines (832/13 and 832/15) also contained very low levels of glucagon mRNA, in keeping with their well differentiated phenotype, and are hereafter referred to as class 3 cells.
Fig. 2.
Differential expression of glucagon (A), Nkx 2.2 (B), and Nkx 6.1 (C) in INS-1-derived cell lines. Shown are representative gels and bar graph summaries, with the latter representing the mean ± SEM for three independent experiments per cell line, each performed in duplicate. #, Class 1 cells have more glucagon and Nkx2.2 mRNA, respectively, than the other two classes of cells, with P < 0.008. *, Class 2 cells have more Nkx6.1 mRNA than class 1 cells, with P < 0.05; **, class 3 cells have more Nkx6.1 mRNA than the other two classes, with P < 0.001.
The other two differentially expressed candidate genes emerging from this study are members of the Nkx transcription factor family. Nkx2.2 was most highly expressed in the glucagon-positive, poorly glucose-responsive class 1 cells (relative RNA levels of 3, 1.0, and 1.0, in class 1, class 2, and class 3 cells, respectively) (Fig. 2B). Interestingly, the inverse pattern was seen for Nkx6.1, which was most abundant in class 3 cells and least abundant in class 1 cells (relative RNA levels of 1.0, 2.6, and 6.4 in class 1, class 2, and class 3 cells, respectively) (Fig. 2C).
Expression of Nkx6.1 in Class 1 Cells Suppresses Glucagon Gene Expression. The expression pattern of Nkx6.1 shown in Fig. 2 suggests a potential role for this transcription factor in the regulation of glucagon expression and/or other differentiated functions of the beta cell, such as GSIS. Treatment of the class 1 cell line 834/105 with AdCMV-Nkx6.1 adenovirus resulted in increases in Nkx6.1 mRNA (Fig. 3A) and protein (Fig. 3B). Moreover, overexpression of Nkx6.1 in these cells suppressed glucagon mRNA levels in a dose-dependent manner (Fig. 3 A and C). Also, at a dose of AdCMV-Nkx6.1 virus that caused a 50% decrease in glucagon mRNA over 24 h, a 71% decrease in glucagon peptide content was observed 48 h after viral treatment (134 ± 24 versus 39 ± 3 ng of glucagon per mg of protein in AdCMV-βGAL- versus AdCMV-Nkx6.1-treated cells, respectively; P = 0.0014).
Fig. 3.
Overexpression of Nkx6.1 in class 1 cells suppresses glucagon expression. Class 1 cells were treated with AdCMV-Nkx6.1 or AdCMV-βGAL. (A) Semiquantitative multiplex PCR analysis of hamster Nkx6.1 and glucagon mRNAs, with α-tubulin as an internal control. (B) Immunoblot analysis of Nkx6.1 protein levels for the same range of viral titers as used in A. (C) The dose-dependent effects of Nkx6.1 overexpression on glucagon mRNA levels quantified by real-time PCR. Data represent the mean ± SEM for four independent experiments per viral dose, with each measurement in duplicate.
Importantly, at a dose of AdCMV-Nkx6.1 adenovirus that caused a 62% suppression of glucagon mRNA levels, no effect on expression of prominent beta cell transcription factors, such as Pdx1, IB1, E47, HNF-1α, Nkx2.2, PAX4, PAX6, HNF3α (FOXO1), or endogenous rat Nkx6.1 (data not shown) were observed. Overexpression of Nkx6.1 also did not affect insulin mRNA levels. Overexpression of Nkx6.1 did cause a small (10%) but statistically significant decrease in expression of NeuroD (P = 0.049). These data suggest that Nkx6.1 overexpression is largely sufficient to suppress glucagon gene expression in INS-1-derived cell lines.
Nkx6.1 Interacts with the Glucagon Promoter and Suppresses Its Function. We next examined the ability of Nkx6.1 to interact with the G1 element of the rat glucagon promoter in EMSAs, because the G1 element contains a potential Nkx6.1-binding site (5′-TAAT-3′) (6, 22). Nuclear extracts from class 3 cells contain significant Nkx6.1 protein, as demonstrated by supershift of a specific protein from the G1/nuclear extract complex in response to treatment with an Nkx6.1-specific antibody (Fig. 4A, lane 4). Adenovirus-mediated overexpression of Nkx6.1 caused a large increase in the amount of supershifted protein (Fig. 4A, lane 2), demonstrating that the overexpressed hamster Nkx6.1 protein binds to elements found within the rat glucagon promoter.
