Abstract
While signal transducers and activators of transcription (STATs) were originally discovered as intracellular effectors of normal signaling by cytokines, increasing evidence also points to a role for STAT transcription factors in oncogenesis. Previous studies have demonstrated that one STAT family member, Stat3, possesses constitutively elevated tyrosine phosphorylation and DNA-binding activity in fibroblasts stably transformed by the Src oncoprotein. To determine if this Stat3 activation by Src could induce Stat3-mediated gene expression, luciferase reporter constructs based on synthetic and authentic promoters were transfected into NIH 3T3 cells. Activation of endogenous cellular Stat3 by the Src oncoprotein induced gene expression through a Stat3-specific binding element (TTCCCGAA) of the C-reactive protein gene promoter. A naturally occurring splice variant of human Stat3 protein, Stat3β, with a deletion in the C-terminal transactivation domain abolished this gene induction in a dominant negative manner. Expression of Stat3β did not have any effect on a reporter construct based on the c-fos serum response element, which is not dependent on Stat3 signaling, indicating that Stat3β does not nonspecifically inhibit other signaling pathways or Src function. Transfection of vectors expressing Stat3β together with Src blocked cell transformation by Src as measured in a quantitative focus formation assay using NIH 3T3 cells. By contrast, Stat3β had a much less pronounced effect on focus formation induced by the Ras oncoprotein, which does not activate Stat3 signaling. In addition, three independent clones of NIH 3T3 cells stably overexpressing Stat3β were generated and characterized, demonstrating that Stat3β overexpression does not have a toxic effect on cell viability. These Stat3β-overexpressing clones were shown to be deficient in Stat3-mediated signaling and refractory to Src-induced cell transformation. We conclude that Stat3 activation by the Src oncoprotein leads to specific gene regulation and that Stat3 is one of the critical signaling pathways involved in Src oncogenesis. Our findings provide evidence that oncogenesis-associated activation of Stat3 signaling is part of the process of malignant transformation.
Signal transducers and activators of transcription (STATs) are latent cytoplasmic transcription factors that were first identified as mediators of cellular responses to interferons (reviewed in references 12, 16 and 35). Signaling induced by the interaction of interferons and other cytokines with their cognate receptors is initiated by a cascade of events, including receptor aggregation and activation of Janus protein tyrosine kinases (JAKs) associated with the receptors. Subsequently, STAT proteins are recruited to the receptor-JAK complexes and activated by tyrosine phosphorylation, which promotes the formation of homodimers or heterodimers of STAT family members. Activated STATs, in turn, translocate to the nucleus and bind to specific DNA response elements that regulate gene expression. There are at least seven genes in the mammalian genome known to encode different STAT family members, which are activated in various combinations in response to stimulation by numerous cytokines (12, 16, 35).
It has become evident that, in addition to cytokines, mitogenic growth factors, such as platelet-derived growth factor and epidermal growth factor, also induce STAT signaling, particularly Stat1, Stat3, and Stat5 (21, 35). An emerging concept is that normal signaling by STAT proteins is involved in control of diverse biological processes regulated by cytokines and growth factors, including cell differentiation, proliferation, development, and apoptosis (2, 4, 10, 13, 19, 20, 22, 23, 26, 29, 31, 37, 39, 40, 50). Increasingly, evidence that indicates an association between abnormal activation of STAT signaling and oncogenesis has also been accumulating. For example, we and other investigators have demonstrated constitutive activation of Stat1, Stat3, Stat5, and Stat6 in cells transformed by Src, Abl, and various other oncoproteins and tumor viruses (7, 8, 11, 14, 24, 25, 47, 51). In the context of human cancer, there is a high frequency of activation of Stat1, Stat3, and Stat5 in breast carcinoma cells (14, 32, 42) and in lymphoid malignancies, including lymphomas and leukemias (15, 43, 49). Although these findings suggest a role for STAT signaling in cell transformation as well as in human cancer, direct evidence for the obligatory requirement of STAT signaling in oncogenesis has not been reported previously.
Because transformation of mammalian fibroblasts by viral Src (v-Src) specifically induces constitutive activation of one STAT family member, Stat3, this system is ideal for investigating the role of Stat3 signaling in oncogenesis (7, 47). The embryonic lethality of targeted disruption of the Stat3 gene (39), however, precludes generation of viable Stat3-deficient mouse models for these studies. An alternative approach for disrupting Stat3 function is to make use of Stat3 dominant negative proteins that interfere with Stat3 signaling. One such variant of Stat3, known as Stat3β, has been shown to block Stat3 function in response to interleukin 5 (IL-5) stimulation (5). Stat3β is a naturally occurring splice variant with a deletion in the C-terminal portion that harbors the transcriptional activation domain and the Ser-727 residue, phosphorylation of which is required for efficient transcriptional activation (44, 45). As a result of this C-terminal deletion, dimers formed with human Stat3β lack transcriptional activity (5). In some studies, however, mouse Stat3β has been shown to activate the promoters of certain genes in a cell type-dependent manner (33, 34), suggesting that the dominant negative effect of Stat3β may be cell type specific.
To investigate Stat3-mediated gene regulation by v-Src and the role of Stat3 in oncogenesis, we disrupted Stat3 signaling by using human Stat3β. We report that in NIH 3T3 fibroblasts transiently expressing luciferase reporter constructs, v-Src induced Stat3-specific transcriptional activation. As a potent dominant negative modulator of Stat3-mediated signaling, Stat3β effectively blocked Stat3-specific gene expression induced by v-Src in these cells. Furthermore, cotransfection of expression vectors encoding Stat3β and v-Src suppressed cell transformation of NIH 3T3 fibroblasts, and stable cell lines overexpressing Stat3β were found to be resistant to transformation by v-Src. Our findings establish that activation of Stat3 by v-Src induces specific gene regulation and provide evidence for the requirement of Stat3 signaling in cell transformation by the Src oncoprotein.
MATERIALS AND METHODS
Construction of plasmids.
