Abstract
DNA polymerase theta () is an error-prone translesion polymerase that becomes crucial for DNA double-strand break repair when cells are deficient in homologous recombination or non-homologous end joining. In some organisms, also promotes tolerance of DNA interstrand crosslinks. Due to its importance in DNA damage tolerance, is an emerging target for treatment of cancer and disease. Prior work has characterized the functions of the helicase-like and polymerase domains, but the roles of the linker domain are largely unknown. Here, we show that the Drosophila melanogaster linker domain promotes egg development and is required for tolerance of DNA double-strand breaks and interstrand crosslinks. While a linker domain with scrambled amino acid residues is sufficient for DNA repair, replacement of the linker with part of the Homo sapiens linker or a disordered region from the FUS RNA-binding protein does not restore function. These results demonstrate that the linker domain is not simply a random tether between the helicase-like and polymerase domains. Furthermore, they suggest that intrinsic amino acid residue properties, rather than protein interaction motifs, are more critical for linker functions in DNA repair.
Introduction
DNA double-strand breaks (DSBs) are harmful DNA lesions that can be induced by exogenous agents such as ionizing radiation (IR) or normal cellular processes such as DNA replication and transcription (1). To prevent genomic catastrophe, metazoans frequently use two pathways to repair DSBs: non-homologous end joining (NHEJ) and homologous recombination (HR). NHEJ is favored when DNA resection is inhibited, involves limited processing and ligation of DSB ends, and can result in small mutations (2–4). HR requires DNA resection, strand invasion and fill-in synthesis, and is typically error-free (5). Since NHEJ and HR account for the majority of DSB repair, deficiencies in either pathway can lead to cell death or pathogenic mutations.
HR or NHEJ-deficient cells often utilize an alternative pathway for DSB repair and survival called theta-mediated end joining (TMEJ) (6–11). The most essential factor in the TMEJ pathway is DNA polymerase theta (, POLQ). Like HR, TMEJ is promoted by DNA resection, but in contrast to HR, TMEJ regularly causes deletions and insertions in the genome (12–14). TMEJ-induced mutations and elevated levels of frequently occur in cancers and have been associated with poor prognoses for cancer patients (13,15–21).
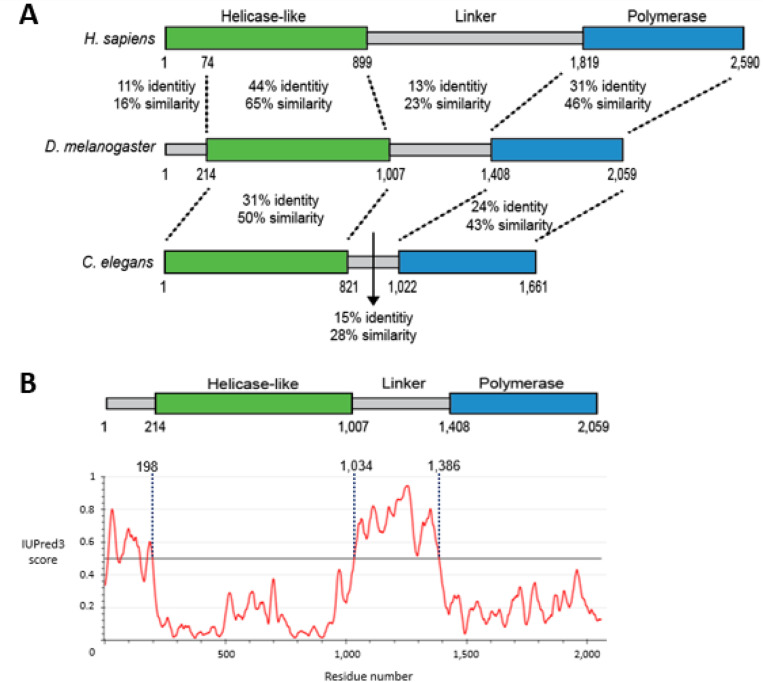
possesses three domains: helicase-like, linker, and polymerase. The helicase-like and polymerase domains are largely conserved in metazoans, while linker domains are highly divergent (Figure 1A, Supplementary Figures S1 and S2). Prior work has shown that the helicase-like domain dissociates RAD51 and RPA from single-stranded DNA (ssDNA) and promotes microhomology synapsis needed for TMEJ (22–24). Following synapsis, the polymerase domain stabilizes synapsed ends and synthesizes new DNA to fill in gaps (25,26).
Figure 1.
The linker domain is mostly not conserved among metazoans and has a disordered structure. (A) Percent identity and similarity scores for pairwise alignments between domains (Needleman-Wunsch global alignments, Blosum62 cost matrix, penalties: gap open=12, gap extension=3, similarity threshold=1). Alignments with previously defined H. sapiens helicase-like (1–899) and polymerase (1,819–2,590) domains (37,38) were used to determine domain boundaries in D. melanogaster and C. elegans sequences: domain ends coincide with the end of grouped identical residues (not shown, Geneious global alignments with free end gaps, Blosum62 cost matrix, penalties: gap open=12, gap extension=3). (B) Prediction of D. melanogaster disorder using IUPred3 long disorder and strong smoothing tools (39). Residues with scores above the 0.5 threshold (horizontal line) are predicted to be disordered (40).
Mammalian cells deficient in demonstrate sensitivity to IR-induced DNA breaks (27,28). HR-deficient Drosophila require polymerase activity, but not helicase-like ATPase activity to resolve IR-induced breaks (29). In addition to repairing DSBs, promotes resolution of interstrand crosslinks (ICLs) in Mus musculus embryonic fibroblasts, Caenorhabditis elegans, Arabidopsis thaliana, and Drosophila melanogaster (30–35). In Drosophila, both polymerase and helicase-like ATPase activities are required for organismal resistance to interstrand crosslinks (29). The mechanism for how supports ICL tolerance is unknown, but in vitro evidence suggests facilitates bypass of ICLs (29), possibly to encourage processive synthesis during DNA replication.
