Abstract
Speech-sound disorder (SSD) is a complex behavioral disorder characterized by speech-sound production errors associated with deficits in articulation, phonological processes, and cognitive linguistic processes. SSD is prevalent in childhood and is comorbid with disorders of language, spelling, and reading disability, or dyslexia. Previous research suggests that developmental problems in domains associated with speech and language acquisition place a child at risk for dyslexia. Recent genetic studies have identified several candidate regions for dyslexia, including one on chromosome 3 segregating in a large Finnish pedigree. To explore common genetic influences on SSD and reading, we examined linkage for several quantitative traits to markers in the pericentrometric region of chromosome 3 in 77 families ascertained through a child with SSD. The quantitative scores measured several processes underlying speech-sound production, including phonological memory, phonological representation, articulation, receptive and expressive vocabulary, and reading decoding and comprehension skills. Model-free linkage analysis was followed by identification of sib pairs with linkage and construction of core shared haplotypes. In our multipoint analyses, measures of phonological memory demonstrated the strongest linkage (marker D3S2465, P=5.6×10-5, and marker D3S3716, P=6.8×10-4). Tests for single-word decoding also demonstrated linkage (real word reading: marker D3S2465, P=.004; nonsense word reading: marker D3S1595, P=.005). The minimum shared haplotype in sib pairs with similar trait values spans 4.9 cM and is bounded by markers D3S3049 and D3S3045. Our results suggest that domains common to SSD and dyslexia are pleiotropically influenced by a putative quantitative trait locus on chromosome 3.
Introduction
Speech-sound disorder (SSD) is a complex behavioral disorder characterized by deficits in motor control of the articulatory mechanism and/or deficits in the general processing, organization, and cognitive representation of linguistic information. Thus, children’s speech-sound productions may reflect deficits in underlying phonological processes, such as phonological memory and speech-sound coding. SSD has a high prevalence in preschool children, estimated at ∼16% at age 3 years (Shriberg 2002), with 3.8% of children continuing to present with speech delay at age 6 years (Shriberg et al. 1999). More than half of these children encounter later academic difficulties in language, reading, and spelling (Shriberg and Kwiatkowski 1988; Aram and Hall 1989; Bishop and Adams 1990; Menyuk et al. 1991; Felsenfeld et al. 1995; Shriberg and Austin 1998; Lewis et al. 2000). The residual effects of preschool SSD may be lifelong, and yet, for the majority of individuals, the etiological basis of this disorder is unknown.
There is a growing body of literature suggesting that susceptibility to SSD is genetic, including twin studies (Lewis and Thompson 1992; Bishop et al. 1996; Tomblin and Buckwalter 1998), familial aggregation studies (Lewis et al. 1989; Lewis 1992; Felsenfeld et al. 1995) and segregation and linkage analyses (Lewis et al. 1993; Schick et al. 2002). Recently, studies of specific language impairment (SLI), defined as a failure to acquire language within normal limits in the absence of a generalized developmental delay, have reported linkage to chromosome 16q for nonword repetition (SLI Consortium 2002) and to chromosome 19q for expressive language (SLI Consortium 2002). There has also been a report of linkage to chromosome 7q31 for verbal dyspraxia in a large family (Fisher et al. 1998; Vargha-Khadem et al. 1998). Multiple genetic influences could also be at work, with different genetic factors contributing to different types of domains in language and speech-sound acquisition.
Pennington and colleagues (Pennington and Lefly 2001; Tunick and Pennington 2002; Raitano et al., in press) proposed that early developmental problems in spoken language predict the later emergence of dyslexia in children from high-risk families. The cognitive linguistic deficits associated with dyslexia include impairments in phonological awareness (explicit knowledge of the sound system of a language), phonological memory (as measured by the repetition of nonsense words), and the ability to decode unfamiliar words (reading decoding), each of which underlie spoken and written language. Although SSD and dyslexia are assessed through different test batteries, possibly at different ages, the similar cognitive linguistic bases of these disorders suggest that they share some common genetic etiology (see fig. 1).
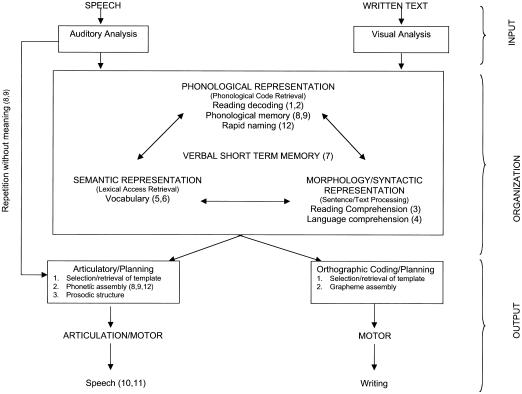
Figure 1.
Shared and unshared processes involved in production of speech and written text. The tasks of processing and producing speech-sound and written text share some neural processes. The processing of speech and written text begins with modality-specific sensory and perceptual analyses of the stimuli—that is, auditory and visual analyses. The analysis of speech and written text relies on shared phonological representations for the conversion of phonetic speech units to phoneme classes and on the conversion of written graphemes to the corresponding phonemes. Phonological memory is a key process in these conversions. Next, meaning is attached to the utterance or text through shared semantic and morphologic/syntactic representations. The output segment of speech production or writing again requires modality-specific processes, including the selection and retrieval of a template for the intended word, the assembly and sequencing of phonetic units or graphemes, and the execution of the motor program. There are also processes contributing to speech and written text output that bypass the phonological representation segment. These processes include repetition of words without meaning and reading by sight vocabulary rather than by reading decoding. The numbers in parentheses in the figure correspond to the measures listed in the key below. Note that a single measure may tap multiple processes. Although the figure depicts processing as linear, we recognize that many of these processes are overlapping and occur simultaneously. Key: 1 = WRMT-ID; 2 = WRMT-AT; 3 = WIAT-RC; 4 = WIAT-LC; 5 = PPVT-III; 6 = EOWPVT-R; 7 = SI; 8 = MSW; 9 = NSW; 10 = PCC; 11 = GFTA; 12 = RAN-C.
Whereas there have been few molecular genetic data reported for SSD, there is an extensive genetic literature on dyslexia. The genetic mechanisms proposed to underlie the behavioral manifestation of dyslexia are complex (see Fisher and DeFries 2002). Various components of dyslexia have been analyzed, and linkages have been found to chromosome 1 (Rabin et al. 1993; Grigorenko et al. 2001); word recognition and reading decoding linkage has been found on chromosome 2 (Fagerheim et al. 1999; Francks et al. 2002; Petryshen et al. 2002; Kaminen et al. 2003); phonological awareness, rapid naming, and verbal short term memory linkage has been found on chromosome 3 (Nopola-Hemmi et al. 2001, 2002); phonemic awareness, reading decoding, single-word reading, orthographic coding, vocabulary, rapid naming, and spelling linkage has been found on chromosome 6 (Cardon et al. 1994, 1995; Grigorenko et al. 1997; Gayan et al. 1999; Fisher et al. 1999, 2002a; Nothen et al.1999; Petryshen et al. 2001; Kaplan et al. 2002); word recognition and spelling linkage has been found on chromosome 15 (Grigorenko et al. 1997; Schulte-Körne et al. 1998; Nothen et al. 1999); and single-word reading and phonological and orthographic processing linkage has been found on chromosome 18 (Fisher et al. 2002a). However, recent evidence suggests that some of these chromosomal regions harbor genes with pleiotropic effects (Marlow et al. 2003).
A recent study by Nopola-Hemmi et al. (2001) analyzed a large Finnish pedigree segregating for developmental dyslexia in an autosomal dominant fashion and found linkage to the pericentromeric region of chromosome 3. On the basis of the hypothesis that SSD and dyslexia share genetic determinants, we genotyped markers in the dyslexia candidate region on chromosome 3 to determine whether linkage would be observed in families ascertained through a proband with SSD. This is the first study, to our knowledge, to examine the common genetic basis of SSD and dyslexia on chromosome 3.