An annealed oligonucleotide corresponding to the −35 to +11 region (relative to the transcriptional start site at +1) of the herpes simplex virus thymidine kinase (TK) promoter was cloned between the KpnI and BglII sites of the basic luciferase reporter pLuc (pGL2; Promega) to construct pLucTK (see Fig. 1 for structures of constructs). The Stat3 reporter pLucTKS3 was constructed from pLucTK by inserting seven copies of an annealed oligonucleotide corresponding to a Stat3-specific binding site from the human C-reactive protein (CRP) gene (48), called the CRP acute-phase response element (cAPRE) (nucleotides −123 to −85), into the SmaI site upstream of the TK minimal promoter. Another reporter, pLucTKSIE, contains two copies of an annealed oligonucleotide corresponding to a high-affinity mutant of the c-fos sis-inducible element (hSIE [mutant m67]) (41) inserted into the SmaI site of pLucTK. To construct pLucCRP, the authentic CRP promoter (nucleotides −123 to +3) was excised from plasmid −123/+3CRP-CAT (48) (a generous gift from D. Samols) by BamHI-XhoI restriction digestion, blunt-ended with Klenow, and inserted into the SmaI site of pLuc. The human Stat3β expression vector pSG5hStat3β is driven by the simian virus 40 promoter contained in pSG5 (Stratagene) and has been previously described (5). The Stat3 expression vector pVRStat3 was constructed by excising the mouse Stat3 cDNA from pBSStat3 by EcoRI and DraIII digestion, blunt-ending with T4 DNA polymerase, and cloning into a vector driven by the cytomegalovirus immediate early gene promoter (2a, 50). The reporter pLucSRE, which contains a serum response element (SRE) in the context of the c-fos promoter driving luciferase expression, as well as the N17-Ras and NT-Raf vectors have been described previously (30, 46). Expression vectors for v-Src, pMvSrc (17), and oncogenically activated c-H-Ras have been described previously (28).
FIG. 1.
Schematic representations of Stat3 proteins and reporter constructs. (A) Full-length Stat3 and Stat3β, which is a naturally occurring splice variant with a deletion in the C-terminal transactivation domain, are diagrammed with various protein domains and the major sites of phosphorylation shown. (B) The reporter construct pLucTK contains only the TK minimal promoter driving luciferase (LUC) gene expression, while pLucTKS3 has seven copies of a Stat3-specific binding site (cAPRE) from the CRP gene inserted upstream of the TK minimal promoter. pLucCRP contains an authentic CRP promoter fragment driving expression of the luciferase gene. pLucTKSIE has two copies of a high-affinity mutant (hSIE) of the c-fos SIE inserted upstream of the TK minimal promoter, whereas pLucSRE contains a c-fos promoter fragment harboring the SRE inserted upstream of TK promoter sequences. Not shown is the basic pLuc backbone vector, which contains the luciferase gene without promoter sequences cloned in pUC19. See Materials and Methods for additional details of constructs.
Cell culture and transfections.
NIH 3T3 fibroblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 5% iron-supplemented bovine calf serum (BCS). Transfections were carried out by the standard calcium phosphate method (9). NIH 3T3 fibroblasts were seeded at 5 × 105 cells/100-mm-diameter plate in DMEM plus 5% BCS at 18 to 24 h prior to transfection. Total DNA for transfections was typically 20 μg per plate, including 4 μg of luciferase reporter construct (pLucTK, pLucTKS3, pLucCRP, pLucTKSIE, or pLucSRE), 0.2 μg of β-galactosidase (β-Gal) internal control vector, and the amounts of expression vector indicated in Results. Transfection was terminated 15 h later by aspirating the medium, washing the cells with 1× phosphate-buffered saline (PBS), and adding fresh DMEM. Where gamma interferon (IFN-γ) was present, it was added 5 h prior to harvest of transfected cells.
Preparation of cytosolic and nuclear extracts.
For transient expression assays, cytosolic extracts were prepared from cells at 48 h posttransfection. Briefly, after two washes with 1× PBS and equilibration for 5 min with 0.5 ml of PBS–0.5 mM EDTA, cells were scraped off of the dishes and the cell pellet was obtained by centrifugation (4,500 × g, 2 min, 4°C). Cells were resuspended in 0.4 ml of low-salt HEPES buffer (10 mM HEPES [pH 7.8], 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol) for 15 min, lysed by the addition of 20 μl of 10% Nonidet P-40, and centrifuged (10,000 × g, 30 s, 4°C) to obtain the cytosolic supernatant, which was used for luciferase assays (Promega) with a luminometer and for β-Gal activity detection by colorimetric assay at A570. As an internal control for transfection efficiency, results were normalized to β-Gal activity. For electrophoretic mobility shift assays (EMSA), nuclear extracts were prepared from transiently transfected NIH 3T3 cells and volumes containing equal amounts of total protein were incubated with 32P-labeled hSIE oligonucleotide probe, as previously reported (47). Supershift assays were performed by using rabbit polyclonal antibodies against the C-terminal amino acid residues (750 to 769) of full-length Stat3 that are absent in Stat3β (Santa Cruz Biotechnology).
Western blot analysis.
Whole-cell lysates were prepared in boiling sodium dodecyl sulfate (SDS) sample buffer in order to extract total Stat3 proteins from the cytoplasm and nucleus. Equivalent amounts of total cellular protein were electrophoresed on an SDS–7.5% polyacrylamide gel and transferred to a nitrocellulose membrane. Probing of nitrocellulose membranes with primary antibodies and detection of horseradish peroxidase-conjugated secondary antibodies by enhanced chemiluminesence (Amersham) were performed as previously described (14, 47). Probes used were rabbit polyclonal antibodies against the Stat3 C-terminal amino acids that are specific for full-length Stat3 (Santa Cruz Biotechnology) or mouse monoclonal antibody against the Stat3 N-terminal amino acid residues (1 to 178) that recognizes both full-length Stat3 and Stat3β (Transduction Laboratories).