Roles of the linker domain in DNA repair are largely unexplored. Previous reports showed that the Homo sapiens linker domain is not required for TMEJ in vitro (22), but phosphorylated serine residues within the linker promote a interaction with TOPBP1 and mitotic DSB repair in vivo (36). It is currently unclear how well the respective phosphorylation sites are conserved within Drosophila and other eukaryotes and if other regions in linker domains may also influence DNA repair in vivo.
linker domains contain intrinsically disordered regions (IDRs) (Figure 1B and Supplementary Figure S3). IDRs can encourage nuclear localization, cell cycle signaling, autoinhibition of protein activity, and protein-protein interactions (41,42). These functions are fine-tuned by post-translational modifications (PTMs) that alter domain architecture and respective enzyme activity or protein binding interfaces (43,44). Previous reports suggest IDRs influence the functions or recruitment of RAD52, p53, FUS, and several other DNA repair proteins (45–48). Accordingly, we postulated that the IDRs in the linker domain may also be important for DNA repair.
To test this hypothesis, we performed mutagen sensitivity assays, measured egg hatching, and explored linker sequence requirements for repair using replacement IDRs. We found that the linker domain is critical for repair of DNA double-strand breaks and tolerance to interstrand crosslinks and promotes normal Drosophila egg development. Interestingly, a randomly scrambled Drosophila linker, but not the Homo sapiens linker sequence or a disordered region from the FUS protein, is sufficient for DNA damage tolerance. Overall, our findings establish that the linker domain is required for -dependent repair of DNA damage in vivo and its functions are likely independent of amino acid residue arrangement.
Materials and Methods
Non-transgenic flies
All flies were raised at 25°C on a standard cornmeal diet with a 12:12 hour light/dark cycle. Wild-type flies were (BDSC 3605) unless otherwise noted.
The null allele contains a piggyBac transposon inserted by CRISPR-mediated knock-in at the endogenous POLQ locus (technique described in https://flycrispr.org/scarless-gene-editing/) (49,50). An 825 bp homology arm containing most of the POLQ promoter and an 800 bp homology arm containing most of POLQ exon 1 were amplified by PCR and inserted into pHD-w+ (Addgene 80927, gift from Kate O’Connor-Giles) (NEBuilder HiFi Assembly). BbsI restriction overhangs were incorporated on the ends of an oligo template (5’ GAAACAGCACCTTAATGGCGC 3’) for a gRNA that targets the beginning of the POLQ coding sequence. The gRNA oligo was inserted into pU6-Bbs1_chiRNA (Addgene 45946, gift from Melissa Harrison, Kate O’Connor-Giles, and Jill Wildonger) (51). The respective gRNA PAM site was mutagenized in a POLQ homology arm within pHD-w+ by Q5 site-directed mutagenesis (New England BioLabs). Another fragment containing PBac{3XP3-DsRed} was amplified (Addgene 80820, gift from Kate O’Connor-Giles) and inserted in between POLQ homology arms in pHD-w+. pHD-w+ and pU6-Bbs1_chiRNA were injected into Cas9-expressing embryos (BDSC 51324) to generate a transformant (BestGene). The piggyBac insertion created an early stop codon at the beginning of the endogenous POLQ locus and the allele was validated by PCR (Primers 1 and 2 in Supplementary Table S1).
The null mutant was made using CRISPR-induced mutagenesis. Cas9 (BDSC 79006) and a spn-A gRNA (BDSC 76440) were expressed in the male germline. In a male offspring, a four-nucleotide deletion and early stop codon were recovered at the beginning of the spn-A coding sequence. The allele was characterized by sequencing and validated in flies by PCR (Primers 3 and 4).
Transgenic flies
To make the transgene, a 4.572 kb fragment, which included 1.287 kb upstream of the POLQ translation start site (endogenous POLQ promoter and 5’ UTR) and 3.252 kb downstream of the translation start site, was amplified using genomic DNA and primers containing BglII and NotI restriction sites. The restriction fragment was ligated into pattB (DGRC 1420). A 2.756 kb fragment, which included 2.329 kb upstream of the stop codon, 3’ UTR, and 242 bp downstream of the 3’ UTR, was ligated into pattB using primers with NotI and Acc65I restriction sites. 953 bp within exon 4 (the linker domain) were excluded from 5’ and 3’ POLQ restriction fragments. was validated by sequencing and injected into -expressing embryos (BestGene). Transgenes were inserted at ZH-68E-attP on chromosome 3L (BDSC 24485). was validated in Drosophila by PCR (Primers 6 and 7).
To make the 3xFLAG-tagged transgene, and primers containing AvrII and SacI restriction sites were used to amplify 1.287 kb upstream of the POLQ translation start site. The restriction fragment was ligated into pAFW-Ago2 (Addgene 50554, gift from Yukihide Tomari) (52). Using pAFW-Ago2, a 1.395 kb fragment (promoter, 5’ UTR, Kozak sequence, and 3xFLAG) was amplified using primers with BamHI and BglII restriction sites and ligated into pattB. The transgene was amplified downstream of the translation start site using as a template and primers with BglII and Acc65I restriction sites, then ligated into pattB. was validated by sequencing and was validated in Drosophila by PCR (Primers 6 and 7).
was made in the same way as except genomic DNA was used to amplify POLQ. was validated by sequencing and was validated in Drosophila by PCR (Primers 5 and 2).
To make replacement-linker transgenes, gBlocks (Integrated DNA Technologies) were inserted into . The gBlocks coded for randomly scrambled Drosophila melanogaster linker residues 1,025–1,342, residues 1,483–1,800 from Homo sapiens , and residues 2–214 from Homo sapiens FUS (Supplementary Table S2). NotI restriction sites and adapters for sequence stabilization were added to gBlock ends. Sequences were codon optimized for Drosophila melanogaster (Integrated DNA Technologies). gBlocks were ligated into pminiT2.0 for propagation (New England BioLabs PCR Cloning) and digested by NotI for insertion into to make , , and . gBlock orientation and composition within each plasmid were validated by PCR and sequencing (Primers 6 and 7). Transgenes were validated in Drosophila by PCR (: primers 8 and 7, : 6 and 9, : 10 and 7).
Quantitative PCR
To extract RNA, 9–15 second/third instar larvae or flies were homogenized in TRIzol (Thermo Fisher Scientific). The homogenate was vigorously mixed with chloroform and centrifuged (15 minutes, 12,000 × g at 4°C). RNA in the upper aqueous layer was removed, mixed with isopropanol, and incubated for 10 minutes at −20°C. After centrifugation (10 minutes, 12,000 × g at 4°C), the pellet was washed with 75% ethanol and allowed to dry. Pellets were resuspended in nuclease-free water and stored at −80°C.