Fig. 4.
EMSA analysis of Nkx6.1 binding to the G1 element of the glucagon promoter. (A) Nuclear extracts were prepared from 832/13 cells treated with AdCMV-Nkx6.1 (lanes 1 and 2) or no virus (lanes 3 and 4) and mixed with a radiolabeled oligonucleotide corresponding to the G1 element of the glucagon promoter. (B) Effects of Ad-siNkx6.1 (Left) and Ad-siPdx1 (Right) viruses on their target proteins in 832/13 cells. Lanes 1, 2, 5, and 6 are from Ad-siLuc-treated cells, lanes 3 and 4 are from Ad-siNkx6.1-treated cells, and lanes 7 and 8 are from Ad-siPdx1-treated cells. (C) Effects of Ad-siPdx1 on Nkx6.1 protein level (Left) and of Ad-siNkx6.1 on Pdx1 protein level (Right) in 832/13 cells. Lanes 1, 2, 5, and 6 are from Ad-siRNA control-treated cells, lanes 3 and 4 are from Ad-siPdx1-treated cells, and lanes 7 and 8 are from Ad-siNkx6.1-treated cells. Before gel electrophoresis, samples were untreated (-) or treated (+) with anti-Nkx6.1 or anti-Pdx1 antibodies as indicated to cause supershift of the specific Nkx6.1/G1 (SS Nkx6.1) or Pdx1/G1 (SS Pdx1) complexes. Data are representative of two or three independent experiments.
To assess the functional consequence of interaction of Nkx6.1 with the glucagon promoter, we performed cotransfection studies in the alpha cell line αTC1.6. Overexpression of Nkx6.1 caused a 9.4 ± 1.1-fold repression of the activity of a glucagon promoter-driven luciferase reporter relative to control cells transfected with empty vector (Fig. 5A). By contrast, repression of control reporters by overexpressed Nkx6.1 was in the 1.5- to 2.5-fold range, suggesting that repression of the glucagon promoter-driven luciferase reporter by Nkx6.1 is specific. Moreover, adenovirus-mediated expression of Nkx6.1 caused a 58% suppression of endogenous glucagon mRNA in αTC1.6 cells, demonstrating that Nkx6.1 can suppress expression of an intact glucagon gene (Fig. 5B).
Fig. 5.
Suppression of glucagon promoter function by Nkx6.1. (A) αTC1.6 cells were cotransfected with the pBAT12.Nkx6.1 plasmid expressing Nkx6.1 and plasmids containing the luciferase reporter under control of the glucagon promoter, CMV promoter, TK promoter, or no promoter (promoterless). Luciferase activity is expressed as fold repression relative to activity in cells cotransfected with the glucagon/luciferase reporter construct and the pBAT12 plasmid lacking a cDNA insert (empty vector). Data represent the mean ± SEM for three independent cotransfections. (B) Suppression of endogenous glucagon expression in αTC1.6 cells in response to adenovirus-mediated overexpression of Nkx6.1. Data represent the mean ± SEM for two independent experiments, each performed in triplicate. (C) ChIP assay of Nkx6.1 binding to the endogenous glucagon promoter in a class 3 cell line (832/3). Immunoprecipitation reactions were performed with nonspecific rabbit serum or anti-Nkx6.1 antibody, followed by real-time PCR analysis of associated DNA with primers specific for the glucagon or myoD genes. Data are expressed as fold increase in recovery of input glucagon or myoD DNA by immunoprecipitation with the respective antibodies and represent the mean ± SEM for five independent experiments, each performed in triplicate. Primary data for these experiments (real-time PCR amplification curves) are available upon request from the authors. *, P < 0.01.