Focus formation assays.
NIH 3T3 fibroblasts were transfected as described above and then harvested by trypsinization at 48 h posttransfection. Cytosolic extracts were prepared from 25% of the transfected cells to measure β-Gal activity, which was used for determination of transfection efficiency. The remaining 75% of the cells were seeded into new dishes and fed 1× DMEM every 3 days. Foci were counted at 16 or 21 days posttransfection by using phase-contrast microscopy.
RESULTS
Stat3-mediated gene regulation is induced by Src.
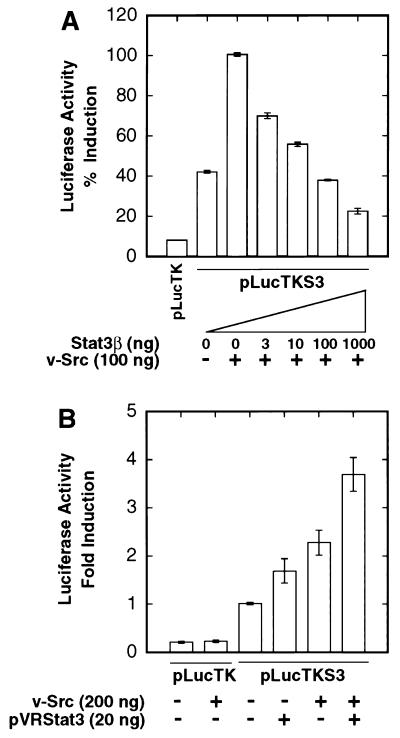
We previously demonstrated that Stat3 is constitutively activated in Src-transformed fibroblasts, as measured by enhanced tyrosine phosphorylation and DNA-binding activity of Stat3 (47). To determine whether Stat3 activation by Src could lead to regulation of gene expression, we transiently transfected NIH 3T3 cells with v-Src expression vector and a reporter construct, pLucTKS3 (Fig. 1B), containing multimerized Stat3-specific binding sites inserted upstream of a TK minimal promoter. This Stat3-specific binding site, which is derived from the human CRP gene and is called cAPRE here, contains the core sequence TTCCCGAA (36, 48). Assays for luciferase reporter gene expression (Fig. 2A) show dose-dependent gene induction, up to 15-fold over basal levels, mediated through activation of endogenous cellular Stat3 by increasing amounts of transfected v-Src expression vector. To confirm that this gene induction is dependent on Stat3, we used an expression vector encoding the Stat3 splice variant, Stat3β (Fig. 1A) (5), to disrupt Stat3 signaling. Figure 2B shows that induction of pLucTKS3 by v-Src requires the presence of cAPRE and is reduced to basal levels by cotransfection with Stat3β expression vector. To further characterize the dominant negative properties of Stat3β, Fig. 3A shows that increasing amounts of transfected Stat3β vector results in dose-dependent inhibition of Src-induced Stat3 reporter expression. In contrast to Stat3β, cotransfection of an expression vector encoding full-length Stat3 results in increased levels of gene induction over that observed with v-Src alone (Fig. 3B). Together, these results establish that activation of Stat3 by v-Src leads to Stat3-mediated gene regulation.
FIG. 2.
Induction of Stat3-specific gene expression by v-Src. NIH 3T3 cells were transiently transfected with the indicated plasmid vectors, and luciferase reporter activity in cytosolic extracts was measured as light emission, with a luminometer. (A) Cells were transfected with pLucTK reporter alone or pLucTKS3 reporter plus increasing concentrations of the v-Src expression vector, pMvsrc. (B) Cells were transfected with pLucTK or pLucTKS3 reporters in the presence or absence of vectors encoding v-Src, Stat3β, or both. Values shown in each panel are means plus standard deviations of at least four independent transfections, each performed in triplicate. For each transfection, luciferase activity was normalized to transfection efficiency, with β-Gal activity as an internal control.
FIG. 3.
Stat3β disrupts, and Stat3 augments, Src-induced gene expression. NIH 3T3 cells were transiently transfected and luciferase reporter activities were assayed as described for Fig. 1. (A) Cells were transfected with the Stat3 reporter, pLucTKS3, and v-Src vector together with increasing concentrations of vector encoding Stat3β. (B) Cells were transfected with pLucTKS3 reporter and vectors encoding v-Src, full-length Stat3 (pVRStat3), or both. Values are means plus standard deviations of at least three independent experiments, each performed in triplicate and normalized to β-Gal activity.
Specificity of Stat3-mediated gene regulation.
An authentic promoter construct, pLucCRP (Fig. 1B), harboring cAPRE embedded in the natural context of the CRP gene’s promoter (48), was used to confirm the results obtained with the chimeric pLucTKS3 reporter. Figure 4A shows that v-Src induces expression from pLucCRP and that Stat3β functions as a dominant negative modulator of transcription from this promoter, in agreement with the results presented above. As a control, Fig. 4B shows that Stat3β overexpression has no effect on the ability of v-Src to induce another reporter construct, pLucSRE (Fig. 1B), containing the c-fos SRE that is dependent on Ras and Raf-1 signaling for activation (46). In similar experiments with 20 μg of Stat3β vector and 200 ng of v-Src vector, the same conditions used in subsequent focus formation assays (see below), there was no effect of Stat3β on the ability of Src to induce pLucSRE expression (data not shown). These results demonstrate that Stat3β acts through Stat3-binding sites and that Stat3β overexpression does not nonspecifically inhibit v-Src function or Stat3-independent signaling pathways leading to SRE induction.
FIG. 4.
Stat3β specifically blocks Stat3 but not Ras or Raf-1 signaling induced by v-Src. NIH 3T3 cells were transiently transfected and luciferase reporter activities were assayed as described for Fig. 1. (A) Cells were transfected with pLucCRP reporter together with or without vectors encoding v-Src, Stat3β, or both. (B) Cells were transfected with pLucSRE reporter together with v-Src in the presence or absence of vectors encoding Stat3β, N17-Ras, or NT-Raf. The N17-Ras and NT-Raf proteins are dominant negative mutants of c-H-Ras and Raf-1, respectively. Values are means plus standard deviations of at least three independent transfections, each performed in triplicate and normalized to β-Gal activity.