A portion of each RNA sample was treated with DNase I in the respective buffer for 30 minutes at 37°. DNase I activity was inactivated in 4mM EDTA and by heating for 10 minutes at 72°C. DNaseI-treated 1 μg RNA aliquots were immediately used in cDNA synthesis and no-reverse transcriptase control (no-RT) reactions with random primers (New England BioLabs Protoscript II kit).
For qPCR reactions, cDNA and no-RT samples were diluted 1:3. The following reagents were loaded into each well: 10 μL of 2X SYBR Green Fast master mix (ABclonal), 0.4 μL of forward and reverse primers (0.2 μM of each), 7.2 μL of nuclease-free H2O, and 2 μL of the diluted sample. Biological replicates were run in triplicate for each genotype and primer set. One primer set amplified POLQ, but did not amplify (Primers 11 and 12, Supplementary Figure S4 and Supplementary Table S3). The second primer set amplified an endogenous control gene, rp49 (Primers 13 and 14). No-template controls for both primer sets were run in triplicate. Samples were analyzed in a QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems) using the following cycling conditions: 50°C for 2 minutes, 95°C for 6 minutes, (95°C for 15 seconds, 60°C for 20 seconds) × 40 cycles, and a continuous melt curve stage (95°C for 15 seconds, 60°C for 20 seconds, 95°C for 15 seconds). rp49 Ct triplicate averages were subtracted from POLQ Ct triplicate averages to calculate for each biological replicate and calibrator values were subtracted from biological replicate values to calculate and relative quantification .
RQmin and RQmax confidence intervals were calculated using the following adapted equations (Applied Biosystems):
T=student’s two-tailed T factor for mean where and .
SE=standard error of where Ct SD=triplicate standard deviation for a target.
Immunoprecipitation and western blotting
, - or , -homozygous flies were loaded into separate sets of 1.3-liter fly cages containing grape juice agar plates. 500 mg of embryos were collected for each genotype and frozen in liquid nitrogen. Embryos were suspended in lysis buffer (20 mM HEPES pH 7.6, 500 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, EDTA-free Pierce protease inhibitor (Thermo Fisher Scientific)) and sonicated for three 10 second bursts at 20% duty cycle with 10 second rests between bursts. Lysates were centrifuged to remove debris (4°C, 24,000 × g). For embryos carrying wild-type POLQ (BDSC 463), proteins were previously extracted using other established methods (53). Lysates were snap-frozen in liquid nitrogen and stored at −80°C.
For 3xFLAG immunoprecipitation, aliquots of ANTI-FLAG M2 Magnetic Beads (20 μL of packed gel volume per aliquot, MilliporeSigma) were washed twice with TBS (50 mM Tris-HCl, 150 mM NaCl) using a magnetic separator. 1 mL lysates were cleared by centrifugation (4°C, 14,000 × g) and added to the bead aliquots. Samples were rotated overnight at 4°C and supernatant was discarded. Beads were washed three times with TBS. FLAG-tagged protein was eluted twice using 100 μL of FLAG peptide (150 ng/μL, gift from Stephen Fuchs) during 30-minute rotations at 4°C.
Proteins were electrophoresed in a 4–20% SurePAGE gel and Tris-MOPS-SDS running buffer (GenScript). Proteins were transferred to a nitrocellulose membrane in wet conditions (25 mM Tris base, 25 mM bicine, 10% ethanol). The membrane was blocked (PBS, 5% non-fat milk) and probed with rabbit anti-DYDDDDK primary antibody (GenScript, 1:2,500 ; PBS-0.05% Tween 20, 1% non-fat milk) and rabbit IgG (HRP) secondary antibody (Abcam,1:10,000 ; PBS-0.05% Tween 20, 1% non-fat milk) with washes after antibody incubations (PBS-0.05% Tween 20). Horseradish peroxidase signal was induced using Prometheus ProSignal ECL (Genesee Scientific).
Mutagen sensitivity assays
In IR assays, each cross included 20–60 females from one stock and 10–30 males from a separately maintained stock that were heterozygous for the same alleles. Crosses were housed at 25°C in 100 mL cages that included grape juice agar plates coated with yeast paste. Grape juice agar plates were replaced in cages every 24 hours. Once third instar larvae emerged, plates were either treated with Ce-137 radiation in a Gammator 1000 irradiator or left untreated. Larvae were transferred to bottles containing standard cornmeal food. Eclosed flies in each bottle were scored as homozygous or heterozygous and relative survival was calculated: . Progeny from 3–4 cross replicates were counted for each genotype. Crosses typically yielded hundreds of flies and not fewer than 32.
We anticipated that unmapped mutations, either randomly inherited during stock construction or caused by DNA repair deficiencies, could result in variable or misleading survival results in sensitivity assays. Accordingly, when possible, female and male parents were taken from separately constructed stocks of the same genotype to increase the likelihood that random mutations would be complemented in trans-heterozygous progeny. Reciprocal crosses, which contained reversed male and female stocks, were used to detect survival variation caused by uncomplemented random mutations on sex chromosomes in male progeny. Survival data for each reciprocal cross was analyzed separately (Supplementary Figure S6) before reciprocal data was combined for each genotype and dose (Figures 3 and 5).
Figure 3.
The linker domain is required for DNA damage tolerance and promotes egg hatching. (A) Survival of irradiated larvae to adulthood relative to untreated larvae. Each point represents mean relative survival from 3–4 replicate crosses and sample standard deviation is shown. Significance symbols denote comparisons between , , and , , (two-way ANOVA and Sidak’s multiple comparisons test, all comparisons listed in Supplementary Table S5). (B) Mean relative survival of larvae treated with 0.007% nitrogen mustard from 11–35 crosses per genotype. Each point represents results for one cross. None=no transgene. Shown are sample standard deviation and comparisons using Kruskal-Wallis ANOVA and Dunn’s multiple comparisons test. (C) Mean egg hatching rates from 3–6 replicate crosses per homozygous genotype. Shown are standard deviations and statistical comparisons using Kruskal-Wallis ANOVA and Dunn’s multiple comparisons test. All panels: *p=0.01 to 0.05, **p=0.001 to 0.01, ****p<0.0001, ns=not significant.
Figure 5.