Subjects and Methods
Collection of Family Data
The sample consisted of 77 families, ascertained through a preschool child with SSD of unknown etiology, who donated samples for DNA analysis. The protocol for collection of buccal smears or blood samples and for participant testing was approved by the institutional review board of University Hospitals, Cleveland, OH. Probands were enrolled in speech/language therapy for a moderate to severe SSD of unknown origin and were recruited from case loads of speech-language pathologists in the greater Cleveland area. Siblings of the probands were recruited and assessed at the same time as the probands. Although historical information on affection was collected from parents, the focus of this study was on quantitative traits; hence, we did not utilize parental data in the analyses.
Measures
Measures of articulation
The Goldman-Fristoe Test of Articulation (GFTA) (Goldman and Fristoe 1986) was used to assess production of consonant sounds in singleton and cluster contexts in the beginning, middle, and final positions of words and blends. Participants were asked to name pictures, and their responses were audiotape recorded and phonetically transcribed by speech-language pathologists. A percentile score was used as the quantitative trait for data analysis. The GFTA is standardized for ages 2–16+ years and was administered to 188 probands and siblings; when it was used in conjunction with the Khan-Lewis Phonological Analysis (KLPA) (Khan and Lewis 1986), 15 common error patterns found in children’s speech were categorized. These error patterns, or “phonological processes,” contribute to a composite percentile rank for age, a speech simplification rating, and an age equivalent score. The KLPA is based on responses to the GFTA and was used in the present study only to identify probands and siblings with moderate to severe affection status; it was not employed in linkage analysis.
The Percentage of Consonants Correct (PCC) (Shriberg et al. 1997) score is the percentage of intended consonant sounds that were articulated correctly in a sample of conversational speech. Conversational speech samples were obtained by examiners using linguistic sampling and audiorecording techniques (Shriberg 1993). The samples were transcribed using computer-assisted narrow phonetic transcription (Shriberg 2002; Shriberg and Kent 2003). PCC data were obtained only for children aged 4–12 years, which yielded a total of 177 observations.
Measures of phonological memory
The Multisyllabic Word Test (MSW) (Catts 1986) and Nonsense Word Repetition Test (NSW) (Kamhi and Catts 1986) were used to assess phonological short-term memory skills. Participants aged 4–18 years were asked to repeat 20 multisyllabic real and 15 nonsense words in response to audiotaped presentations of the words. Responses were audiotaped and transcribed, indicating the percentage of words correctly repeated. In prior studies, MSW and NSW have discriminated individuals with histories of speech and language disorders who no longer demonstrate overt speech production errors in conversational speech from individuals without such histories (Lewis and Freebairn 1998). Data were available for 196 children for MSW and 191 children for NSW. We refer to either “speech-sound coding” or “phonological memory” when discussing results associated with MSW or NSW.
Measure of rapid naming
The Rapid Auditory Naming test (RAN) (Denckla and Rudel 1976) is a timed task in which participants are asked to name letters, objects, numbers, or colors as rapidly as possible. The colors subtest (RAN-C) was used to assess speeded naming. The RAN-C assesses phonological retrieval and processing speed (Denckla and Cutting 1999). The RAN-C test is appropriate for children aged 5–10 years. Data were available for 151 children in the sample.
Measure of verbal short-term memory
Measures of verbal short-term memory included the Sentence Imitation subtest of the Test of Language Development–Primary, Second Edition (TOLD-P2) (Newcomer and Hammill 1988), for participants aged 5–7 years, or the corresponding Recalling Sentences subtest from the Clinical Evaluation of Language Fundamentals–Revised (CELF-R) (Semel et al. 1987), for older participants. Both subtests assess verbatim recall of sentences of varying syntactic complexity. Scores were combined into a single measure, referred to as SI, which was available for 193 children.
Measures of vocabulary and language comprehension
Measures of receptive and expressive vocabulary and language comprehension included the Peabody Picture Vocabulary–Third Edition (PPVT-III) (Dunn and Dunn 1997), the Expressive One Word Picture Vocabulary Test–Revised (EOWPVT-R) (Gardner 1990), and the Wechsler Individual Achievement Test (WIAT) (Wechsler 1992) Listening Comprehension subtest (WIAT-LC). These measures were given to children aged 5–18 years. PPVT-III scores were available for 157 children, EOWPVT-R scores for 142 children, and WIAT-LC scores for 137 children.
Reading measures
We administered the Woodcock Reading Mastery Tests–Revised (Woodcock 1987) Word Identification (WRMT-ID) and Word Attack (WRMT-AT) subtests to assess reading decoding, as well as the WIAT Reading Comprehension (WIAT-RC) subtest to assess reading comprehension. These reading tests are appropriate for children aged 7–18 years, and age-standardized scores were used in analysis. WRMT-ID scores were available for 137 children, WRMT-AT scores for 135 children, and WIAT-RC scores for 136 children.
Screening Measures for Probands
Probands were ascertained for moderate to severe SSD if they scored at the 10th percentile or lower on the GFTA (Goldman and Fristoe 1986) prior to enrollment in speech-language therapy and if they displayed at least three common phonological error types on the KLPA (Khan and Lewis 1986). These children were also required to have a normal peripheral speech mechanism, as documented by z scores within 1 SD of the normative reference point on both the Total Function and Total Structure subscales of the Oral and Speech Motor Control Protocol (Robbins and Klee 1987). Other inclusionary criteria included at least low average intelligence, as defined by a Performance IQ of at least 80 on the Wechsler Intelligence Scale for Children–Third Edition (WISC-III) (Wechsler 1991) or the Wechsler Preschool and Primary Scale of Intelligence Test–Revised (WPPSI-R) (Wechsler 1989), as well as normal hearing, fewer than six episodes of otitis media prior to age 3 years, and absence of neurological disorders or developmental delays, as reported by the parents. Stuttering and other dysfluencies were not included in defining affection status. Siblings of probands were required to meet the same criteria to be classified as affected. This qualitative categorization was used only for generating descriptive statistics in the present study. The remainder of the analyses were conducted on the age-appropriate quantitative traits through use of the previously described test battery. Comorbid language disorder was diagnosed for scores 1 SD below the mean on two or more subtests of a standardized language measure (TOLD-P2, CELF-P, or the CELF-R), and comorbid reading disorder was diagnosed for scores 1 SD below the mean on either the WRMT or the WIAT-RC. All other comorbidities were based on historical report.
Genotyping
High-molecular-weight DNA was isolated from an aliquot of blood through use of the Puregene Kit (Gentra Systems) or from buccal swabs through use of the BioRad InstaGene Matrix protocol (BioRad Laboratories). DNA was arrayed in a 96-well format, and PCR was performed in a MJ Tetrad thermocycler. To examine the candidate region on chromosome 3, we used standard methods to genotype 15 markers spanning a 56-cM region (table 1). Of these 15 markers, 12 were selected from the markers studied in the developmental dyslexia research reported by Nopola-Hemmi et al. (2001).
Table 1.
Markers Analyzed, with Genetic Map Locations
| Marker Namea | Location(cM) |
| D3S1766 | 78.6 |
| D3S1285 | 91.1 |
| D3S2454* | 97.7 |
| D3S2406* | 102.6 |
| D3S3681 | 109.2 |
| D3S3049* | 109.2 |
| D3S2465* | 111.9 |
| D3S1595* | 112.4 |
| D3S1752* | 114.0 |
| D3S2462* | 115.1 |
| D3S3716* | 115.7 |
| D3S3655* | 117.7 |
| D3S2459* | 119.1 |
| D3S3045* | 124.1 |
| D3S2460* | 134.6 |
Markers indicated with an asterisk (*) were analyzed by Nopola-Hemmi et al. (2001).