Data in Fig. 5 A and B suggest but do not prove that Nkx6.1 binds directly to the endogenous glucagon promoter in beta cells. To address this issue, we performed ChIP assays. Nkx6.1 is present at relatively low abundance in beta cells; therefore, as a prelude to these studies, we screened multiple cell lines with robust GSIS for their endogenous levels of Nkx6.1. Line 832/3, which was previously identified as a line with robust GSIS (3), contained 4.8 times more Nkx6.1 mRNA than lines 832/13 or 832/15 characterized earlier, and glucagon expression was virtually undetectable in these cells. As shown in Fig. 5C, 2.5 times more input glucagon promoter was recovered (P = 0.001) in nuclear extract samples from 832/3 cells immunoprecipitated with anti-Nkx6.1 antibody compared with samples treated with nonspecific rabbit serum. Moreover, the anti-Nkx6.1 antibody did not preferentially immunoprecipitate a control promoter (myoD). These data demonstrate direct interaction of Nkx6.1 with the glucagon promoter in living beta cells.
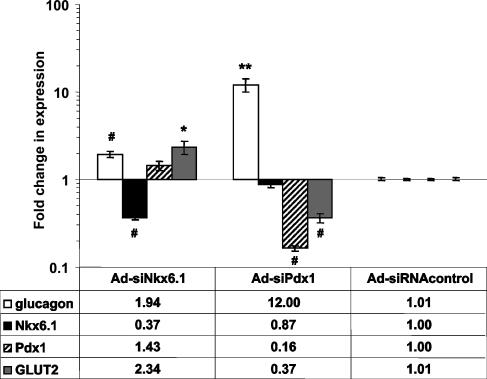
Suppression of Nkx6.1 or Pdx1 Expression Activates Glucagon Expression. A previous study has demonstrated that overexpression of Pdx1 in INS-1 cells causes increased expression of Nkx6.1 and suppression of glucagon expression (23). However, the specific roles played by Nkx6.1 versus Pdx1 in suppression of glucagon expression were not elucidated. To gain more insight into this issue, we prepared recombinant adenoviruses containing siRNAs specific for Nkx6.1 or Pdx1 (Ad-siNkx6.1 and Ad-siPdx1) and used these reagents to suppress the expression of the individual transcription factors in class 3 cells (832/13). Treatment of these cells with Ad-siNkx6.1 resulted in suppression of Nkx6.1 mRNA levels by 65%, whereas treatment with Ad-siPdx1 caused an 85% decrease in Pdx1 mRNA content (Fig. 6). Moreover, treatment of class 3 cells with Ad-siNkx6.1 caused near-complete extinction of the supershifted Nkx6.1 band detected by EMSA analysis with the glucagon promoter G1 element probe (Fig. 4B, compare lane 4 with lane 2). Similarly, treatment of 832/13 cells with the Ad-siPdx1 adenovirus resulted in strong suppression of an anti-Pdx1 antibody-supershifted band (Fig. 4B, compare lane 8 with lane 6).
Fig. 6.
Effects of RNA interference-mediated suppression of Nkx6.1 or Pdx1 expression in 832/13 cells. 832/13 cells were treated with Ad-siNkx6.1 (left group of bars), Ad-siPdx1 (center group of bars), or Ad-siRNAcontrol (right group of bars), and glucagon, Nkx6.1, Pdx1, and GLUT2 mRNA levels were analyzed by real-time PCR. Data are normalized to the levels in Ad-siRNAcontrol-treated cells and represent the mean ± SEM for three to five independent experiments, each performed in triplicate. Numbers below the graph indicate the average fold changes in expression of the indicated mRNAs in response to Nkx6.1 or Pdx1 suppression. Symbols denote statistically significant increases or decreases in mRNA levels: *, P < 0.05; **, P < 0.01; #, P < 0.002.
The level of Nkx6.1 suppression shown in Figs. 4B and 6 resulted in a 2-fold increase in glucagon mRNA levels, further supporting a direct role for Nkx6.1 in control of glucagon expression in beta cells (Fig. 6). Interestingly, Pdx1 suppression caused a much larger increase (12-fold) in glucagon expression than that achieved by Nkx6.1 suppression. Pdx1 suppression resulted in modest decreases in Nkx6.1 mRNA (Fig. 6) and protein levels as judged by EMSA analyses (Fig. 4C). Conversely, suppression of Nkx6.1 by RNA interference treatment had no effect on Pdx1 mRNA (Fig. 6) or protein levels (Fig. 4C). Our data suggest that Nkx6.1 and Pdx1 have independent effects on glucagon expression in beta cells.