The scrambled linker rescues -dependent resolution of DNA damage and the FUS linker rescues egg hatching. (A) Relative survival of irradiated larvae to adulthood. Each point represents mean survival from 3–4 crosses and sample standard deviation is shown. Data for , , and , , are also shown in Figure 3A. Significance symbols denote comparisons between , , and replacement-linker mutants (two-way ANOVA and Sidak’s multiple comparisons test, all comparisons shown in Supplementary Table S7). (B) Relative survival of larvae treated with 0.007% nitrogen mustard. Shown are mean survival from 12–35 crosses per genotype and sample standard deviation. Data for , and , are also shown in Figure 3B. Nonsignificant comparisons are not shown (Kruskal-Wallis ANOVA and Dunn’s multiple comparisons test). (C) Mean hatching rates of eggs homozygous for transgenes and . Standard deviation is shown for 3–6 crosses per genotype. Each cross consisted of two parental fly stocks homozygous for a particular genotype, except for , which was crossed with itself (see methods and Supplementary Figure S6). Data for , and , are also shown in Figure 3C. Most nonsignificant comparisons are not shown (Kruskal-Wallis ANOVA and Dunn’s multiple comparisons test). All panels: *p=0.01 to 0.05, **p=0.001 to 0.01, ***p=0.0001 to 0.001, ****p<0.0001, ns=not significant.
In nitrogen mustard assays, each vial contained 5 females and 3 males heterozygous for specified alleles and followed the same complementation principles as IR assays. On day three, all crosses were transferred to a second set of vials. On day four, the first set of vials was treated with 0.007% nitrogen mustard. On day six, after flies were removed, the second set of vials was treated with water on day seven. Between 10 and 18 days after starting crosses, eclosed flies were scored as homozygous or heterozygous. Relative survival was not calculated for vials containing fewer than 10 flies. Progeny from 11–35 cross replicates were counted for each genotype.
Hatching assays
Crosses included 8–30 females and 3–15 males homozygous for specified alleles. Crosses followed the same complementation principles as sensitivity assays and were housed at 25°C in cages that contained grape juice agar plates coated with yeast paste. Females were allowed to lay eggs on each plate for 24 hours. Hatched and unhatched eggs were counted 48 hours after each plate was removed from a cage. Eggs were counted from 3–6 crosses for each genotype and each cross produced greater than 44 eggs.
Software
Graphs and statistical analyses were made using GraphPad Prism 10.1.2. Sequence analyses and alignments were made using Geneious Prime 2023.2.1. Accession numbers for alignment sequences are listed in Supplementary Table S4. Protein disorder graphs were made using IUPred3 (https://iupred3.elte.hu/).
Results
The Drosophila linker domain is needed for DNA double-strand break repair and tolerance to interstrand crosslinks
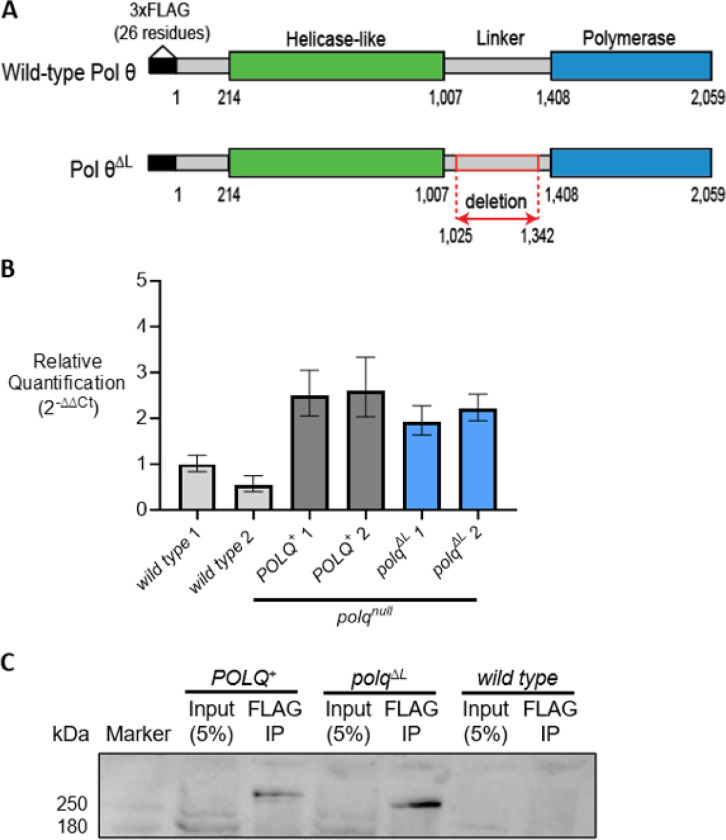
To determine if the linker domain promotes Drosophila resistance to DNA damage, we constructed a wild-type transgene and a -mutant transgene in which most of the disordered linker domain is deleted (Figure 2A). We designed the mutation to avoid removing partially conserved sequences near the helicase-like and polymerase domains. Both transgenes included 1.195 kb of the endogenous POLQ promoter. The transgenes were separately integrated into the left arm of chromosome 3 to create and fly stocks. Each transgene was recombined onto a chromosome containing a allele. and transgenes are sufficiently expressed in larvae and adult flies relative to endogenous POLQ (Figure 2B, Supplementary Figure S4, and Supplementary Table S3) and the respective proteins can be detected via immunoprecipitation from Drosophila embryos (Figure 2C and Supplementary Figure S5).
Figure 2.
POLQ transgenes are expressed and their respective proteins are present in vivo. (A) Protein products from and transgenes. Transgenes contain the endogenous POLQ promoter (not shown) and encode a N-terminal 3xFLAG tag. An in-frame deletion in subtracts linker residues 1,025–1,342. Transgenes were integrated on chromosome 3L and flies carried an endogenous allele. (B) POLQ mRNA expression in larvae homozygous for endogenous POLQ (wild type) or , or , alleles. 1 and 2 represent separately derived biological samples. Quantification was computed relative to endogenous control, rp49 expression, and the calibrator sample, wild type 1. Error bars represent RQmin and RQmax confidence intervals (see methods). (C) Immunoprecipitated 3xFLAG-tagged and from Drosophila embryo lysates. Proteins were detected using an anti-FLAG primary antibody.
We first analyzed roles of the linker domain in DSB repair. Previously, it was shown that HR-deficient Drosophila lacking the Rad51 ortholog, spn-A, depend on to repair IR-induced damage (54). Therefore, we put the transgenic flies into a spn-A mutant background and measured IR sensitivity. We found that the wild-type transgene provided similar tolerance to IR-induced DSBs as endogenous POLQ at doses of 50, 125, and 250 rads, but not 500 rads (Figure 3A, Supplementary Figure S6A, and Supplementary Table S5). Interestingly, mutants were hypersensitive to IR relative to larvae and not significantly different from mutants. These findings demonstrate that the linker domain is required for -dependent DSB repair.