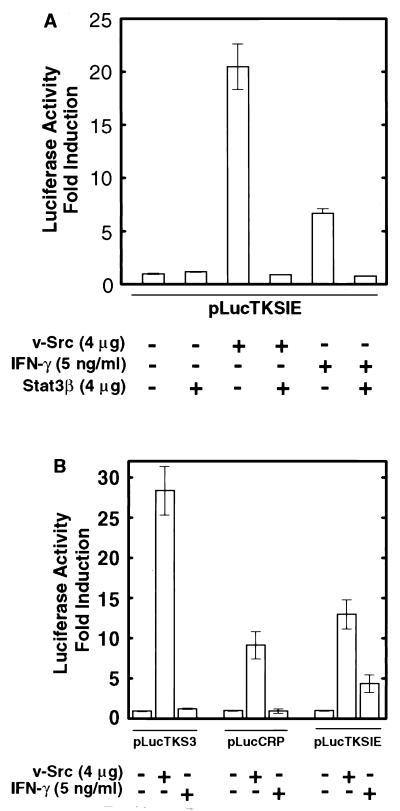
To further characterize the specificity of Src-induced STAT signaling and Stat3β function, we used a reporter construct based on the c-fos SIE inserted upstream of the TK promoter. This reporter, pLucTKSIE (Fig. 1B), contains a high-affinity mutant of the SIE (hSIE) that binds both Stat1 and Stat3 (41, 47). Figure 5A shows that expression from pLucTKSIE was induced by v-Src, which activates Stat3 but not Stat1 (47), and to a lesser extent by IFN-γ, which activates Stat1 but not Stat3 (4, 47). In both cases, gene induction was blocked by expression of Stat3β, indicating that Stat3β can disrupt Stat1 and Stat3 signaling. For comparison of the specificity of the various reporter constructs for Stat1 and Stat3, pLucTKS3, pLucCRP, and pLucTKSIE were activated by either v-Src or IFN-γ. Results shown in Fig. 5B demonstrate that the pLucTKS3 and pLucCRP constructs, both of which harbor cAPRE, are induced specifically by v-Src and not by IFN-γ. This finding confirms that cAPRE responds only to Stat3 signaling and not to Stat1 signaling. Together, our results demonstrate that activation of endogenous cellular Stat3 signaling by v-Src leads to Stat3-specific induction of gene expression, which is disrupted in a dominant-negative manner by Stat3β.
FIG. 5.
Specificity of promoter elements for STAT signaling induced by v-Src or IFN-γ. NIH 3T3 cells were transiently transfected and luciferase reporter activities were assayed as described for Fig. 1. (A) Cells were transfected with pLucTKSIE together with expression vectors for v-Src, Stat3β, or both. Transfectants with reporter alone or reporter and Stat3β vector were treated with IFN-γ for 5 h prior to harvest of cells. (B) Cells were transfected with the indicated reporters in the presence or absence of v-Src expression vector. Cells transfected with reporter alone were treated with IFN-γ for 5 h prior to harvest. Values are means plus standard deviations of at least three independent experiments, each performed in triplicate and normalized to β-Gal activity.
Induction of Stat3 and Stat3β DNA-binding activity by Src.
Because Stat3β retains an intact DNA-binding domain as well as the SH2 domain and tyrosine required for dimerization, activation of Stat3β by v-Src is predicted to induce dimerization and DNA binding. To measure DNA-binding activities, we prepared nuclear extracts from transiently transfected NIH 3T3 cells and performed EMSA with the 32P-labeled SIE probe that binds both Stat1 and Stat3 with high affinity. Consistent with earlier studies of fibroblasts stably transformed by v-Src (47), Fig. 6A (lanes 1, 2, and 11 to 15) shows that specific DNA-binding activity of endogenous Stat3, but not Stat1, is induced in an Src-dependent manner in transiently transfected NIH 3T3 cells. Moreover, the Src-induced levels of specific DNA-binding activity detected in transiently transfected cells overexpressing Stat3 or Stat3β are greater than 10-fold higher than that of cells containing only endogenous Stat3 (Fig. 6A, lanes 3 to 10). As reported earlier (5, 34), Stat3β appears to have a higher basal level of DNA-binding activity than does Stat3 in the absence of any external stimulus.
FIG. 6.
Induction of SIE binding activity by v-Src in transfected cells. Nuclear extracts were prepared from transiently transfected NIH 3T3 cells, and volumes containing equal amounts of total protein were subjected to EMSA by using 32P-labeled hSIE. (A) Cells were transfected with v-Src vector alone (NIH 3T3) or v-Src vector together with either Stat3 or Stat3β vector, as indicated. Lanes 7 to 10 are 1:10 dilutions of the samples loaded in lanes 3 to 6, respectively. Competitions of endogenous hSIE binding activity present in nuclear extracts of NIH 3T3 cells transfected with v-Src vector alone (lanes 12 and 13) were performed with a 100-fold molar excess of unlabeled hSIE or the unrelated c-fos intragenic regulatory element (FIRE) oligonucleotides. Supershifts (lanes 14 and 15) were performed with antibodies recognizing either amino acids 688 to 710 of Stat1 (αST1) or amino acids 750 to 769 of full-length Stat3 (αST3). (B) Nuclear extracts from cells transfected with v-Src vector plus Stat3 and/or Stat3β vector were subjected to EMSA, with competitions and supershifts performed as described for panel A. ST3, ST3β, and ST3/ST3β indicate migration of complexes containing Stat3 homodimers, Stat3β homodimers, and Stat3-Stat3β heterodimers, respectively. Asterisks indicate positions of supershifted complexes.