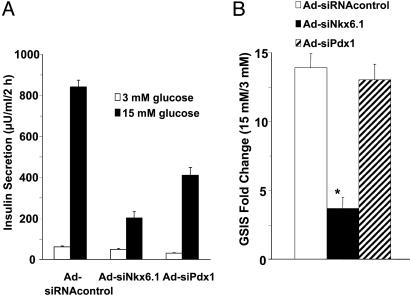
Nkx6.1 Suppression Causes Impairment of GSIS in Class 3 Cells. The expression pattern of Nkx6.1 in INS-1-derived cell lines suggests a possible role for this transcription factor in the robust GSIS exhibited by class 3 cells. To test this possibility directly, we used Ad-siNkx6.1 to suppress Nkx6.1 mRNA levels by 65% in 832/13 cells. As shown in Fig. 7, suppression of Nkx6.1 caused a significant impairment in GSIS, with a decline from a 13.9-fold response in cells treated with the Ad-siRNA control virus to a 3.7-fold response in cells treated with Ad-siNkx6.1. Similar effects of Nkx6.1 suppression were observed in a second class 3 cell line, 832/3 (data not shown). In contrast, suppression of Pdx1 expression caused a decrease in basal and stimulated insulin secretion, but did not affect the fold response. The differential effects of Nkx6.1 and Pdx1 suggests that these transcription factors control distinct sets of genes involved in insulin secretion. Consistent with this idea, suppression of Pdx1 caused a 63% decrease, whereas Nkx6.1 suppression caused a 2.3-fold increase, in GLUT-2 mRNA levels (Fig. 6). Importantly, neither Pdx-1 suppression nor Nkx6.1 suppression affected insulin content (16.8 ± 3.5, 14.3 ± 1.9, and 15.5 ± 3.5 microunits of insulin per μg of protein in Ad-siRNAcontrol-, Ad-siNkx6.1-, and Ad-siPdx1-treated cells, respectively).
Fig. 7.
Effects of Nkx6.1 and Pdx1 suppression on GSIS in class 3 cells. 832/13 cells were treated with the indicated siRNA-containing recombinant adenoviruses, followed by assay of insulin secretion in response to 3 or 15 mM glucose. (A) Insulin secretion expressed as microunits per ml per every 2 h. (B) Insulin secretion expressed as fold response. Data represent the mean ± SEM for three to five independent experiments, each performed in triplicate. *, Fold response was decreased in Ad-siNkx6.1-treated cells, with P < 0.001.
We also investigated whether increasing the levels of Nkx6.1 in class 1 or class 2 cells would result in improved GSIS in these lines. AdCMV-Nkx6.1 was used to raise Nkx6.1 mRNA levels in class 1 or class 2 cells to a level approximating the endogenous levels of class 3 cells. This maneuver did not improve GSIS relative to AdCMV-βGAL-treated class 1 or class 2 cells (data not shown). These data and the data of Fig. 7 suggest that Nkx6.1 is a necessary but not sufficient factor for conferring GSIS in INS-1-derived cell lines.
Suppression of Nkx6.1 Expression Impairs GSIS in Rat Islets. To determine whether our findings also apply to normal rat islets, we used the Ad-siNkx6.1 virus to suppress Nkx6.1 mRNA levels by 46% in primary cells. This maneuver resulted in a decrease in GSIS from 7.1 ± 0.2-fold in Ad-siRNAcontrol-treated islets to 5.2 ± 0.5-fold in Ad-siNkx6.1-treated islets (P = 0.012) and a 56% decrease in insulin secretion at stimulatory glucose (16.7 mM) (Fig. 8). Suppression of Nkx6.1 did not affect insulin content in the primary islet experiments.
Fig. 8.
Suppression of Nkx6.1 expression impairs GSIS in rat islets. Rat islets were treated with Ad-siNkx6.1 or Ad-siRNAcontrol viruses, resulting in a 46% decrease in Nkx6.1 mRNA in the Ad-siNkx6.1-treated cells. Data represent the mean ± SEM for three independent experiments, with data normalized to insulin content. *, Fold response in Ad-siNkx6.1-treated cells was significantly reduced (P = 0.012), and insulin secretion at stimulatory glucose was decreased by 56% (P = 0.001).