In HR-competent Drosophila, is critical for tolerance of ICLs induced by nitrogen mustard (29,30). We tested the ability of larvae lacking the linker domain to survive to adulthood following nitrogen mustard treatment. We found that mutants were hypersensitive to nitrogen mustard and their survival was not significantly different from (Figure 3B and Supplementary Figure S6B), indicating that the linker domain is required for either bypass or repair of ICLs.
We also wondered whether the linker domain is required for repair of endogenously derived DNA damage in Drosophila. Drosophila eggs lacking POLQ have thin eggshells and severe hatching defects, likely because they are unable to repair DNA breaks produced during gene amplification of the DAFCs (Drosophila Amplicons in Follicle Cells) (55–57). To assess if the linker domain has a role in repair of these breaks, we measured hatching rates of eggs homozygous for . We compared eggs to eggs from a new cross (), since eggs had atypical hatching rates relative to other stocks, possibly caused by randomly accumulated mutations (Supplementary Figures S6 and S7). eggs had substantially lower, albeit not significant, hatching frequencies than eggs and significantly lower hatching frequencies than wild-type eggs (Figure 3C and Supplementary Figure S6C). Interestingly, egg hatching rates were not significantly different from , suggesting that the linker domain promotes resolution of endogenously derived DNA breaks during egg development.
We were curious if the N-terminal 3xFLAG tag in reduced the fitness of mutants in sensitivity and hatching assays. However, no significant differences were identified between and mutants (Supplementary Figure S7 and Supplementary Table S6). Taken together, our results strongly support that the linker domain is needed for -dependent repair of DNA damage.
Linker residue properties are important for functions in DNA repair and damage tolerance
Since linkers are not well-conserved among Drosophila species (Supplementary Figure S2), we hypothesized that amino acid residue properties may be more critical for linker functions than protein-binding motifs or catalytic subdomains. At the primary sequence level, linker domains are disordered and composed mostly of hydrophilic polar or charged residues (Table 1, Supplementary Figures S3 and S8).
Table 1.
Linker domain properties within homologs.
| Organism | Predicted boundaries | Length (residues) | Overall charge | % Polar uncharged | % Charged | % Hydrophobic |
|---|---|---|---|---|---|---|
| H. sapiens | 900–1,818 | 919 | −16 | 36 | 27 | 37 |
| R. norvegicus | 898–1,777 | 880 | −27 | 34 | 28 | 39 |
| M. musculus | 896–1,773 | 878 | −18 | 34 | 27 | 39 |
| D. rerio | 1,101–1,827 | 727 | −38 | 35 | 31 | 35 |
| C. elegans | 822–1,021 | 200 | −23 | 26 | 36 | 39 |
| A. thaliana | 1,320–1,601 | 282 | −7 | 31 | 25 | 46 |
| D. melanogaster | 1,008–1,407 | 400 | +2 | 35 | 26 | 39 |
| D. melanogaster helicase-like | 214–1,007 | 794 | +1 | 25 | 25 | 51 |
| D. melanogaster polymerase | 1,408–2,059 | 652 | +14 | 29 | 25 | 47 |
Pairwise alignments with human helicase-like and polymerase domains were used to predict linker boundaries. D. melanogaster helicase-like and polymerase domains are listed for comparison to disordered linker domains. Properties were calculated in Geneious Prime. Charge was determined at pH 7.
To explore residue properties of the linker domain that are required for repair, we designed three replacement IDRs (Table 2 and Figure 4A). The first consists of randomly scrambled D. melanogaster linker residues. Length, disorder, overall charge, hydrophilic distribution, and residue composition are similar between the scrambled and wild-type linkers, while potential protein-binding motifs are likely interrupted in the scrambled sequence (Table 2 and Supplementary Figures S8, S9, and S10). The second includes part of the human linker domain that is also disordered and has a similar length, hydrophilic distribution, and residue composition as the Drosophila linker (Table 2 and Supplementary Figures S8, S9, and S10). Notably, the human linker has an overall negative charge that contrasts with the positive charge of the Drosophila linker. The third consists of the N-terminal low-complexity domain from Homo sapiens FUS, which promotes aggregation at DNA damage for NHEJ and HR repair (58–60). The FUS low-complexity domain is highly disordered (Supplementary Figure S10), but has fewer residues and is more polar and hydrophilic than the Drosophila linker (Table 2, Supplementary Figures S8 and S9).
Table 2.
Replacement-linker properties.
| D. melanogaster linker | Length (residues) | Overall charge | % Polar uncharged | % Charged | % Hydrophobic |
|---|---|---|---|---|---|
| Wild type (deleted in mutant) | 318 | +7 | 35 | 27 | 39 |
| Scrambled (SCR) | 324 | +7 | 34 | 26 | 40 |
| Human (HUM) | 324 | −26 | 33 | 27 | 40 |
| FUS (FUS) | 220 | −4 | 63 | 3 | 35 |
Figure 4.
polq-mutant transgenes with replacement linkers are expressed in vivo. (A) The deleted linker in was replaced with the D. melanogaster linker sequence scrambled in a random order (), or residues 1483–1800 from the linker domain of human (), or residues 2–214 from the N-terminal low-complexity domain of human FUS () (58,59). Transgenes were integrated at the same chromosome 3 site as and put into a background. (B) POLQ mRNA expression in wild-type larvae or those homozygous for a transgene and . 1 and 2 represent separately derived biological samples. The endogenous control, rp49 expression, and the calibrator sample, wild type 1, were used to calculate relative quantification. Error bars represent RQmin and RQmax. Wild-type, , and data are also shown in Figure 2B.
Replacement linker DNA sequences were codon optimized for Drosophila and separately inserted at the deletion site in to make new transgenes that were integrated into the same chromosomal location as the transgene. Replacement-linker transgenes had similar expression patterns as and transgenes (Figure 4B, Supplementary Figure S4, and Supplementary Table S3).
In a spn-A null mutant background, we observed that and larvae had similar sensitivities to IR, while and mutants were hypersensitive (Figure 5A and Supplementary Table S7). Interestingly, mutants were significantly more resistant than larvae at 250 rads and and mutants were significantly more sensitive than mutants at 125 rads. These observations suggest that the scrambled linker promotes efficient -dependent DSB repair, while human and FUS linkers are not sufficient and may even inhibit DSB repair.