The multiplexed markers were run on an ABI 3700 capillary machine (Applied Biosystems). Five-percent blind replicates and two CEPH controls were included on each gel to serve as internal controls. The ABI ROX 500 standard (present in every lane) was used to estimate size of alleles. Inconsistencies in the segregation of the genotypes within families were examined using MARKERINFO (S.A.G.E. [2003], version 4.4). Individuals demonstrating Mendelian inconsistencies at multiple markers that could not be resolved by retyping were treated as missing for the purpose of this analysis. In total, 6% of the data were treated as missing. We also checked the marker data for any significant departures from Hardy-Weinberg proportions. We then established the allele frequencies for each genetic marker by simple gene counting (disregarding relationships).
Statistical Analysis
Measures
For most probands and siblings, test scores on several variables were available at multiple time points during childhood. For all such data, we analyzed the first available observation, hypothesizing that information at the earliest time point would be most sensitive to SSD and associated verbal traits and least influenced by uncontrolled developmental processes and experiences. We examined the effects of age, age squared, sex, socioeconomic status (SES) based on the Hollingshead Four Factor Index of Social Class (Hollingshead 1975), and first-order interaction terms on these quantitative scores. Covariates and interaction terms significant at the α=0.05 level were adjusted for in a linear regression model, and standardized residuals were obtained for further statistical analyses. To adjust the distributions of scores for severity differences, preschool test scores were age-adjusted to 4 years (i.e., the mean value at age 4 years was added to each residual), whereas school-age test scores for single-word decoding and for reading and listening comprehension were adjusted to age 18 years (i.e., the mean value at age 18 years was added to each residual). We compared the mean values of affected sibs (not including the proband) and unaffected sibs through use of a t test. We also estimated correlations for the quantitative traits through use of FCOR (S.A.G.E. [2003], version 4.4).
Factor analysis
To reduce the battery of speech/language measures to a smaller set of test constructs, principal axis factor analysis with varimax rotation was conducted on the adjusted values for tests that were administered to both younger and older participants (i.e., age ⩾4 years). To identify distinct factors, scores with the high cross loadings were excluded after initial analyses, and factor analysis was repeated. The final analysis yielded three factors, accounting for 64% of the variance in scores: an articulation factor (ARTF) with loadings on GFTA and PCC, a phonological short-term memory factor (PHONF) with loadings on MSW and NSW, and a vocabulary factor (VOCF) with loadings on the PPVT-III and EOWPVT-R. The primary loadings of the tests on their respective factors were uniformly high (all >.65), with low cross loadings (all <.26). Separate analyses of data from the younger and older participants yielded similar results. For purposes of analysis, the two z scores that loaded on each factor were averaged to create three factor score composites.
Linkage analysis
We conducted model-free multipoint linkage analyses of six reading and language scores (WRMT-ID, WRMT-AT, WIAT-RC, WIAT-LC, SI, and RAN-C) and three factor scores (ARTF, PHONF, and VOCF) to determine whether component traits of SSD were influenced by QTLs in the chromosome 3 region identified by Nopola-Hemmi et al. (2001). The quantitative traits comprising the three factors were examined individually when the factors demonstrated evidence for linkage in the significant range (Lander and Kruglyak 1995).
Genotypes from parents and offspring were used to calculate identity by descent (IBD) allele sharing distributions through use of the GENIBD program of S.A.G.E. version 4.4 (S.A.G.E. 2003). Evidence for linkage was evaluated by a Haseman-Elston regression (Haseman and Elston 1972; Elston et al. 2000), as implemented in SIBPAL (S.A.G.E., version 4.4), using either the original Haseman-Elston regression (Haseman and Elston 1972) or the newest adaptation of the method, which parameterizes the sib pair’s trait value as a weighted combination of the squared trait difference and squared mean corrected trait sum adjusted for the nonindependence of sib pairs (W4 option [Shete et al. 2003]). Although the latter is more powerful asymptotically, in a finite sample the weights may not be optimally estimated, with the result that the former method outperforms it. Therefore, the smaller P values are reported. In all cases, the alternate method also gave evidence for linkage, albeit with a slightly larger P value. Asymptotic P values achieving nominal statistical significance were confirmed by empirically deriving a null permutation distribution through use of a sample of 50,000 replicate permutations of the data, as implemented in SIBPAL. These empirical P values were uncorrected for multiple testing.
Identification of sib pairs with linkage
We sought to identify the sib pairs that were contributing to the linkage results for both MSW and NSW, since separate linkage analyses of these traits suggested they were influenced independently by the putative QTL. For each sibling pair, we computed a score based on the squared sib-pair difference and the estimated sib-pair marker allele sharing, on which to rank the sib pairs for linkage informativity:
where y1 and y2 are the trait values for sibs 1 and 2,  is the average of (y1-y2)2 over the whole sample, and
is the average of (y1-y2)2 over the whole sample, and 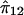 is the estimated mean allele sharing for the two sibs. This score is large (positive) either when the squared sib pair difference is small and
is the estimated mean allele sharing for the two sibs. This score is large (positive) either when the squared sib pair difference is small and 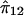 is large or when the squared sib-pair difference is large and
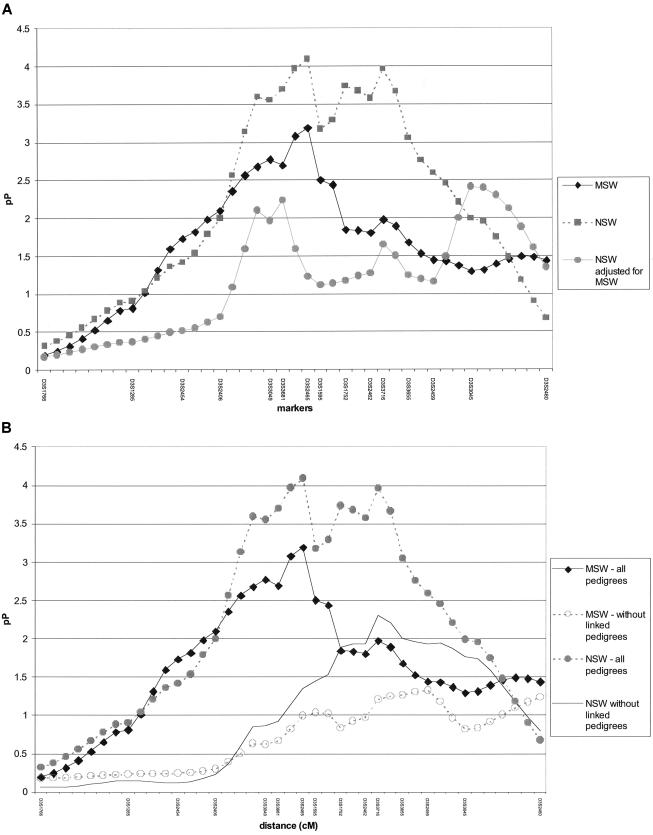
is large or when the squared sib-pair difference is large and  is small—that is, when the sibs are similarly alike, in terms of deviation from the mean, for both their traits and allele-sharing; otherwise the score will tend to be small (negative). Sib pairs with values in the upper 30th percentile of the distribution for both NSW and MSW were considered to be linked to the locus. To verify that these sib pairs indeed contributed most to the linkage effect, the pedigrees containing these sib pairs were removed from the data, and linkage analysis of both NSW and MSW was conducted again. The resulting linkage results were much less significant (fig. 3B).
is small—that is, when the sibs are similarly alike, in terms of deviation from the mean, for both their traits and allele-sharing; otherwise the score will tend to be small (negative). Sib pairs with values in the upper 30th percentile of the distribution for both NSW and MSW were considered to be linked to the locus. To verify that these sib pairs indeed contributed most to the linkage effect, the pedigrees containing these sib pairs were removed from the data, and linkage analysis of both NSW and MSW was conducted again. The resulting linkage results were much less significant (fig. 3B).
Figure 3.