Discussion
INS-1 cells (4) have been used to study many aspects of beta cell biology (5, 21, 23-31). The parental INS-1 cell line is now recognized as a mixed population of cells with different phenotypic features, because subclones derived from these cells have varying degrees of glucose responsiveness (3, 5, 28, 32). Microarray analyses of rodent insulinoma cell lines with differing capacities for GSIS (33, 34) or as a function of exposure to inflammatory cytokines (35-37) have been reported, with hundreds of differentially expressed genes appearing in each of these studies. However, follow-up functional analysis aimed at determining which of the differentially expressed genes actually contribute to the phenotype under study have been rare. This report represents our first attempt at defining differentially expressed genes that contribute to beta cell function in INS-1-derived cell lines.
The candidate gene strategy reported here has led to the identification of three classes of INS-1-derived subclones. Class 1 cells were found to coexpress insulin and glucagon and were poorly glucose-responsive, class 2 cells had very low glucagon expression and were also poorly glucose-responsive, and class 3 cells had low glucagon expression and robust GSIS. These phenotypes were negatively and positively correlated with levels of expression of Nkx2.2 and Nkx6.1, respectively.
Coexpression of insulin and glucagon has been reported previously in the parental INS-1 cell line (38). In addition Wang et al. (23) described two INS-1-derived clones, INSrαβ and INSrβ; INSrαβ coexpressed glucagon and insulin, whereas glucagon expression was absent in INSrβ cells. GSIS was modest in both of these clones (3- to 4-fold), but a clear understanding of this aspect of their phenotype was complicated by the fact that the cells were transfected with either a Pdx1 transgene (INSrβ) or a dominant negative Pdx1 construct (INSrαβ). Overexpression of Pdx1 in INSrαβ cells resulted in stimulation of Nkx6.1 expression in concert with suppression of glucagon expression. However, these studies did not determine whether the suppression of glucagon expression was mediated directly by Nkx6.1 or by another mechanism.
The current study shows that Nkx6.1 maintains the mature beta cell phenotype in part through participation in suppression of glucagon expression. Several lines of evidence support this conclusion: (i) Overexpression of Nkx6.1 decreases glucagon mRNA and peptide levels in a dose-dependent fashion in class 1 cells; (ii) Lowering of Nkx6.1 levels in class 3 cells results in an increase in glucagon mRNA; (iii) EMSA demonstrates Nkx6.1 interaction with the G1 element of the rat glucagon promoter; (iv) Nkx6.1 suppresses activity of a glucagon promoter/reporter construct in cotransfection experiments in αTC-1 cells and suppresses endogenous glucagon expression in these cells; (v) ChIP analysis demonstrates direct interaction of Nkx6.1 with the glucagon promoter in class 3 cells; and (vi) Nkx6.1 expression does not affect the expression of other transcription factors that have been prominently implicated in beta cell development. Our findings contrast with a previous study in a different alpha cell line (MSL-G-AN) in which no suppression of endogenous glucagon expression was reported in response to stable expression of Nkx6.1 (22). However, in that study, changes in glucagon expression were only compared in Nkx6.1-transfected and control cells after prolonged in vivo passage to allow tumor formation, whereas the use of adenovirus vectors in the current study allowed us to examine the acute effects of modulation of Nkx6.1 levels directly in cultured cells. Our results also show that Nkx6.1 and Pdx1 have independent suppressive effects on glucagon expression in beta cell lines.
We also report that siRNA-mediated silencing of Nkx6.1 in class 3 cells causes a dramatic decrease in GSIS. siRNA-mediated suppression of Pdx1 expression, in contrast, caused a decrease in the amount of insulin secreted at basal and stimulatory glucose concentrations but did not affect fold response. These effects on insulin secretion were not attributable to changes in insulin content. Importantly, suppression of Nkx6.1 expression also impairs GSIS in primary rat islets.