Following treatment with nitrogen mustard, the transgene rescued survival relative to the transgene, although not to the level of the wild-type transgene (Figure 5B). and mutants rarely survived and were not significantly different from mutants. This indicates that, in addition to DSB repair, the scrambled linker can promote tolerance to interstrand crosslinks, while the human and FUS linkers are unable to substitute for this function.
Lastly, we measured hatching rates of eggs homozygous for replacement-linker transgenes. Surprisingly, -mutant hatching was not significantly different from hatching while and eggs had low hatching frequencies (Figure 5C). These findings suggest that the scrambled domain sequence is unable to support normal egg development, while the FUS low-complexity domain may share properties with the Drosophila linker that are required for DNA repair during egg development.
Discussion
-mediated DNA repair is critical for genome maintenance and cellular survival. promotes tolerance of exogenously induced ICLs and is often required for resolution of DSBs in NHEJ- and HR-deficient cells. However, TMEJ frequently causes insertions and deletions in the genome that can elicit human disease. Cancer cells with a variety of genetic backgrounds require to survive, but comprehensive roles of domains in cell survival mechanisms are not entirely understood.
While prior investigations identified multiple functions for the helicase-like and polymerase domains, the linker domain has been largely ignored. In this study, we have shown that the Drosophila linker domain is required for DNA damage tolerance and has a specific sequence composition that promotes its functions. Importantly, flies possessing a deletion of the disordered linker region are unable to support any of the functions of wild-type that we tested, including DSB repair, ICL tolerance, and egg viability.
Functions of the linker domain in DSB and ICL repair
The importance of the linker domain in DSB repair was initially surprising, given it is disordered, mostly not conserved in Drosophila species, and the human linker domain is largely dispensable for in vitro (22) and in vivo DSB repair (61). Since linkers are mostly not conserved, we postulated that the linker functions may depend on residue composition instead of binding motifs or catalytic subdomains. We explored this hypothesis by designing replacement linkers that have similar properties to the Drosophila linker domain. Interestingly, the scrambled Drosophila melanogaster linker fully rescued DSB repair and substantially rescued ICL tolerance. In contrast, part of the human linker domain and the entire FUS low-complexity domain did not rescue survival in sensitivity assays. Since the human and FUS replacement linkers did not rescue repair, we think it is unlikely that the linker is simply acting as a nonspecific tether between the helicase-like and polymerase domains.
Damage tolerance promoted by the scrambled linker suggests that the ratio of linker residues is important, while linker-mediated protein interactions are unlikely needed for resolution of exogenously induced DNA damage in Drosophila. Prior work showed that the human linker interacts with TOPBP1 to promote mitotic DSB repair (36). It has also been reported that the human linker interacts with RAD51 to suppress HR (6), although this remains to be validated (35,62,63). At the moment, it is unclear if Drosophila linker domains interact with other proteins. Protein interactions may be more common in mammalian linker domains since they are more conserved than Drosophila linker domains (Supplementary Figure S1 and S2). However, some regions in Drosophila linker domains are partially conserved and may interact with proteins in specific DNA repair or developmental contexts to promote functions.
Based on our findings, we propose that the Drosophila linker could promote damage tolerance in at least two ways. First, the slightly positive, but overall neutral charge of the linker may allow interactions between the negatively charged N-terminus and positively charged polymerase domain (Supplementary Figure S11). These interactions could be critical for the helicase-like domain to be situated near the polymerase domain during microhomology synapsis and subsequent DNA synthesis. The scrambled linker may have rescued repair because it has the same overall charge and polarity as the Drosophila linker, while the strong negative charge of the human linker and strong polarity of the FUS linker could have disrupted termini interactions.
Second, the linker domain may regulate multimerization at DNA damage. When in vitro and not bound to DNA, the human linker suppresses from forming multimers that promote TMEJ (22). The wild-type and scrambled linkers may function similarly in vivo and prevent unnecessary multimerization prior to DNA damage.
Potential roles of the linker domain in Drosophila egg development
Re-replication of the DAFC genomic regions within Drosophila follicle cells is beneficial for eggshell development, but also causes DNA breaks, some of which require for repair (55,56). Like mutants in our study, eggs had low hatching frequencies. Surprisingly, and eggs also had low hatching frequencies while the transgene partially rescued hatching. Since the transgene did not rescue tolerance in sensitivity assays, repair of re-replication-induced DNA damage and exogenously induced damage likely require different linker-mediated mechanisms.
The FUS low-complexity domain normally encourages FUS aggregation at DNA damage (58–60), which is promoted by PARylation and discouraged by phosphorylation (64,65). Thus, one possibility is that FUS PTMs mimic Drosophila linker PTMs, which are required for -mediated DNA repair during re-replication but are not sufficient to repair exogenously induced damage.
Conclusion
Overall, we establish that residue specificity plays an important role in how the linker domain regulates -dependent DNA damage tolerance in Drosophila. Our findings highlight that future studies should consider how the linker domain may influence overall functions and the efficacy of inhibitors for cancer treatment.
Supplementary Material
Acknowledgements
We thank Anna Joseph for creating the mutant and Alice Miller for assistance with embryo lysis and fly counting.
Funding
This work was supported by the National Science Foundation (MCB1716039); and the National Institutes of Health (R01GM125827) to M.M.
Footnotes
Supplementary Data
Supplementary Data are available at NAR Online.
Conflict of interest statement. None declared.
Data Availability
Primary data that underlie the main and supplementary figures can be found in the supplementary Excel file.