A, Decomposition of the phonology factor into its component traits, multisyllabic word repetition and nonsense-word repetition. The linkage results for the phonology factor and the two component tests, NSW and MSW, are plotted as pP = −log10(asymptotic P value) on the Y-axis against genetic distance (in cM) on the X-axis. B, Comparison of linkage results for NSW and MSW after removal of sib pairs contributing to linkage based on IBD sharing for marker D3S2465.
Construction of core haplotypes in affected sib pairs
To characterize the smallest region of allele sharing, we constructed haplotypes in the concordantly affected sib pairs with linkage, defined as being in the upper 30th percentile of the score distribution and both sibs having low scores for NSW and MSW. The most likely haplotypes for each individual were constructed using Merlin (Abecasis et al. 2002). The procedure used the genotype data from the 15 markers in the parents and sibs of the linked sib pairs (which were identified as described above). Merlin uses sparse gene flow trees to estimate the most likely haplotypes according to a Markov process, under the assumption that no recombination occurs between consecutive markers.
Results
We examined speech-sound phenotypes in 77 pedigrees ascertained through a proband with SSD (table 2). Previous research has shown that SSD is more prevalent in males than in females (Shriberg et al. 1999), and this was indeed the case in our sample, since there were more than twice the number of affected males than females (P=.019). Language disorder was present in 23.4% of the children, and reading disorder was present in 21.6% (table 2). Other comorbidities were reported by the parents, but each represented <15% of the sample (table 2). The majority of the sample was white, and most of the sample was from the middle- to upper-class SES strata.
Table 2.
Description of Sample Ascertained through a Proband with Speech-Sound Disorder
| Characteristic of Sample | Value |
| Pedigrees (N) | 77 |
| Children (N) | 205 |
| Mothers (N) | 77 |
| Fathers (N) | 103 |
| Sex of children (no. of females/no. of males) | 80/125 |
| Sex of affected individuals (no. of females/no. of males) | 41/87 |
| Informative sib pairs (N) | 200 |
| Age of child at first visit (years) | Range 3–16; mean 7.35 |
| Performance IQ | Range 60–142; mean 101.76 |
| SES (Hollingshead Four-Factor Index)a: | |
| Category 1 | 6.3% |
| Category 2 | 11.3% |
| Category 3 | 20.5% |
| Category 4 | 35.0% |
| Category 5 | 26.8% |
| Race: | |
| White | 85.8% |
| African American | 9.7% |
| Hispanic | 2.1% |
| Asian | 2.4% |
| Comorbidities tested: | |
| Language disorder | 23.4% |
| Reading disorder | 21.6% |
| Comorbidities by historical report: | |
| Apraxia of speech | 5.9% |
| Stuttering | 3.2% |
| Voice disorder | 0.5% |
| Attention deficit disorder | 12.6% |
| Learning disability | 11.5% |
Category 1 is the lowest SES stratum, and category 5 is the highest.
Conceptualization and Analysis of Quantitative Traits
Figure 1 provides a schema conceptualizing hypothetical associations among the multiple speech-sound and dyslexia/reading phenotypes considered in this study. As illustrated in the figure, the specific role of each quantitative trait is embedded within a far more complex cognitive-linguistic network, including the subdomains of auditory and visual perception of words and letters underlying the production of spoken and written language. Thus, a deficit in phonological representation may underlie both SSD and dyslexia. Measures such as MSW and NSW assess the integrity of these representations. MSW and NSW test batteries can be presented either auditorily for preschoolers or as printed words for school-age children who can read; the latter invokes the use of visual processes, whereas the former relies on auditory mechanisms. Because we have used both types of measures, we refer to the auditorily presented measure as “phonological memory” or “speech-sound coding”; tests invoking visual processes are referred to as “reading decoding.”
Prior to conducting linkage analysis, adjustments were made for covariates and first-order interaction terms that accounted for significant (α=0.05) variance in scores. As shown in table 3, SES was significantly associated with most of the traits. After covariate adjustment, significant differences between affected and unaffected sibs remained on multiple measures. To characterize associations between affection status and the quantitative traits, table 4 presents mean scores for probands, affected siblings, and unaffected siblings. Statistically significant differences between the groups were most marked for GFTA, MSW, NSW, WRMT-ID, and RAN-C.
Table 3.
Covariate Adjustments Performed on the Quantitative Traits
| Variable | Adjustment(s) |
| GFTA | Age, sex |
| PCC | Age, age2, SES, age × SES, age2 × SES |
| MSW | Age, age2, SES, age × SES, age2 × SES |
| NSW | Age, age2, SES |
| EOWPVT-R | Age, age2, SES |
| PPVT-III | Age, age2, SES |
| SI | Age, age2, SES, sex |
| RAN-C | SES |
| WRMT-AT | Age, SES |
| WRMT-ID | Age2, SES |
| WIAT-LC | SES |
| WIAT-RC | Age, SES |
Table 4.
Mean Test Scores and SEs for Probands and Siblings Affected and Unaffected with SSD
|
Mean Test Score (SE) in |
||||
| Trait | Probands(N=77) | Affected Sibs(N=51) | Unaffected Sibs(N=77) | P Valuea |
| GFTA | 28.84 (2.55) | 58.45 (4.26) | 80.93 (2.48) | 2×10-5 |
| PCC | 83.98 (.94) | 92.01 (.95) | 94.74 (.58) | .027 |
| MSW | 25.91 (1.99) | 44.70 (3.14) | 66.00 (2.62) | 9×10-8 |
| NSW | 25.82 (2.03) | 40.92 (2.98) | 58.19 (2.22) | 6×10-6 |
| EOWPVT-R | 102.56 (1.81) | 109.17 (2.21) | 114.81 (1.46) | .110 |
| PPVT-III | 97.42 (1.62) | 100.54 (1.94) | 106.99 (1.34) | .033 |
| SI | 7.31 (.32) | 9.16 (.41) | 10.38 (.26) | .116 |
| WRMT-AT | 87.31 (1.74) | 92.85 (2.34) | 105.62 (1.12) | .022 |
| WRMT-ID | 89.27 (1.49) | 97.43 (1.89) | 105.98 (1.09) | 4×10-6 |
| WIAT-RC | 93.79 (1.49) | 100.27 (2.02) | 106.96 (1.25) | .030 |
| WIAT-LC | 100.77 (1.73) | 106.28 (2.08) | 108.15 (1.25) | .418 |
| RAN-C | .07 (.12) | .03 (.17) | −.41 (.10) | 4×10-5 |
Difference between affected and unaffected sibs calculated by use of the t test.
Table 5 summarizes associations between the quantitative measures. These data show that articulation skills (GFTA and PCC) and phonological memory (NSW and MSW) were highly intercorrelated. Despite the different demands of the tests of phonological memory compared with the reading measures (the latter tests involving visual processes and letter-to-sound conversion), robust correlations were also found between these domains. Correlations of measures of articulation and phonological processing with tests of vocabulary (PPVT-III and EOWPVT-R) were of only moderate magnitude. The fact that the PPVT-III scores were more variable than the EOWPVT-R among children who were old enough to take the reading tests may help to account for the stronger association of the PPVT-III with reading.
Table 5.