Glucagon-expressing alpha cells and insulin-expressing beta cells arise from a common neurogenin3-expressing precursor cell (39). Differentiation from these common precursors into alpha or beta cells is directed by an array of transcription factors that are expressed in a specific temporal and spatial context (40). Nkx2.2 is expressed early in pancreatic differentiation (embryonic day 9.5), later restricted to endocrine cells (embryonic day 15.5), and expressed in alpha, beta-, and PP cells in mature islets. Disruption of the Nkx2.2 gene results in a decrease in islet mass with an absence of insulin-expressing cells and a reduction in glucagon- and PP-expressing cells (41). Nkx6.1 can first be detected at embryonic day 10.5, depends on Nkx2.2 for its expression, and is tightly restricted to the beta cell lineage (42), consistent with the idea that Nkx6.1 could function as a glucagon suppressor. Islets from Nkx6.1 knockout animals are slightly smaller than littermate controls, with 94% reduction in the number of beta cells but with no change in the number of glucagon-positive cells (42). The surfeit of beta cell mass in Nkx6.1-/- animals is due in part to an absence of mature beta cells produced from the so-called “secondary transition.” These features of the model prevent a full assessment of the role of Nkx6.1 in maintenance of mature beta cell function because of an insufficient number of beta cells for study. Thus, our work with overexpression and RNA interference-mediated suppression of Nkx6.1 in well differentiated insulinoma cells and primary rat islets has allowed us to uncover roles for the transcription factor that were not discernable earlier.
Our findings may shed light on development of beta cell dys-function in diabetes. Of interest in this context is the finding that impaired GSIS induced by partial pancreatecomy is accompanied by reduced Nkx6.1 expression (43). Similarly, Nkx6.1 mRNA levels are decreased by 60% in islets of Zucker diabetic fatty rats relative to islets from lean control animals (J.C.S., D.L., and C.B.N., unpublished observations). Nkx6.1 usually functions as a transcriptional repressor; therefore, a decrease in its expression in islets during development of diabetes might lead to an increase in expression of other genes that cause impairment of beta cell function. Further study, including microarray analysis of class 3 cells and normal islets with and without Nkx6.1 suppression, will be required to identify Nkx6.1 target genes that mediate GSIS and to determine whether such genes play a significant role in beta cell failure of diabetes.
Acknowledgments
We thank Lisa Poppe, Paul Anderson, and Theresa Eversole for expert technical assistance; Drs. Rafael Nesher and Ludivina Robles of the Joslin Diabetes Center for assistance in identification of siPdx1 targeting sequences; Dr. Suzanne Ziesmann for help with the luciferase studies; and Dr. Christopher Wright (Vanderbilt University, Nashville, TN) for provision of the anti-Pdx1 antibody. The work was supported by National Institutes of Health Grants U01-DK-56047 and RO1-DK-58398 (to C.B.N.), R01-DK-60581 (to R.G.M.), and U19DK61251 (to G.C.W.).
Author contributions: J.C.S., P.B.J., R.G.M., and C.B.N. designed research; J.C.S., P.B.J., D.G.T., T.C.B., F.K.K., S.T., D.L., and R.G.M. performed research; J.C.S., P.B.J., D.G.T., T.C.B., F.K.K., M.G., G.C.W., and R.G.M. contributed new reagents/analytic tools; J.C.S., P.B.J., M.G., R.G.M., and C.B.N. analyzed data; and J.C.S., G.C.W., R.G.M., and C.B.N. wrote the paper.
Abbreviations: ChIP, chromatin immunoprecipitation; GLUT2, glucose transporter-2; GSIS, glucose-stimulated insulin secretion; siRNA, small interfering RNA.
References
- 1.Shapiro, A. M., Lakey, J. R., Ryan, E. A., Korbutt, G. S., Toth, E., Warnock, G. L., Kneteman, N. M. & Rajotte, R. V. (2000) N. Engl. J. Med. 343, 230-238. [DOI] [PubMed] [Google Scholar]
- 2.Lebovitz, H. E. (2004) in Diabetes Mellitus, eds. LeRoith, D., Taylor, S. I. & Olefsky, J. M. (Lippincott, Philadelphia), pp. 1107-1138.