References
- 1.Cannan W.J. and Pederson D.S. (2016) Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J Cell Physiol, 231, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhargava R., Sandhu M., Muk S., Lee G., Vaidehi N. and Stark J.M. (2018) C-NHEJ without indels is robust and requires synergistic function of distinct XLF domains. Nat Commun, 9, 2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D. and Boulton S.J. (2013) RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell, 49, 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stinson B.M., Moreno A.T., Walter J.C. and Loparo J.J. (2020) A Mechanism to Minimize Errors during Non-homologous End Joining. Mol Cell, 77, 1080–1091 e1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scully R., Panday A., Elango R. and Willis N.A. (2019) DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol, 20, 698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceccaldi R., Liu J.C., Amunugama R., Hajdu I., Primack B., Petalcorin M.I., O’Connor K.W., Konstantinopoulos P.A., Elledge S.J., Boulton S.J. et al. (2015) Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature, 518, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng W., Simpson D.A., Carvajal-Garcia J., Price B.A., Kumar R.J., Mose L.E., Wood R.D., Rashid N., Purvis J.E., Parker J.S. et al. (2019) Genetic determinants of cellular addiction to DNA polymerase theta. Nat Commun, 10, 4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mateos-Gomez P.A., Gong F., Nair N., Miller K.M., Lazzerini-Denchi E. and Sfeir A. (2015) Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature, 518, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schimmel J., Kool H., van Schendel R. and Tijsterman M. (2017) Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J, 36, 3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyatt D.W., Feng W., Conlin M.P., Yousefzadeh M.J., Roberts S.A., Mieczkowski P., Wood R.D., Gupta G.P. and Ramsden D.A. (2016) Essential Roles for Polymerase Theta-Mediated End Joining in the Repair of Chromosome Breaks. Mol Cell, 63, 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J., Gelot C., Pantelidou C., Li A., Yucel H., Davis R.E., Farkkila A., Kochupurakkal B., Syed A., Shapiro G.I. et al. (2021) A first-in-class Polymerase Theta Inhibitor selectively targets Homologous-Recombination-Deficient Tumors. Nat Cancer, 2, 598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvajal-Garcia J., Cho J.E., Carvajal-Garcia P., Feng W., Wood R.D., Sekelsky J., Gupta G.P., Roberts S.A. and Ramsden D.A. (2020) Mechanistic basis for microhomology identification and genome scarring by polymerase theta. Proc Natl Acad Sci U S A, 117, 8476–8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luedeman M.E., Stroik S., Feng W., Luthman A.J., Gupta G.P. and Ramsden D.A. (2022) Poly(ADP) ribose polymerase promotes DNA polymerase theta-mediated end joining by activation of end resection. Nat Commun, 13, 4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truong L.N., Li Y., Shi L.Z., Hwang P.Y., He J., Wang H., Razavian N., Berns M.W. and Wu X. (2013) Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A, 110, 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allera-Moreau C., Rouquette I., Lepage B., Oumouhou N., Walschaerts M., Leconte E., Schilling V., Gordien K., Brouchet L., Delisle M.B. et al. (2012) DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis, 1, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies H., Glodzik D., Morganella S., Yates L.R., Staaf J., Zou X., Ramakrishna M., Martin S., Boyault S., Sieuwerts A.M. et al. (2017) HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med, 23, 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins G.S., Harris A.L., Prevo R., Helleday T., McKenna W.G. and Buffa F.M. (2010) Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget, 1, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamura K., Bahar R., Seimiya M., Chiyo M., Wada A., Okada S., Hatano M., Tokuhisa T., Kimura H., Watanabe S. et al. (2004) DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer, 109, 9–16. [DOI] [PubMed] [Google Scholar]
- 19.Lemee F., Bergoglio V., Fernandez-Vidal A., Machado-Silva A., Pillaire M.J., Bieth A., Gentil C., Baker L., Martin A.L., Leduc C. et al. (2010) DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci U S A, 107, 13390–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillaire M.J., Selves J., Gordien K., Gourraud P.A., Gentil C., Danjoux M., Do C., Negre V., Bieth A., Guimbaud R. et al. (2010) A ‘DNA replication’ signature of progression and negative outcome in colorectal cancer. Oncogene, 29, 876–887. [DOI] [PubMed] [Google Scholar]
- 21.Shinmura K., Kato H., Kawanishi Y., Yoshimura K., Tsuchiya K., Takahara Y., Hosokawa S., Kawase A., Funai K. and Sugimura H. (2019) POLQ Overexpression Is Associated with an Increased Somatic Mutation Load and PLK4 Overexpression in Lung Adenocarcinoma. Cancers, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black S.J., Ozdemir A.Y., Kashkina E., Kent T., Rusanov T., Ristic D., Shin Y., Suma A., Hoang T., Chandramouly G. et al. (2019) Molecular basis of microhomology-mediated end-joining by purified full-length Pol theta. Nat Commun, 10, 4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mateos-Gomez P.A., Kent T., Deng S.K., McDevitt S., Kashkina E., Hoang T.M., Pomerantz R.T. and Sfeir A. (2017) The helicase domain of Pol theta counteracts RPA to promote alt-NHEJ. Nat Struct Mol Biol, 24, 1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaub J.M., Soniat M.M. and Finkelstein I.J. (2022) Polymerase theta-helicase promotes end joining by stripping single-stranded DNA-binding proteins and bridging DNA ends. Nucleic Acids Res, 50, 3911–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arana M.E., Seki M., Wood R.D., Rogozin I.B. and Kunkel T.A. (2008) Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res, 36, 3847–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maga G., Shevelev I., Ramadan K., Spadari S. and Hubscher U. (2002) DNA polymerase theta purified from human cells is a high-fidelity enzyme. J Mol Biol, 319, 359–369. [DOI] [PubMed] [Google Scholar]
- 27.Higgins G.S., Prevo R., Lee Y.F., Helleday T., Muschel R.J., Taylor S., Yoshimura M., Hickson I.D., Bernhard E.J. and McKenna W.G. (2010) A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res, 70, 2984–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yousefzadeh M.J., Wyatt D.W., Takata K., Mu Y., Hensley S.C., Tomida J., Bylund G.O., Doublie S., Johansson E., Ramsden D.A. et al. (2014) Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet, 10, e1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beagan K., Armstrong R.L., Witsell A., Roy U., Renedo N., Baker A.E., Schärer O.D. and McVey M. (2017) Drosophila DNA polymerase theta utilizes both helicase-like and polymerase domains during microhomology-mediated end joining and interstrand crosslink repair. PLoS Genet, 13, e1006813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyd J.B., Sakaguchi K. and Harris P.V. (1990) mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics, 125, 813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inagaki S., Suzuki T., Ohto M.A., Urawa H., Horiuchi T., Nakamura K. and Morikami A. (2006) Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell, 18, 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muzzini D.M., Plevani P., Boulton S.J., Cassata G. and Marini F. (2008) Caenorhabditis elegans POLQ-1 and HEL-308 function in two distinct DNA interstrand cross-link repair pathways. DNA Repair (Amst), 7, 941–950. [DOI] [PubMed] [Google Scholar]