Previous studies of cells stimulated with IL-5 demonstrated that Stat3β can form homodimers, which migrate more slowly in EMSA than Stat3 homodimers, as well as heterodimers with Stat3, which exhibit an intermediate degree of migration (5). By supershift analysis with an antibody that recognizes a C-terminal epitope present in full-length Stat3 but not in Stat3β, we confirmed that activation of Stat3 or Stat3β by v-Src induces mostly homodimers when either one is overexpressed alone (Fig. 6B, lanes 1 to 10) and predominantly Stat3-Stat3β heterodimers when both proteins are overexpressed together (lanes 11 to 16). Combined with our results presented above, these data suggest that Stat3β disrupts Stat3-specific gene regulation by binding to Stat3 response elements as either a homodimer or a heterodimer and preventing transcriptional activation.
Cotransfection of Stat3β vector blocks Src transformation.
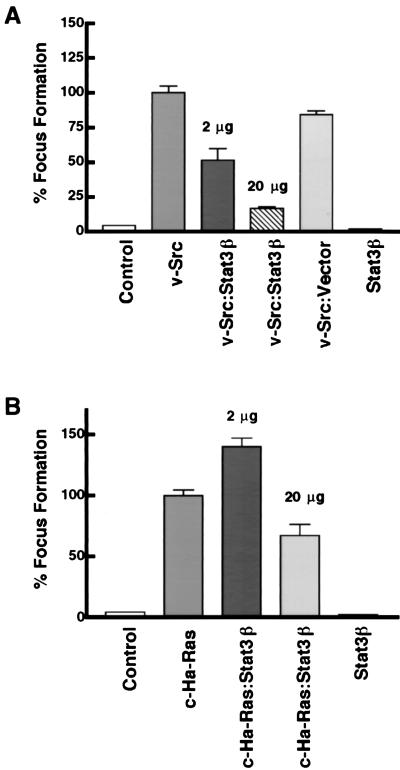
To investigate the role of Stat3 signaling in cell transformation, we tested the effect of Stat3β on transformation of NIH 3T3 cells by v-Src. As a sensitive and quantitative measure of cell transformation by v-Src, we used a focus formation assay, which in the case of v-Src correlates very well with growth in soft agar and tumorigenesis (18). Focus formation assays were performed by using cells transfected with expression vectors for v-Src alone or v-Src together with Stat3β (Fig. 7A). Results show that Stat3β consistently inhibited Src-induced focus formation by 50% with small amounts (2 μg) of Stat3β expression vector, with greater than 80% inhibition observed in cotransfections with larger amounts (20 μg) of Stat3β expression vector. As a control, cotransfection of empty vector alone did not significantly affect focus formation by v-Src, whereas expression of Stat3β alone resulted in levels of focus formation comparable to the background of spontaneous transformation. For comparison with v-Src, the effect of Stat3β overexpression on focus formation induced by the activated c-H-Ras oncoprotein, which does not activate Stat3 signaling (14), was also examined. Stat3β overexpression had either no effect (at 2 μg of vector) or much-less-pronounced effects (at 20 μg of vector) on Ras-induced transformation compared to Src-induced transformation (compare Fig. 7A and B). These results indicate that Stat3β inhibits transformation by Src to a significantly greater extent than it does transformation by Ras.
FIG. 7.
Cotransfection of Stat3β vector blocks transformation of NIH 3T3 cells induced by Src. (A) Cells were transfected with carrier DNA alone (control), with v-Src vector (200 ng) in the presence or absence of Stat3β vector (2 μg or 20 μg, as indicated) or empty vector, or with Stat3β vector (20 μg) alone. (B) Cells were transfected as described for panel A, except that activated c-H-Ras vector was used instead of v-Src vector. At least three independent sets of transfections were analyzed for Src and Ras focus formation assays. Values are means plus standard deviations of transfections from each experiment; percent focus formation is relative to that induced by Src or Ras alone (100%) within each of the independent sets of experiments.
Cell lines stably overexpressing Stat3β are resistant to Src transformation.
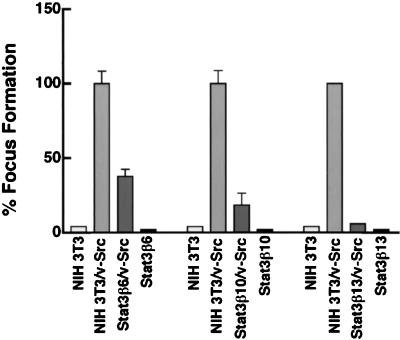
To confirm the results obtained in the cotransfection experiments described above, and to exclude a possible toxic effect of Stat3β overexpression on cell viability, a different approach was taken with cell lines stably overexpressing Stat3β. Following transfection with Stat3β expression vector, G418-resistant colonies were selected, expanded, and further characterized. Western blot analysis with antibodies directed against the N-terminal portion of Stat3 identified three independent clones that stably overexpressed Stat3β compared to control NIH 3T3 cells (Fig. 8A). Transient transfection of these cells with pLucTKS3 reporter together with v-Src vector confirmed that Stat3-mediated signaling was abrogated by Stat3β overexpression, while Stat3-independent signaling leading to pLucSRE induction was not affected (Fig. 8B). In agreement with the results presented in Fig. 2 to 4, these results indicate that Stat3β specifically disrupts Stat3 signaling without impairing v-Src function or other signaling pathways. These findings further demonstrate that cells which stably overexpress Stat3β and are deficient in Stat3-mediated signaling remain viable and proliferate. To assess whether these Stat3β overexpressing clones could be transformed by Src, focus formation assays were performed following transfection with v-Src expression vector. Results presented in Fig. 9 show that all three Stat3β-overexpressing clones were refractory to Src-induced cell transformation, with up to 90% inhibition of focus formation. Together, the data shown in Fig. 7 to 9 demonstrate that overexpression of Stat3β blocks cell transformation by v-Src, presumably by disrupting Stat3-dependent regulation of the cellular gene expression that is required for v-Src transformation.