Cross-Trait Correlations for Quantitative Traits Related to SSD
|
Correlation |
|||||||||||
| PCC | GFTA | MSW | NSW | EOWPVT-R | PPVT-III | SI | RAN-C | WRMT-AT | WRMT-ID | WIAT-RC | |
| PCC | … | … | … | … | … | … | … | … | … | … | … |
| GFTA | .707 | … | … | … | … | … | … | … | … | … | … |
| MSW | .650 | .693 | … | … | … | … | … | … | … | … | … |
| NSW | .524 | .552 | .794 | … | … | … | … | … | … | … | … |
| EOWPVT-R | .373 | .462 | .458 | .420 | … | … | … | … | … | … | … |
| PPVT-III | .208 | .348 | .442 | .400 | .591 | … | … | … | … | … | … |
| SI | −.287 | −.368 | −.475 | −.462 | −.534 | −.501 | … | … | … | … | … |
| RAN-C | −.033 | −.118 | −.219 | −.261 | −.124 | −.182 | .300 | … | … | … | … |
| WRMT-AT | .179 | .131 | .423 | .468 | .084 | .430 | −.437 | −.354 | … | … | … |
| WRMT-ID | .117 | .193 | .342 | .428 | .211 | .431 | −.436 | −.291 | .652 | … | … |
| WIAT-RC | .102 | .162 | .416 | .430 | .177 | .424 | −.448 | −.238 | .633 | .634 | … |
| WIAT-LC | .128 | .202 | .200 | .226 | .377 | .381 | −.447 | −.187 | .273 | .527 | .520 |
Linkage Analysis
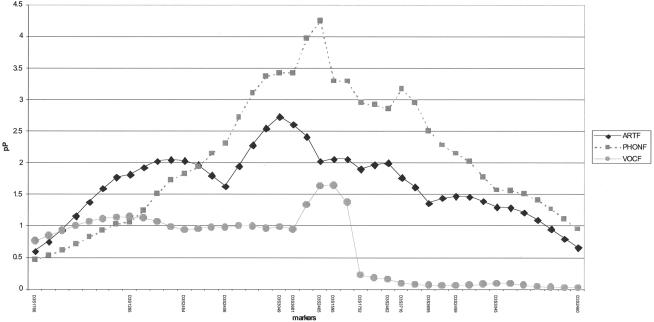
We conducted a model-independent linkage analysis of the chromosome 3p12-q13 region for each of the nine quantitative traits. Because the three factor scores summarize major domains of SSD (articulation, phonological memory, and vocabulary), only these linkage results are presented (fig. 2). We found significant evidence for linkage to PHONF (D3S2465, P=5.6×10-5, empirical P=2×10-5; and D3S3716, P=6.8×10-4, empirical P=4.6×10-4) and suggestive evidence for linkage to ARTF (D3S3049, P=1.8×10-3, empirical P=1.5×10-3), SI (D3S2454, P=6×10-3, empirical P=9×10-3), and the reading decoding variable WRMT-ID (D3S2465, P=.0040, empirical P=.0034) (fig. 2; table 6). For brevity, variables with little or no evidence of linkage are omitted from the tabular and graphic summaries. These findings suggest that the susceptibility gene in the pericentromeric region of chromosome 3 predisposes children for deficits in phonological processing and speech-sound production.
Figure 2.
Results of the model-free linkage analyses for the phonology, articulation, and vocabulary factors, plotted as pP = −log10(asymptotic P value) on the Y-axis against genetic distance (in cM) on the X-axis.
Table 6.
Linkage Analysis Results for Traits with Nominal Significance
|
P Value from Method |
||
| Trait andMarker | OriginalHaseman-Elston | W4 |
| WRMT-AT: | ||
| D3S1595 | .046 | .0054 |
| WRMT-ID: | ||
| D3S2465 | .069 | .0040 |
| WIAT-LC: | ||
| D3S2406 | .022 | .110 |
| D3S2460 | .0008 | .003 |
| SI: | ||
| D3S1595 | .028 | .017 |
| D3S2454 | .064 | .006 |
We further analyzed the linkage result for the PHONF by examining the component traits MSW and NSW individually. Each individually demonstrated substantial evidence of linkage to these markers, although the NSW trait gave stronger evidence of linkage (fig. 3A). The 1-LOD support interval for MSW spanned from D3S2406 to D3S1595, whereas the 1-LOD support interval for NSW was larger, spanning from D3S3049 to D3S3655. As indicated in table 5, these traits are highly correlated (r=.79). Thus, the use of a single trait may have been sufficient to explain the linkage results. Alternatively, it is feasible that the uncorrelated residual variability may have contributed uniquely to the genetic susceptibility, and it is important to examine both phenotypes. To assess the unique contributions of each variable to the linkage findings, we adjusted NSW for MSW in a linear regression model and analyzed the new residuals. We found that the evidence for linkage at our original peak (D3S2465) had diminished, although residual evidence for linkage with NSW remained at the region around D3S3045 (fig. 3A). This result suggests that this putative QTL influences NSW independently of MSW.
We identified 34 sib pairs contributing to linkage effects seen for both MSW and NSW, through use of the allele sharing estimates from the most significant marker (D3S2465). Of these, 9 pairs were concordantly affected (both having low scores for MSW and NSW), 2 were concordantly unaffected (both with high scores for MSW and NSW), and the remaining sib pairs were discordant for these phonological short-term memory measures. When the families containing these 34 sib pairs were removed from the linkage analysis, evidence for linkage disappeared at D3S2465 for the MSW/NSW joint signal, but nominal evidence for linkage with NSW persisted in the region around D3S3716 (fig. 3B). We analyzed D3S2465 and D3S3716 in a multiple regression model and found only D3S2465 to be marginally significant, and there appeared to be no interaction effect (data not shown). However, since the markers are relatively close (∼13 cM apart) and the sample size is moderate, we may not have had sample of sufficient size to accurately test a one-locus versus a two-locus model. Alternatively, the two markers may be too highly correlated, given their close proximity, resulting in confounding. Because a weak linkage signal for NSW persisted both after adjustment for MSW and after removal of the sib pairs with linkage, a second distant locus with independent effects on NSW cannot be ruled out. To examine the hypothesis of pleiotropy, we created scores for WRMT-AT and WRMT-ID through use of equation (1). Sample size for these analyses was reduced because of the inappropriateness of the reading tests for the younger children in the sample. Nevertheless, of the seven concordantly affected sib pairs with linkage to MSW and NSW who had data available for WRMT-AT and WRMT-ID, all showed linkage to WRMT-AT and WRMT-ID as well.
Haplotype Analysis of Sib Pairs with Linkage
Comparison of haplotypes in the 56-cM region in concordantly affected linked sib pairs (fig. 4) indicated extensive retention of shared chromosomal segments. The markers demonstrating maximal sharing extended from D3S3049 to D3S3045 and overlapped significantly with the Nopola-Hemmi et al. (2001) conserved disease haplotype. Most of these markers are near the centromere, where recombination is less likely to occur. In contrast to affected individuals sharing a single haplotype, as was described for the autosomal dominant form of dyslexia in the Nopola-Hemmi et al. (2001) article, affected sibs in the present study shared both maternal and paternal haplotypes—that is, diplotypes. This may be because of our selection scheme for sib pairs with linkage who have excess IBD sharing at marker D3S2465 among sibs with low trait values. However, for a dominant locus, one would anticipate that any excess in IBD sharing among cosibs with similarly low trait values would be equally distributed among those with only one allele shared IBD and those with two alleles shared IBD, similarly influencing the haplotypic outcome. To further examine mode of inheritance, we conducted a multiple regression to test the additive and dominance components of variance at D3S2465. We found that only the additive component of variance was significant, whereas the dominance component was not (P=.8).
Figure 4.
Shared haplotype blocks in sibling pairs who have concordantly low values for MSW and NSW and who are linked on the basis of expression (1). “NI” refers to genotypes that were not informative for resolving phase.
Discussion
SSD is a common multifactorial disorder with characteristics that overlap with those of developmental dyslexia (see fig. 1). Consistent with the possibility that these disorders share susceptibility loci, we report, in an independent sample, linkage of intermediate phenotypes for SSD to a locus on chromosome 3 that was originally identified as predisposing to developmental dyslexia in a single Finnish family (Nopola-Hemmi et al. 2001). Scores on factors and tasks that quantify a speaker’s skills in phonological short-term memory (PHONF, MSW, and NSW) showed strong evidence of linkage. Other verbal trait variables, including those indexed by the articulation factor (ARTF), verbal short term memory (SI), and single-word decoding (WRMT-AT and WRMT-ID) also showed some degree of evidence of linkage, suggesting that this putative QTL may act in a pleiotropic fashion, influencing both SSD and developmental dyslexia. The present findings expand the scope of developmental influence of the 3p12-q13 dyslexia locus from the Nopola-Hemmi et al. (2001) study indicating monogenic linkage in an extended family to consequences including SSD. More generally, our linkage finding for speech-sound production (ARTF), typically not considered a correlate of dyslexia or SLI, adds to the genetics of cognitive linguistic processes.