- 33.Ramsden D.A., Carvajal-Garcia J. and Gupta G.P. (2022) Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining. Nat Rev Mol Cell Biol, 23, 125–140. [DOI] [PubMed] [Google Scholar]
- 34.Schrempf A., Slyskova J. and Loizou J.I. (2021) Targeting the DNA Repair Enzyme Polymerase theta in Cancer Therapy. Trends Cancer, 7, 98–111. [DOI] [PubMed] [Google Scholar]
- 35.Wood R.D. and Doublie S. (2022) Genome Protection by DNA Polymerase theta. Annu Rev Genet, 56, 207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gelot C., Kovacs M.T., Miron S., Mylne E., Haan A., Boeffard-Dosierre L., Ghouil R., Popova T., Dingli F., Loew D. et al. (2023) Pol theta is phosphorylated by PLK1 to repair double-strand breaks in mitosis. Nature, 621, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman J.A., Cooper C.D.O., Aitkenhead H. and Gileadi O. (2015) Structure of the Helicase Domain of DNA Polymerase Theta Reveals a Possible Role in the Microhomology-Mediated End-Joining Pathway. Structure, 23, 2319–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahn K.E., Averill A.M., Aller P., Wood R.D. and Doublie S. (2015) Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat Struc Mol Biol, 22, 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erdos G., Pajkos M. and Dosztanyi Z. (2021) IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res, 49, W297–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dosztanyi Z., Csizmok V., Tompa P. and Simon I. (2005) The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol, 347, 827–839. [DOI] [PubMed] [Google Scholar]
- 41.Cermakova K. and Hodges H.C. (2023) Interaction modules that impart specificity to disordered protein. Trends Biochem Sci, 48, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright P.E. and Dyson H.J. (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol, 16, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grofte M., Rask M.D., Streicher W., Jungmichel S., Nielsen M.L. et al. (2015) Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun, 6, 8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bah A. and Forman-Kay J.D. (2016) Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J Biol Chem, 291, 6696–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamagata K., Kanbayashi S., Honda M., Itoh Y., Takahashi H., Kameda T., Nagatsugi F. and Takahashi S. (2020) Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci Rep, 10, 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levone B.R., Lenzken S.C., Antonaci M., Maiser A., Rapp A., Conte F., Reber S., Mechtersheimer J., Ronchi A.E., Muhlemann O. et al. (2021) FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J Cell Biol, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oshidari R., Huang R., Medghalchi M., Tse E.Y.W., Ashgriz N., Lee H.O., Wyatt H. and Mekhail K. (2020) DNA repair by Rad52 liquid droplets. Nat Commun, 11, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spegg V. and Altmeyer M. (2021) Biomolecular condensates at sites of DNA damage: More than just a phase. DNA Repair (Amst), 106, 103179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gratz S.J., Goel P., Bruckner J.J., Hernandez R.X., Khateeb K., Macleod G.T., Dickman D. and O’Connor-Giles K.M. (2019) Endogenous Tagging Reveals Differential Regulation of Ca(2+) Channels at Single Active Zones during Presynaptic Homeostatic Potentiation and Depression. J Neurosci, 39, 2416–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruckner J.J., Zhan H., Gratz S.J., Rao M., Ukken F., Zilberg G. and O’Connor-Giles K.M. (2017) Fife organizes synaptic vesicles and calcium channels for high-probability neurotransmitter release. J Cell Biol, 216, 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gratz S.J., Cummings A.M., Nguyen J.N., Hamm D.C., Donohue L.K., Harrison M.M., Wildonger J. and O’Connor-Giles K.M. (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics, 194, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwasaki S., Kobayashi M., Yoda M., Sakaguchi Y., Katsuma S., Suzuki T. and Tomari Y. (2010) Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell, 39, 292–299. [DOI] [PubMed] [Google Scholar]
- 53.Yang L., Paul S., DuBois-Coyne S., Kyriakakis P. and Veraksa A. (2017) Medium-scale Preparation of Drosophila Embryo Extracts for Proteomic Experiments. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan S.H., Yu A.M. and McVey M. (2010) Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet, 6, e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alexander J.L., Barrasa M.I. and Orr-Weaver T.L. (2015) Replication fork progression during re-replication requires the DNA damage checkpoint and double-strand break repair. Curr Biol, 25, 1654–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexander J.L., Beagan K., Orr-Weaver T.L. and McVey M. (2016) Multiple mechanisms contribute to double-strand break repair at rereplication forks in Drosophila follicle cells. Proc Natl Acad Sci U S A, 113, 13809–13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Claycomb J.M. and Orr-Weaver T.L. (2005) Developmental gene amplification: insights into DNA replication and gene expression. Trends Genet, 21, 149–162. [DOI] [PubMed] [Google Scholar]
- 58.Mastrocola A.S., Kim S.H., Trinh A.T., Rodenkirch L.A. and Tibbetts R.S. (2013) The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem, 288, 24731–24741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murray D.T., Kato M., Lin Y., Thurber K.R., Hung I., McKnight S.L. and Tycko R. (2017) Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell, 171, 615–627 e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singatulina A.S., Hamon L., Sukhanova M.V., Desforges B., Joshi V., Bouhss A., Lavrik O.I. and Pastre D. (2019) PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep, 27, 1809–1821 e1805. [DOI] [PubMed] [Google Scholar]
- 61.Fijen C., Drogalis Beckham L., Terino D., Li Y., Ramsden D.A., Wood R.D., Doublie S. and Rothenberg E. (2024) Sequential requirements for distinct Pol theta domains during theta-mediated end joining. Mol Cell, 84, 1460–1474 e1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang T., Reh S., Dunbayev Y., Zhong Y., Takata Y., Shen J., McBride K.M., Murnane J.P., Bhak J., Lee S. et al. (2020) Defining the mutation signatures of DNA polymerase theta in cancer genomes. NAR Cancer, 2, zcaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zelensky A.N., Schimmel J., Kool H., Kanaar R. and Tijsterman M. (2017) Inactivation of Pol theta and C-NHEJ eliminates off-target integration of exogenous DNA. Nat Commun, 8, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monahan Z., Ryan V.H., Janke A.M., Burke K.A., Rhoads S.N., Zerze G.H., O’Meally R., Dignon G.L., Conicella A.E., Zheng W. et al. (2017) Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J, 36, 2951–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rulten S.L., Rotheray A., Green R.L., Grundy G.J., Moore D.A., Gomez-Herreros F., Hafezparast M. and Caldecott K.W. (2014) PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res, 42, 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary data that underlie the main and supplementary figures can be found in the supplementary Excel file.