FIG. 8.
Characterization of NIH 3T3 cell lines stably overexpressing Stat3β. (A) Western blot analysis of whole-cell lysates prepared from three independent NIH 3T3 cell lines stably overexpressing Stat3β. Lanes 1 to 4 were probed with antibodies against the N-terminal portion of Stat3 which recognize both full-length Stat3 and Stat3β. Lanes 5 to 8 were probed with antibodies to the Stat3 C terminus which recognize full-length Stat3 but not Stat3β. (B) Clone Stat3β10 was transiently transfected with either pLucSRE or pLucTKS3 reporter in the presence or absence of vector encoding v-Src, as indicated. Values for luciferase activity are means plus standard deviations of at least two independent transfections, each performed in triplicate. For each transfection, luciferase activity was normalized to transfection efficiency by using β-Gal activity as an internal control.
FIG. 9.
Cell lines stably overexpressing Stat3β are resistant to cell transformation by Src. Focus formation assays were performed with normal NIH 3T3 cells or the three independent clones overexpressing Stat3β represented in Fig. 8. In each experiment, cells were transfected with 200 ng of v-Src expression vector, with the exception of Stat3β13, in which 20 μg of v-Src vector was used. Values are means plus standard deviations of three independent transfections, except in the case of Stat3β13, which was tested once.
DISCUSSION
The initial discoveries of STAT proteins as mediators of intracellular signaling in response to cytokines and growth factors have launched an intensive investigation into the diverse biological functions of STATs (12, 16, 21). Activation of STAT signaling pathways, including Stat3, has been increasingly associated with cell transformation and human cancer (7, 11, 14, 15, 24, 25, 32, 42, 43, 47, 49). Cell transformation by v-Src is an ideal system for evaluating the role of Stat3 in oncogenesis because Stat3 is constitutively activated in mammalian fibroblasts stably transformed by v-Src (7, 47), suggesting a requirement for continuous signaling through Stat3 in order to maintain transformation. Moreover, we have determined that Stat3 is the predominant STAT family member activated in Src-transformed NIH 3T3 cells, with very little or no detectable activation of other STATs (14, 47), simplifying the analysis of Stat3’s role without the complexity of signaling by other STAT family members. To evaluate the role of Stat3 signaling in Src transformation, it was important to first establish that activation of Stat3 by v-Src leads to regulation of specific gene expression which could be disrupted in a dominant-negative manner by Stat3β. Our results demonstrate that activation of endogenous cellular Stat3 by the v-Src oncoprotein induces Stat3-mediated gene regulation that is specifically abrogated by overexpression of Stat3β in NIH 3T3 cells. The findings reported here establish the transcriptional potential of the frequently observed activation of Stat3 in transformed cells and provide evidence that this Stat3 signaling participates in oncogenesis. Because Stat3 is a constitutively activated transcription factor in the context of oncogenesis, these findings further imply a permanent alteration in the genetic program that contributes to oncogenesis of transformed cells harboring activated Stat3.
While the precise mechanism of Stat3 activation by Src is not completely defined, it has been shown that v-Src associates in a complex with Stat3 (7), consistent with the possibility that v-Src directly phosphorylates and activates Stat3. Other studies have provided evidence that JAK family kinases are constitutively activated in Src-transformed fibroblasts, suggesting that v-Src may indirectly activate Stat3 through JAKs (6). These two mechanisms are not mutually exclusive, and it is possible that both contribute to activation of Stat3 signaling by v-Src. In addition, we analyzed the interactions of Stat3 and Stat3β with each other to explore the mechanism by which overexpression of Stat3β could disrupt Stat3 signaling activated by Src. We detected DNA-binding activity by overexpressed Stat3β homodimers and Stat3-Stat3β heterodimers in nuclear extracts prepared from cells transiently transfected with v-Src, consistent with two possible mechanisms. One potential mechanism is the occupation of Stat3 binding sites by Stat3β homodimers, thereby titrating sites available for binding Stat3 homodimers. On the other hand, another possibility is that Stat3β could form heterodimers with Stat3 which may be transactivation deficient. Either or both of these mechanisms may contribute to the disruption of Stat3 signaling in NIH 3T3 cells under conditions in which Stat3 is constitutively activated by v-Src.
At the amino acid level, Stat3β is identical to full-length Stat3 for most of the protein but diverges at the carboxyl terminus with an internal deletion in the transactivation domain (5, 34). In the case of mouse Stat3, 55 amino acids are replaced by 7 different residues in Stat3β (34), whereas in the case of human Stat3, 17 amino acids are replaced by 7 different residues in Stat3β (5). Significantly, Stat3β retains the critical tyrosine residue at position 705 (Tyr-704 in human Stat3β), the phosphorylation of which is required for dimerization and DNA binding, but lacks the serine residue at position 727 (Ser-726 in human Stat3β). Phosphorylation of Ser-727 in full-length Stat3 is not required for DNA binding (44), although it is required for efficient transcriptional activation (45). However, apparently conflicting reports in the literature regarding the transactivation potential of Stat3β underscore the complexity of STAT signal transduction pathways. While an earlier report showed that coexpression of mouse Stat3β together with c-Jun transactivates an AP-1-dependent promoter (34), recent evidence demonstrates that mouse Stat3β acts as a transactivator or dominant negative modulator of transcription in a cell-type-specific manner (33). Although the basis for the variable transactivation potentials of Stat3β is unknown, it may involve interactions of Stat3β with other cell-type-specific transcription factors or coactivators. It is also possible that the structural differences between mouse and human Stat3β in the C-terminal portion or elsewhere account for the different transactivation potentials that have been observed (5, 34).