These results provide the first molecular genetic data supporting Pennington and Lefly’s (2001) hypothesis that early deficits in phonological processing skills and ability to repeat unfamiliar words, as in the NSW task, are predictors of subsequent dyslexia. High correlation among numerous dyslexia spectrum phenotypes has previously been reported (Fisher et al. 2002a; Grigorenko et al. 2003). In the present analysis, we found that similarly high correlations exist for traits measured in children ascertained for SSD (table 6), and the measures that demonstrated the greatest evidence for linkage were presented auditorily (MSW and NSW) rather than visually, with many of the participant children not yet readers. These measures identify early deficits in phonological memory and speech-sound coding of unfamiliar words. Nopola-Hemmi et al. (2002) observed that the neurocognitive type of dyslexia segregating in the Finnish family with linkage to 3p12-q13 consisted of deficits in phonological awareness, verbal short-term memory, and rapid naming. Findings for our phonology variables (PHONF and SI) are consistent with their findings, but, unlike in their findings, analyses of rapid naming (RAN-C) did not provide evidence for linkage. The rapid naming task in the Nopola-Hemmi et al. (2002) study required reading, which may have invoked visual abilities associated with other phonological representational skills, such as phoneme-grapheme correspondence. The linkage findings for the two measures that require reading skills (WRMT-AT and WRMT-ID) but assess processing constructs similar to that invoked in the auditorily presented measures further strengthen the support for this hypothesis. Results from both studies together suggest that the putative locus on chromosome 3 influences phonological processing.
Another genome scan in samples ascertained for dyslexia found that phenotypes for single-word reading, phonemic awareness, reading decoding, and orthographic coding were linked to markers located between 90 cM and 136.6 cM on chromosome 3 (Fisher et al. 2002a). The location of a second dyslexia linkage signal within 10–25 cM of our maximum lends further support for the possibility that at least one gene for phonological processing is located in this region of chromosome 3. However, the investigators did not provide sufficient detail for us to be able to fully compare the results. A genome scan for SLI found potential evidence of linkage to 3p24 but not to the 3p12-q13 region described in the present article (SLI Consortium 2002). This genome scan was conducted using the receptive and expressive scales of the CELF-R and a test for nonword repetition, a measure of phonological short-term memory. Extrapolating from our current study that cognitive domains common to both SSD and dyslexia are both influenced by a locus on chromosome 3, we anticipate that other loci linked to dyslexia and or SLI may also contribute to SSD susceptibility. We have not addressed these hypotheses but envisage that future studies will elucidate these relationships.
In analyzing NSW and MSW, we found that the putative chromosome 3 locus contributes not only to their shared variance but also uniquely to NSW (fig. 3A). Although both purportedly test phonological memory, they likely assess cognitively different processes. MSW uses words that may be in the subject’s lexicon, possibly relying on long-term memory. In contrast, NSW uses novel stimuli, which makes it a less confounded and possibly more sensitive test of phonological working memory. ARTF also shows suggestive linkage to this region (fig. 2), and its component traits, GFTA and PCC, are highly correlated with NSW and MSW (table 5). The GFTA measures the articulation of consonant sounds in familiar single words, and the PCC measures the articulation of consonant sounds in spontaneous conversational speech. Although both the GFTA and PCC assess articulation, they also invoke phonological memory, because the child is required to code information phonologically for temporary storage in short-term memory while performing each task. Finally, vocabulary measures (PPVT-III and EOWPVT-R) were included in the study to detect comorbid language impairment (LI). Vocabulary scores emerged as a significant factor in our analyses, possibly because of the high rate of comorbid LI in our sample (∼50%). Another explanation may be found in the emerging literature on the relationship between speech-sound development and vocabulary (Walley and Metsala 1990; Metsala 1997). Phonological information in long-term memory may play an active role in recall of short-term memory (Roodenrys et al. 2002). Thus, lexical familiarity may influence articulation accuracy. For example, children may repeat real multisyllabic words more accurately than nonsense words, because real words may be associated with phonological information stored in long-term memory. Thus, as with many cognitive test batteries, a single task relies on an array of neural processes.
The original article by Nopola-Hemmi et al. (2001) identified a core haplotype extending from D3S3049 to D3S1291 (spanning 16 markers over 41 cM), which was shared by 19 of 21 affected family members. Our evidence for linkage is strongest in this same region and is consistent for several of the traits analyzed. Using a simple method, we have identified sibs that are the source of our linkage signal and have examined the extent of haplotype sharing in these sibs. Our region of sharing overlaps with the conserved haplotype identified in the Nopola-Hemmi et al. (2001) article. In contrast to their results, however, both our haplotype analyses and our variance components analysis suggest a recessive or additive mode of inheritance for SSD.
To fully comprehend the biological basis of a multifactorial phenotype such as SSD, any binary classification, which is likely a function of multiple quantitative traits (Risch 2000), is probably too broad to adequately dissect the phenotypic overlap. Further, the lack of consensus on the etiological basis for SSD and its comorbidity with language and reading disorders makes the use of a standard qualitative affection phenotype less reliable. In contrast, well-chosen quantitative traits may be more closely tied to gene expression and therefore biologically more informative compared with the sensitivity of a clinical diagnosis or a threshold scale (Rice et al. 2001; Shriberg 1993). This approach also reduces misclassification of individuals into incorrect categories (Elston 1979). Moreover, as was done in the present analyses, utilizing appropriate quantitative traits for complex disorders may increase power to detect linkage (Duggirala et al. 1997) and may illustrate common quantitative QTLs that underlie these related disorders. Use of severity indices for gene mapping has been successful for dyslexia (Cardon et al. 1994, 1995; Grigorenko et al. 1997, 2003; Fisher and DeFries 2002; Fisher et al. 2002a; Kaminen et al. 2003; Marlow et al. 2003), as well as for other related traits such as specific language impairment (SLI Consortium 2002) and attention-deficit/hyperactivity disorder (Fisher et al. 2002b).
As pointed out by Fisher and DeFries (2002), interpretation of univariate QTL mapping data like that reported for the present study is challenging. In this study, we found at least nominal evidence of linkage to a region on chromosome 3 for about half of the SSD traits that we examined. Although we cannot say with certainty that this chromosomal region specifically affects deficits common to both SSD and developmental dyslexia, the reports reviewed and findings from the present results support that conclusion. Further, our linkage analysis suggests that this locus on chromosome 3 affects a core domain of SSD, articulation, which, to our knowledge, we are the first to examine. Since our variables for phonological memory (NSW/MSW) are correlated with the articulation measures (see table 5), we are unable to specify whether the linkage with the ARTF factor is due to articulation alone. We are in the process of testing a larger sample and plan to use multivariate linkage analysis (cf. Marlow et al. 2003; Stein et al. 2003) to address this issue. Our data also cannot be used to distinguish between the presence of one or two genes in this region. A similar pattern was seen in another quantitative trait analysis of dyslexia (Grigorenko et al. 2003). Because we have a moderately sized sample, we may not have the statistical power to resolve accurately a single versus multiple QTL model (Cordell 2001). Such information awaits a multivariate analysis of an independent sample.
Lastly, we acknowledge that additional loci with larger effect sizes associated with the domains depicted in figure 1 can be identified through genomewide scanning of this cohort. We are in the process of assembling a larger cohort, so that we can undertake this task. Because our focus is on speech-sound production, we are especially interested in identifying loci that would not ordinarily be identified through scans for dyslexia and SLI.