- 3.Hohmeier, H. E., Mulder, H., Chen, G., Henkel-Rieger, R., Prentki, M. & Newgard, C. B. (2000) Diabetes 49, 424-430. [DOI] [PubMed] [Google Scholar]
- 4.Asfari, M., Janjic, D., Meda, P., Li, G., Halban, P. A. & Wollheim, C. B. (1992) Endocrinology 130, 167-178. [DOI] [PubMed] [Google Scholar]
- 5.Chen, G., Hohmeier, H. E., Gasa, R., Tran, V. V. & Newgard, C. B. (2000) Diabetes 49, 562-570. [DOI] [PubMed] [Google Scholar]
- 6.Mirmira, R. G., Watada, H. & German, M. S. (2000) J. Biol. Chem. 275, 14743-14751. [DOI] [PubMed] [Google Scholar]
- 7.Gomez-Foix, A. M., Coats, W. S., Baque, S., Alam, T., Gerard, R. D. & Newgard, C. B. (1992) J. Biol. Chem. 267, 25129-25134. [PubMed] [Google Scholar]
- 8.Becker, T. C., Noel, R. J., Coats, W. S., Gomez-Foix, A. M., Alam, T., Gerard, R. D. & Newgard, C. B. (1994) Methods Cell Biol. 43, 161-189. [DOI] [PubMed] [Google Scholar]
- 9.Herz, J. & Gerard, R. D. (1993) Proc. Natl. Acad. Sci. USA 90, 2812-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bain, J. R., Schisler, J. C., Takeuchi, K., Newgard, C. B. & Becker, T. C. (2004) Diabetes 53, 2190-2194. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 12.Bewig, B. & Schmidt, W. E. (2000) BioTechniques 28, 870-873. [DOI] [PubMed] [Google Scholar]
- 13.Clark, S. A., Quaade, C., Constandy, H., Hansen, P., Halban, P., Ferber, S., Newgard, C. B. & Normington, K. (1997) Diabetes 46, 958-967. [DOI] [PubMed] [Google Scholar]
- 14.Hohmeier, H. E., BeltrandelRio, H., Clark, S. A., Henkel-Rieger, R., Normington, K. & Newgard, C. B. (1997) Diabetes 46, 968-977. [DOI] [PubMed] [Google Scholar]
- 15.Jensen, J., Serup, P., Karlsen, C., Nielsen, T. F. & Madsen, O. D. (1996) J. Biol. Chem. 271, 18749-18758. [DOI] [PubMed] [Google Scholar]
- 16.An, J., Muoio, D. M., Shiota, M., Fujimoto, Y., Cline, G. W., Shulman, G. I., Koves, T. R., Stevens, R., Millington, D. & Newgard, C. B. (2004) Nat. Med. 10, 268-274. [DOI] [PubMed] [Google Scholar]
- 17.German, M. S., Moss, L. G., Wang, J. & Rutter, W. J. (1992) Mol. Cell. Biol. 12, 1777-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakrabarti, S. K, Francis, J., Ziesmann, S. M., Garmey, J. C. & Mirmira, R. G. (2003) J. Biol. Chem. 278, 23617-23623. [DOI] [PubMed] [Google Scholar]
- 19.Chakrabarti, S. K., James, J. C. & Mirmira, R. G. (2002) J. Biol. Chem. 277, 13286-13293. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, J. H., Crider, B. P., McCorkle, K., Alford, M. & Unger, R. H. (1990) N. Engl. J. Med. 322, 653-659. [DOI] [PubMed] [Google Scholar]
- 21.Antinozzi, P. A., Segall, L., Prentki, M., McGarry, J. D. & Newgard, C. B. (1998) J. Biol. Chem. 273, 16146-16154. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen, M. C., Vestergard Petersen, H., Ericson, J., Madsen, O. D. & Serup, P. (1999) FEBS Lett. 461, 287-294. [DOI] [PubMed] [Google Scholar]
- 23.Wang, H., Maechler, P., Ritz-Laser, B., Hagenfeldt, K. A., Ishihara, H., Philippe, J. & Wollheim, C. B. (2001) J. Biol. Chem. 276, 25279-25286. [DOI] [PubMed] [Google Scholar]