Despite being one of the most well characterized oncoproteins, the molecular mechanisms by which v-Src subverts normal cellular signaling pathways and transforms cells are not fully defined (1, 38). The present data demonstrate that activation of Stat3 signaling is required for cell transformation by the v-Src oncoprotein, suggesting that constitutive activation of Stat3 is one of the cellular signaling pathways that participates in maintenance of transformation by v-Src. Given the myriad changes that accompany cell transformation (18), it is probable that activation of Stat3 signaling is not sufficient by itself to induce cell transformation and that other signaling pathways are also required for transformation by v-Src. At the same time, it is unlikely that Stat3 is involved in transformation by all oncoproteins, since cell transformation mediated by activated c-H-Ras was not significantly inhibited by coexpression of Stat3β. In addition, because we have been able to generate stable cell lines overexpressing Stat3β, these results demonstrate that enforced overexpression of Stat3β does not severely impair normal cell function or have toxic effects on cell viability. Using a panel of various Stat3 dominant negative mutants and different assays of cell transformation, Bromberg et al. have arrived independently at similar conclusions (3). Together, these findings provide the first direct evidence that, in addition to their signaling functions in normal cells, STATs also participate in oncogenesis. Because activation of Stat3 is associated with human tumors, including breast carcinomas and various lymphoid malignancies (14, 15, 32, 42, 43, 49), our findings further suggest an important role for Stat3 signaling in the development of these cancers.
The biological mechanism by which Stat3 contributes to oncogenesis remains to be determined. We speculate that Stat3 signaling may contribute to transformation induced by v-Src in NIH 3T3 cells through one of two likely biological mechanisms. One possible mechanism is that constitutive activation of Stat3 signaling may directly stimulate cell proliferation. This possibility is consistent with the finding that numerous growth factors, such as platelet-derived growth factor and epidermal growth factor, activate signaling by STATs, including Stat3 (22, 31, 40, 50). In addition, gene knockout studies have demonstrated a requirement for Stat4 and Stat6 signaling in mitogenic responses to cytokine stimulation of immune cells (19, 20). Alternatively, Stat3 may contribute to cell transformation by preventing apoptosis, thereby indirectly increasing cell numbers. This possibility is supported by the finding that activation of Stat3 is required for the anti-apoptosis response to IL-6 stimulation in a murine myeloid cell line (13). Since Stat3 signaling is implicated in control of cell differentiation, proliferation, and apoptosis (10, 13, 27, 39), the biological consequences of constitutive Stat3 activation are likely to be complex and dependent on the specific cell type. Nevertheless, the demonstration that Stat3 is one of the critical signaling pathways required for cell transformation by v-Src implies the existence of Stat3-regulated genes that participate in oncogenesis. Thus, identification and characterization of the nuclear genes regulated by Stat3 should provide new insights into the specific events leading to the loss of normal cell growth control and the process of malignant transformation.
ACKNOWLEDGMENTS
The first two authors (J.T. and T.B.) contributed equally to this work.
We thank D. Samols for the −123/+3CRP-CAT plasmid; N. Sinibaldi for help with constructing pLucCRP; K. Pumiglia for the NT-Raf and N17-Ras vectors; D. Cress, J. Pledger, and J. Wu for advice and comments on the manuscript; members of the lab for stimulating discussions; J. Zeng for technical assistance; and the Moffitt Cancer Center Molecular Biology Core and Molecular Imaging Facility.
This work was supported by NIH grant CA55652 to R.J.
REFERENCES
- 1.Aftab D T, Kwan J, Martin G S. Ras-independent transformation by v-Src. Proc Natl Acad Sci USA. 1997;94:3028–3033. doi: 10.1073/pnas.94.7.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Nishio Y, Inoue M, Wang X-J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 2a.Bowman, T., and R. Jove. Unpublished data.
- 3.Bromberg J F, Horvath C M, Besser D, Lathem W W, Darnell J E., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldenhoven E, van Dijk T B, Solari R, Armstrong J, Raaijmakers J A M, Lammers J W J, Koenderman L, de Groot R P. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J Biol Chem. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 6.Campbell G S, Yu C L, Jove R, Carter-Su C. Constitutive activation of JAK1 in Src-transformed cells. J Biol Chem. 1997;272:2591–2594. doi: 10.1074/jbc.272.5.2591. [DOI] [PubMed] [Google Scholar]
- 7.Cao X, Tay A, Guy G R, Tan Y H. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaturvedi P, Sharma S, Reddy E P. Abrogation of interleukin-3 dependence of myeloid cells by the v-src oncogene requires SH2 and SH3 domains which specify activation of STATs. Mol Cell Biol. 1997;17:3295–3304. doi: 10.1128/mcb.17.6.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C A, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 10.Cressman D E, Greenbaum L E, DeAngelis R A, Ciliberto G, Furth E E, Poli V, Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 11.Danial N N, Pernis A, Rothman P B. Jak-STAT signaling induced by the v-abl oncogene. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 12.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 13.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of Stat3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 14.Garcia R, Yu C-L, Hudnall A, Catlett R, Nelson K L, Smithgall T, Fujita D J, Ethier S P, Jove R. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ. 1997;8:1267–1276. [PubMed] [Google Scholar]
- 15.Gouilleux-Gruart V, Gouilleux F, Desaint C, Claisse J F, Capiod J C, Delobel J, Weber-Nordt R, Dusanter-Fourt I, Dreyfus F, Groner B, Prin L. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood. 1996;87:1692–1697. [PubMed] [Google Scholar]