Acknowledgments
This study was supported by U. S. Public Health National Institute on Deafness and Other Communication Disorders research grants NIDCD DC00528 and DC-004005, National Institute of General Medical Sciences grant GM 28656, and National Heart, Lung, and Blood Institute training grant HL 07567. The results of this study were obtained by using the program package S.A.G.E., release 4.4, which is supported by U. S. Public Health Resource Grant RR03655 from the National Center for Research Resources. We are grateful to the families who participated in this research. We would also like to thank Mrs. Paula Wedig, who rendered technical assistance on the manuscript. Finally, we would like to thank the anonymous reviewers for suggestions they made on the content of this article.
References
- Abecasis G, Cherny S, Cookson W, Cardon L (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Aram DM, Hall NC (1989) Longitudinal follow-up of children with preschool communication disorders: treatment implications. School Psych Rev 18:487–501 [Google Scholar]
- Bishop D, Adams C (1990) A prospective study of the relationship between specific language impairment, phonological disorders, and reading retardation. J Child Psychol Psychiatry 31:1027–1050 [DOI] [PubMed] [Google Scholar]
- Bishop D, North T, Donlan C (1996) Nonword repetition as a behavioural marker for inherited language impairment: evidence from a twin study. J Child Psychol Psychiatry 37:391–403 [DOI] [PubMed] [Google Scholar]
- Cardon L, Smith S, Fulker D, Kimberling W, Pennington B, DeFries J (1994) Quantitative trait locus for reading disability on chromosome 6. Science 266:276–279 [DOI] [PubMed] [Google Scholar]
- ——— (1995) Quantitative trait locus for reading disability: correction. Science 268:1553 [DOI] [PubMed] [Google Scholar]
- Catts H (1986) Speech production/phonological deficits in reading disordered children. J Learn Disabil 19:504–508 [DOI] [PubMed] [Google Scholar]
- Cordell H (2001) Sample size requirements to control for stochastic variation in magnitude and location of allele-sharing linkage statistics in affected sibling pairs. Ann Hum Genet 65:491–502 10.1046/j.1469-1809.2001.6550491.x [DOI] [PubMed] [Google Scholar]
- Denckla M, Cutting L (1999) History and significance of rapid auditory naming. Ann Dyslexia 49:29–42 [Google Scholar]
- Denckla M, Rudel R (1976) Rapid “automatized” naming (R.A.N.): dyslexia differentiated from other learning disabilities. Neuropsychologia 14:471–479 10.1016/0028-3932(76)90075-0 [DOI] [PubMed] [Google Scholar]
- Duggirala R, Williams J, Williams-Blangero S, Blangero J (1997) A variance components approach to dichotomous trait linkage analysis using a threshold model. Genet Epidemiol 14:987–992 [DOI] [PubMed] [Google Scholar]
- Dunn L, Dunn L (1997) Peabody picture vocabulary test–third edition. American Guidance Service, Circle Pines, MN [Google Scholar]
- Elston R (1979) Major locus analysis for quantitative traits. Am J Hum Genet 31:655–661 [PMC free article] [PubMed] [Google Scholar]
- Elston R, Buxbaum S, Jacobs K, Olson J (2000) Haseman and Elston revisited. Genet Epidemiol 19:1–17 [DOI] [PubMed] [Google Scholar]
- Fagerheim T, Raeymaekers P, Tonnessen FE, Pedersen M, Tranebjaerg L, Lubs HA (1999) A new gene (DYX3) for dyslexia is located on chromosome 2. J Med Genet 36: 664–669 [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld S, McGue M, Broen P (1995) Familial aggregation of phonological disorders: results from a 28 year follow-up. J Speech Hear Res 38:1091–1107 [DOI] [PubMed] [Google Scholar]
- Fisher S, DeFries J (2002) Developmental dyslexia: genetic dissection of a complex cognitive trait. Nat Rev Neurosci 3:767–780 10.1038/nrn936 [DOI] [PubMed] [Google Scholar]
- Fisher S, Francks C, Marlow A, MacPhie I, Newbury D, Cardon L, Ishikawa-Brush Y, Richardson AJ, Talcott JB, Gayan J, Olson RK, Pennington BF, Smith SD, Stein JF, Monaco AP (2002a) Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat Genet 30:86–91 10.1038/ng792 [DOI] [PubMed] [Google Scholar]
- Fisher S, Francks C, McCracken J, McGough J, Marlow A, MacPhie I, Newbury D, Crawford LR, Palmer CGS, Woodward JA, Del’Homme M, Cantwell DP, Nelson DP, Monaco AP, Smalley SL (2002b) A genomewide scan for loci involved in attention-deficit/hyperactivity disorder. Am J Hum Genet 70:1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP (1999) A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet 64:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Vargha-Khadem F, Watkins KE, Monaco AP, Pembrey ME (1998) Localisation of a gene implicated in a severe speech and language disorder. Nat Genet 18:168–170 [DOI] [PubMed] [Google Scholar]
- Francks C, Fisher SE, Olson RK, Pennington BF, Smith SD, DeFries JC, Monaco A (2002) Fine mapping of the chromosome 2p12-16 dyslexia susceptibility locus: quantitative association analysis and positional candidate genes SEMA4F and OTX1. Psychiatr Genet 12:35–41 10.1097/00041444-200203000-00005 [DOI] [PubMed] [Google Scholar]
- Gardner M (1990) Expressive one word picture vocabulary test–revised. Academic Therapy Publications, Novato, CA [Google Scholar]
- Gayan J, Smith S, Cherny S, Cardon L, Fulker D, Brower A, Olson R, Pennington BF, DeFries JC (1999) Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet 64:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R, Fristoe M (1986) The Goldman-Fristoe test of articulation. American Guidance Service, Circle Pines, MN [Google Scholar]
- Grigorenko E, Wood F, Golovyan L, Meyer M, Romano C, Pauls D (2003) Continuing the search for dyslexia genes on 6p. Am J Med Genet 118B:89–98 [DOI] [PubMed] [Google Scholar]
- Grigorenko E, Wood F, Meyer M, Hart L, Speed W, Shuster A, Pauls D (1997) Susceptibility loci for distinct components of developmental dyslexia on chromosome 6 and 15. Am J Hum Genet 60:27–39 [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls JE, Hart LA, Pauls DL (2001) Linkage studies suggest a possible locus for developmental dyslexia on chromosome 1p. Am J Med Genet 105: 120–129 [DOI] [PubMed] [Google Scholar]
- Haseman J, Elston R (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav Genet 2:3–19 [DOI] [PubMed] [Google Scholar]
- Hollingshead A (1975) Four factor index of social class. Report #06520, Department of Sociology, Yale University, New Haven, CT [Google Scholar]
- Kamhi A, Catts H (1986) Toward an understanding of developmental language disorders and reading disorders. J Speech Hear Disord 51:337–347 [DOI] [PubMed] [Google Scholar]
- Kaminen N, Hannula-Jouppi K, Kestila M, Lahermo P, Muller K, Kaaranen M, Myllyluoma B, Voutilainen A, Lyytinen H, Nopola-Hemmi J, Kere J (2003) A genome scan for developmental dyslexia confirms linkage to chromosome 2p11 and suggests a new locus on 7q32. J Med Genet 40:340–345 10.1136/jmg.40.5.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DE, Gayan J, Ahn J, Won TW, Pauls D, Olson RK, DeFries JC, Wood F, Pennington BF, Page GP, Smith SD, Gruen JR (2002) Evidence for linkage and association with reading ability on 6p21.3-22. Am J Hum Genet 70: 1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan L, Lewis N (1986) Khan-Lewis phonological analysis. American Guidance Service, Circle Pines, MN [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lewis B (1992) Pedigree analysis of children with phonology disorders. J Learn Disabil 25:586–597 [DOI] [PubMed] [Google Scholar]