- 24.Noel, R. J., Antinozzi, P. A., McGarry, J. D. & Newgard, C. B. (1997) J. Biol. Chem. 272, 18621-18627. [DOI] [PubMed] [Google Scholar]
- 25.Segall, L., Lameloise, N., Assimacopoulos-Jeannet, F., Roche, E., Corkey, P., Thumelin, S., Corkey, B. E. & Prentki, M. (1999) Am. J. Physiol. 277, E521-E528. [DOI] [PubMed] [Google Scholar]
- 26.Wang, H., Maechler, P., Antinozzi, P. A., Herrero, L., Hagenfeldt-Johansson, K. A., Bjorklund, A. & Wollheim, C. B. (2003) J. Biol. Chem. 278, 16622-16629. [DOI] [PubMed] [Google Scholar]
- 27.Chen, G., Hohmeier, H. E. & Newgard, C. B. (2001) J. Biol. Chem. 276, 766-772. [DOI] [PubMed] [Google Scholar]
- 28.Tran, V. V., Chen, G., Newgard, C. B. & Hohmeier, H. E. (2003) Diabetes 52, 1423-1432. [DOI] [PubMed] [Google Scholar]
- 29.Smukler, S. R., Tang, L., Wheeler, M. B. & Salapatek, A. M. (2002) Diabetes 51, 3450-3460. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, S. & Kim, K. H. (1995) FEBS Lett. 377, 237-239. [DOI] [PubMed] [Google Scholar]
- 31.Ahren, B. & Havel, P. J. (1999) Am. J. Physiol. 277, R959-R966. [DOI] [PubMed] [Google Scholar]
- 32.Merglen, A., Theander, S., Rubi, B., Chaffard, G., Wollheim, C. B. & Maechler, P. (2004) Endocrinology 145, 667-678. [DOI] [PubMed] [Google Scholar]
- 33.Lilla, V., Webb, G., Rickenbach, K., Maturana, A., Steiner, D. F., Halban, P. A. & Irminger, J. C. (2003) Endocrinology 144, 1368-1379. [DOI] [PubMed] [Google Scholar]
- 34.Zimmer, Y., Milo-Landesman, D., Svetlanov, A. & Efrat, S. (1999) FEBS Lett. 457, 65-70. [DOI] [PubMed] [Google Scholar]
- 35.Kutlu, B., Cardozo, A. K., Darville, M. I., Kruhoffer, M., Magnusson, N., Orntoft, T. & Eizirik, D. L. (2003) Diabetes 52, 2701-2719. [DOI] [PubMed] [Google Scholar]
- 36.Rasschaert, J., Liu, D., Kutlu, B., Cardozo, A. K., Kruhoffer, M., ORntoft, T. F. & Eizirik, D. L. (2003) Diabetologia 46, 1641-1657. [DOI] [PubMed] [Google Scholar]
- 37.Eizirik, D. L., Kutlu, B., Rasschaert, J., Darville, M. & Cardozo, A. K. (2003) Ann. N.Y. Acad. Sci. 1005, 55-74. [DOI] [PubMed] [Google Scholar]
- 38.Poitout, V., Olson, L. K. & Robertson, R. P. (1996) Diabetes Metab. 22, 7-14. [PubMed] [Google Scholar]
- 39.Schwitzgebel, V. M., Scheel, D. W., Conners, J. R., Kalamaras, J., Lee, J. E., Anderson, D. J., Sussel, L., Johnson, J. D. & German, M. S. (2000) Development (Cambridge, U.K.) 127, 3533-3542. [DOI] [PubMed] [Google Scholar]
- 40.Wilson, M. E., Scheel, D. & German, M. S. (2003) Mech. Dev. 120, 65-80. [DOI] [PubMed] [Google Scholar]
- 41.Sussel, L., Kalamaras, J., Hartigan-O'Connor, D. J., Meneses, J. J., Pedersen, R. A., Rubenstein, J. L. & German, M. S. (1998) Development (Cambridge, U.K.) 125, 2213-2221. [DOI] [PubMed] [Google Scholar]
- 42.Sander, M., Sussel, L., Conners, J., Scheel, D., Kalamaras, J., Dela Cruz, F., Schwitzgebel, V., Hayes-Jordan, A. & German, M. (2000) Development (Cambridge, U.K.) 127, 5533-5540. [DOI] [PubMed] [Google Scholar]
- 43.Jonas, J. C., Sharma, A., Hasenkamp, W., Ilkova, H., Patane, G., Laybutt, R., Bonner-Weir, S. & Weir, G. C. (1999) J. Biol. Chem. 274, 14112-14121. [DOI] [PubMed] [Google Scholar]