- 16.Horvath C M, Darnell J E., Jr The state of the STATs: recent developments in the study of signal transduction to the nucleus. Curr Opin Cell Biol. 1997;9:233–239. doi: 10.1016/s0955-0674(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 17.Johnson P J, Coussens P M, Danko A V, Shalloway D. Overexpressed pp60c-src can induce focus formation without complete transformation of NIH 3T3 cells. Mol Cell Biol. 1985;5:1073–1083. doi: 10.1128/mcb.5.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jove R, Hanafusa H. Cell transformation by the viral src oncogene. Annu Rev Cell Biol. 1987;3:31–56. doi: 10.1146/annurev.cb.03.110187.000335. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan M H, Schindler U, Smiley S T, Grusby M J. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan M H, Sun Y L, Hoey T, Grusby M J. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 21.Leaman D W, Leung S, Li X, Stark G R. Regulation of STAT-dependent pathways by growth factors and cytokines. FASEB J. 1996;10:1578–1588. [PubMed] [Google Scholar]
- 22.Leaman D W, Pisharody S, Flickinger T W, Commane M A, Schlessinger J, Kerr I M, Levy D E, Stark G R. Roles of JAKs in activation of STATs and stimulation of c-fos gene expression by epidermal growth factor. Mol Cell Biol. 1996;16:369–375. doi: 10.1128/mcb.16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Robinson G W, Hennighausen L. Activation of Stat5a and Stat5b by tyrosine phosphorylation is tightly linked to mammary gland differentiation. Mol Endocrinol. 1996;10:1496–1506. doi: 10.1210/mend.10.12.8961260. [DOI] [PubMed] [Google Scholar]
- 24.Lund T C, Garcia R, Medveczky M M, Jove R, Medveczky P G. Activation of STAT transcription factors by herpesvirus saimiri Tip-484 requires p56lck. J Virol. 1997;71:6677–6682. doi: 10.1128/jvi.71.9.6677-6682.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Migone T S, Lin J X, Cereseto A, Mulloy J C, O’Shea J J, Franchini G, Leonard W J. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-1. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 26.Muli A L, Wakao H, Kinoshita T, Kitamura T, Miyajima A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J. 1996;15:2425–2433. [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 28.Parada L F, Tabin C J, Shih C, Weinberg R A. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 29.Planas A M, Berruezo M, Justicia C, Barron S, Ferrer I. Stat3 is present in the developing and adult rat cerebellum and participates in the formation of transcription complexes binding DNA at the sis-inducible element. J Neurochem. 1997;68:1345–1351. doi: 10.1046/j.1471-4159.1997.68041345.x. [DOI] [PubMed] [Google Scholar]
- 30.Pumiglia K, Chow Y H, Fabian J, Morrison D, Decker S, Jove R. Raf-1 N-terminal sequences necessary for Ras-Raf interaction and signal transduction. Mol Cell Biol. 1995;15:398–406. doi: 10.1128/mcb.15.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruff-Jamison S, Chen K, Cohen S. Induction by EGF and interferon-gamma of tyrosine phosphorylated DNA binding proteins in mouse liver nuclei. Science. 1993;261:1733–1736. doi: 10.1126/science.8378774. [DOI] [PubMed] [Google Scholar]
- 32.Sartor C I, Dziubinski M L, Yu C L, Jove R, Ethier S P. Role of epidermal growth factor receptor and STAT-3 activation in autonomous proliferation of SUM-102PT human breast cancer cells. Cancer Res. 1997;57:978–987. [PubMed] [Google Scholar]
- 33.Sasse J, Hemmann U, Schwartz C, Schniertshauer U, Heesel B, Landgraf C, Schneider-Mergener J, Heinrich P C, Horn F. Mutational analysis of acute-phase response factor/Stat3 activation and dimerization. Mol Cell Biol. 1997;17:4677–4686. doi: 10.1128/mcb.17.8.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer T S, Sanders L K, Nathans D. Cooperative transcriptional activity of Jun and Stat3beta, a short form of Stat3. Proc Natl Acad Sci USA. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 36.Seidel H M, Milocco L H, Lamb P, Darnell J E, Stein R B, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens J M, Morrison R F, Pilch P F. The expression and regulation of STATs during 3T3-L1 adipocyte differentiation. J Biol Chem. 1996;271:10441–10444. doi: 10.1074/jbc.271.18.10441. [DOI] [PubMed] [Google Scholar]
- 38.Stofega M R, Yu C L, Wu J, Jove R. Activation of extracellular signal-regulated kinase (ERK) by mitogenic stimuli is repressed in v-Src-transformed cells. Cell Growth Differ. 1997;8:113–119. [PubMed] [Google Scholar]
- 39.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vignais M L, Sadowski H B, Watling D, Rogers N C, Gilman M. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol Cell Biol. 1996;16:1759–1769. doi: 10.1128/mcb.16.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner B J, Hayes T E, Hoban C J, Cochran B H. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson C J, Miller W R. Elevated levels of members of the STAT family of transcription factors in breast carcinoma nuclear extracts. Br J Cancer. 1995;71:840–844. doi: 10.1038/bjc.1995.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber-Nordt R M, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- 44.Wen Z, Darnell J E., Jr Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res. 1997;25:2062–2067. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 46.Yamauchi K, Holt K, Pessin J E. Phosphatidylinositol 3-kinase functions upstream of Ras and Raf in mediating insulin stimulation of c-fos transcription. J Biol Chem. 1993;268:14597–14600. [PubMed] [Google Scholar]
- 47.Yu C L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 48.Zhang D, Sun M, Samols D, Kushner I. STAT3 participates in transcriptional activation of the C-reactive protein gene by interleukin-6. J Biol Chem. 1996;271:9503–9509. doi: 10.1074/jbc.271.16.9503. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Q, Nowak I, Vonderheid E C, Rook A H, Kadin M E, Nowell P C, Shaw L M, Wasik M A. Activation of Jak/STAT proteins involved in signal transduction pathway mediated by receptor for interleukin 2 in malignant T lymphocytes derived from cutaneous anaplastic large T-cell lymphoma and Sezary syndrome. Proc Natl Acad Sci USA. 1996;93:9148–9153. doi: 10.1073/pnas.93.17.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong Z, Wen Z, Darnell J E., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 51.Zong C, Yan R, August A, Darnell J E, Hanafusa H. Unique signal transduction of Eyk: constitutive stimulation of the JAK-STAT pathway by an oncogenic receptor-type tyrosine kinase. EMBO J. 1996;15:4515–4525. [PMC free article] [PubMed] [Google Scholar]