- Lewis B, Cox N, Byard P (1993) Segregation analysis of speech and language disorders. Behav Genet 23:291–297 [DOI] [PubMed] [Google Scholar]
- Lewis B, Ekelman B, Aram D (1989) A familial study of severe phonological disorders. J Speech Hear Res 32:713–724 [DOI] [PubMed] [Google Scholar]
- Lewis B, Freebairn L (1998) Speech production skills of nuclear family members of children with phonology disorders. Lang Speech 41:45–61 [DOI] [PubMed] [Google Scholar]
- Lewis B, Freebairn L, Taylor H (2000) Follow-up of children with early expressive phonology disorders. J Learn Disabil 33:433–444 [DOI] [PubMed] [Google Scholar]
- Lewis B, Thompson L (1992) A study of developmental speech and language disorders in twins. J Speech Hear Res 35:1086–1094 [DOI] [PubMed] [Google Scholar]
- Marlow A, Fisher S, Francks C, MacPhie I, Cherny S, Richardson A, Talcott J, Stein JF, Monaco AP, Cardon LR (2003) Use of multivariate linkage analysis for dissection of a complex trait. Am J Hum Genet 72:561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menyuk P, Chesnick M, Liebergott J, Korngold B, D’Agostino R, Belanger A (1991) Predicting reading problems in at-risk children. J Speech Hear Res 34:893–903 [DOI] [PubMed] [Google Scholar]
- Metsala JL (1997) An examination of word frequency and neighborhood density on development of spoken-word recognition. Mem Cognit 25:47–56 [DOI] [PubMed] [Google Scholar]
- Newcomer P, Hammill D (1988) Test of language development–primary, second edition. Pro-Ed, Austin, TX [Google Scholar]
- Nopola-Hemmi J, Myllyluoma B, Haltia T, Taipale M, Ollikainen V, Ahonen T, Voutilainen A, Kere J, Widen E (2001) A dominant gene for developmental dyslexia on chromosome 3. J Med Genet 38:658–664 10.1136/jmg.38.10.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopola-Hemmi J, Myllyluoma B, Voutilainen A, Leinonen S, Kere J, Ahonen T (2002) Familial dyslexia: neurocognitive and genetic correlation in a large Finnish family. Dev Med Child Neurol 44:580–586 10.1017/S0012162201002614 [DOI] [PubMed] [Google Scholar]
- Nothen MM, Schulte-Korne G, Grimm T, Chichon S, Vogt IR, Muller-Myhsok B, Propping P, Remschmidt H (1999) Genetic linkage analysis with dyslexia: evidence for linkage of spelling disability to chromosome 15. Eur Child Adolesc Psychiatry 8:56–59 10.1007/s007870050084 [DOI] [PubMed] [Google Scholar]
- Pennington B, Lefly D (2001) Early reading development in children at family risk for dyslexia. Child Dev 72:816–833 10.1111/1467-8624.00317 [DOI] [PubMed] [Google Scholar]
- Petryshen TL, Kaplan BJ, Fu Liu M, de French NS, Tobias R, Hughes ML, Field LL (2001) Evidence for a susceptibility locus on chromosome 6q influencing phonological coding dyslexia. Am J Med Genet 105:507–517 10.1002/ajmg.1475 [DOI] [PubMed] [Google Scholar]
- Petryshen TL, Kaplan BJ, Hughes ML, Tzenova J, Field LL (2002) Supportive evidence for the DYX3 dyslexia susceptibility gene in Canadian families. J Med Genet 39:125–126 10.1136/jmg.39.2.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin M, Wen XL, Hepburn M, Lubs HA, Feldman E, Duara R (1993) Suggestive linkage of developmental dyslexia to chromosome 1p34-p36. Lancet 342:178 10.1016/0140-6736(93)91384-X [DOI] [PubMed] [Google Scholar]
- Raitano NA, Pennington BF, Tunick RA, Boada R, Shriberg LD. Pre-literacy skills of subgroups of children with speech sound disorders. J Child Psychol Psychiatr (in press) [DOI] [PubMed] [Google Scholar]
- Rice J, Saccone N, Rasmussen E (2001) Definition of the phenotype. Adv Genet 42:69–76 [DOI] [PubMed] [Google Scholar]
- Risch N (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 10.1038/35015718 [DOI] [PubMed] [Google Scholar]
- Robbins J, Klee T (1987) Clinical assessment of oropharyngeal motor development in young children. J Speech Hear Disord 52:271–277 [DOI] [PubMed] [Google Scholar]
- Roodenrys S, Hulme C, Lethbridge A, Hinton M, Nimmo LM (2002) Word-frequency and phonological-neighborhood effects on verbal short-term memory. J Exp Psychol Learn Mem Cogn 28:1019–1034 10.1037//0278-7393.28.6.1019 [DOI] [PubMed] [Google Scholar]
- S.A.G.E. (2003) Statistical analysis for genetic epidemiology. Statistical Solutions, Cork, Ireland [Google Scholar]
- Schick J, Kundtz A, Tiwari H, Taylor H, Freebairn L, Hansen A, Shriberg L, Lewis BA, Iyengar SK (2002) Broad phenotype of speech disorders shows strong evidence of linkage at candidate region 7q31. Paper presented at the European Human Genetics Conference. Strasbourg, France, May 25–28 [Google Scholar]
- Schulte-Körne G, Grimm T, Nöthen MM, Müller-Myhsok B, Cichon S, Vogt IR, Propping P, Remschmidt H (1998) Evidence for linkage of spelling disability to chromosome 15. Am J Hum Genet 63:279–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semel E, Wiig E, Secord W (1987) Clinical evaluation of language fundamentals–revised. The Psychological Corporation, San Antonio, TX [Google Scholar]
- Shete S, Jacobs K, Elston R (2003) Adding further power to the Haseman and Elston method for detecting linkage in larger sibships: weighting sums and differences. Hum Hered 55:79–85 10.1159/000072312 [DOI] [PubMed] [Google Scholar]
- Shriberg L (1993) Four new speech and prosody voice measures for genetics research and other studies in developmental phonological disorders. J Speech Hear Res 36:105–140 [DOI] [PubMed] [Google Scholar]
- ——— (2002) Classification and misclassification of child speech sound disorders. Paper presented at the Annual Convention of the American Speech-Language-Hearing Association. Atlanta, November 20–24 [Google Scholar]
- Shriberg L, Austin D (1998) Comorbidity of speech-language disorder: implications for a phenotype marker for speech delay. In: Paul R (ed) Exploring the speech/language connection. Brookes, Baltimore, pp 73–118 [Google Scholar]
- Shriberg L, Austin D, Lewis B, McSweeny J, Wilson D (1997) The percentage of consonants correct (PCC) metric: Extensions and reliability data. J Speech Lang Hear Res 40:708–722 [DOI] [PubMed] [Google Scholar]
- Shriberg L, Kent R (2003) Clinical phonetics. 3rd ed. Allyn and Bacon, Boston [Google Scholar]
- Shriberg L, Kwiatkowski J (1988) A follow-up of children of phonological disorders of unknown origin. J Speech Hear Res 53:144–155 [DOI] [PubMed] [Google Scholar]
- Shriberg L, Tomblin J, McSweeny J (1999) Prevalence of speech delay in 6-year-old children and comorbidity with language impairment. J Speech Lang Hear Res 42:1461–1481 [DOI] [PubMed] [Google Scholar]
- Specific Language Impairment Consortium (2002) A genomewide scan identifies two novel loci involved in specific language impairment. Am J Hum Genet 70:384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CM, Song Y, Elston RC, Jun G, Tiwari HK, Iyengar SK (2003) Structural equation model-based genome scan for metabolic syndrome. BMC Genetics 4 Suppl 1:S99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomblin J, Buckwalter P (1998) The heritability of poor language achievement among twins. J Speech Lang Hear Res 41:188–199 [DOI] [PubMed] [Google Scholar]
- Tunick R, Pennington B (2002) The etiological relationship between reading disability and phonological disorder. Ann Dyslexia 52:75–95 [Google Scholar]
- Vargha-Khadem F, Watkins KE, Price CJ, Ashburner J, Alcock KJ, Connelly A, Frackowiak RS, Friston KJ, Pembrey ME, