Abstract
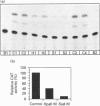
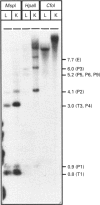
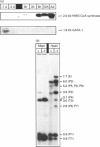
The mitochondrial 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase gene is expressed in a limited set of tissues in the adult rat. Methylation of the 5' flanking region of the gene in vitro leads to its transcriptional inactivation when transfected in hepatoma-derived cell lines. In liver and kidney, expression of the gene correlates inversely with its degree of methylation, indicating that the methylation of the 5' flanking region and the first exon of the gene may be one of the factors responsible for the repression of its transcription. During the fetal/neonatal transition, a process of selective undermethylation of specific sites takes place in the 5' flanking region of the mitochondrial HMG-CoA synthase gene. Moreover, treatment with the hypomethylating agent 5-azacytidine of a hepatoma-derived cell line that presents barely detectable levels of mitochondrial HMG-CoA synthase mRNA leads to a significant increase in the mRNA levels. These results point to methylation as one of the regulatory mechanisms that operate on the mitochondrial HMG-CoA synthase gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990 Aug 10;62(3):503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Ayté J., Gil-Gómez G., Haro D., Marrero P. F., Hegardt F. G. Rat mitochondrial and cytosolic 3-hydroxy-3-methylglutaryl-CoA synthases are encoded by two different genes. Proc Natl Acad Sci U S A. 1990 May;87(10):3874–3878. doi: 10.1073/pnas.87.10.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenisty N., Mencher D., Meyuhas O., Razin A., Reshef L. Sequential changes in DNA methylation patterns of the rat phosphoenolpyruvate carboxykinase gene during development. Proc Natl Acad Sci U S A. 1985 Jan;82(2):267–271. doi: 10.1073/pnas.82.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenisty N., Reshef L. Developmental expression and modification of genes. Biol Neonate. 1987;52(2):61–69. doi: 10.1159/000242685. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Casals N., Roca N., Guerrero M., Gil-Gómez G., Ayté J., Ciudad C. J., Hegardt F. G. Regulation of the expression of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene. Its role in the control of ketogenesis. Biochem J. 1992 Apr 1;283(Pt 1):261–264. doi: 10.1042/bj2830261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Dashti N., Ontko J. A. Rate-limiting function of 3-hydroxy-3-methylglutaryl-coenzyme A synthase in ketogenesis. Biochem Med. 1979 Dec;22(3):365–374. doi: 10.1016/0006-2944(79)90024-3. [DOI] [PubMed] [Google Scholar]
- Dynan W. S. Understanding the molecular mechanism by which methylation influences gene expression. Trends Genet. 1989 Feb;5(2):35–36. doi: 10.1016/0168-9525(89)90016-4. [DOI] [PubMed] [Google Scholar]
- Ferré P., Pégorier J. P., Williamson D. H., Girard J. R. The development of ketogenesis at birth in the rat. Biochem J. 1978 Dec 15;176(3):759–765. doi: 10.1042/bj1760759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Gómez G., Ayté J., Hegardt F. G. The rat mitochondrial 3-hydroxy-3-methylglutaryl-coenzyme-A-synthase gene contains elements that mediate its multihormonal regulation and tissue specificity. Eur J Biochem. 1993 Apr 15;213(2):773–779. doi: 10.1111/j.1432-1033.1993.tb17819.x. [DOI] [PubMed] [Google Scholar]
- Gil G., Goldstein J. L., Slaughter C. A., Brown M. S. Cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme A synthase from the hamster. I. Isolation and sequencing of a full-length cDNA. J Biol Chem. 1986 Mar 15;261(8):3710–3716. [PubMed] [Google Scholar]
- Girard J., Duée P. H., Ferré P., Pégorier J. P., Escriva F., Decaux J. F. Fatty acid oxidation and ketogenesis during development. Reprod Nutr Dev. 1985;25(1B):303–319. doi: 10.1051/rnd:19850221. [DOI] [PubMed] [Google Scholar]
- Jones P. A. Altering gene expression with 5-azacytidine. Cell. 1985 Mar;40(3):485–486. doi: 10.1016/0092-8674(85)90192-8. [DOI] [PubMed] [Google Scholar]
- Khandjian E. W. UV crosslinking of RNA to nylon membrane enhances hybridization signals. Mol Biol Rep. 1986;11(2):107–115. doi: 10.1007/BF00364822. [DOI] [PubMed] [Google Scholar]
- Lewis J., Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991 Jul 22;285(2):155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- McGarry J. D., Foster D. W. Regulation of hepatic fatty acid oxidation and ketone body production. Annu Rev Biochem. 1980;49:395–420. doi: 10.1146/annurev.bi.49.070180.002143. [DOI] [PubMed] [Google Scholar]
- Murray E. J., Grosveld F. Site specific demethylation in the promoter of human gamma-globin gene does not alleviate methylation mediated suppression. EMBO J. 1987 Aug;6(8):2329–2335. doi: 10.1002/j.1460-2075.1987.tb02508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quant P. A., Robin D., Robin P., Ferre P., Brand M. D., Girard J. Control of hepatic mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase during the foetal/neonatal transition, suckling and weaning in the rat. Eur J Biochem. 1991 Jan 30;195(2):449–454. doi: 10.1111/j.1432-1033.1991.tb15724.x. [DOI] [PubMed] [Google Scholar]
- Quant P. A., Tubbs P. K., Brand M. D. Glucagon activates mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase in vivo by decreasing the extent of succinylation of the enzyme. Eur J Biochem. 1990 Jan 12;187(1):169–174. doi: 10.1111/j.1432-1033.1990.tb15291.x. [DOI] [PubMed] [Google Scholar]
- Quant P. A., Tubbs P. K., Brand M. D. Treatment of rats with glucagon or mannoheptulose increases mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase activity and decreases succinyl-CoA content in liver. Biochem J. 1989 Aug 15;262(1):159–164. doi: 10.1042/bj2620159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo T., Pedragosa M. J., Ayté J., Gil-Gómez G., Vilaró S., Hegardt F. G. Testis and ovary express the gene for the ketogenic mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase. J Lipid Res. 1993 Jun;34(6):867–874. [PubMed] [Google Scholar]
- Thompson J. P., Simkevich C. P., Holness M. A., Kang A. H., Raghow R. In vitro methylation of the promoter and enhancer of Pro alpha 1(I) collagen gene leads to its transcriptional inactivation. J Biol Chem. 1991 Feb 5;266(4):2549–2556. [PubMed] [Google Scholar]
- Tsai S. F., Martin D. I., Zon L. I., D'Andrea A. D., Wong G. G., Orkin S. H. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989 Jun 8;339(6224):446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- de Wet J. R., Wood K. V., DeLuca M., Helinski D. R., Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987 Feb;7(2):725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]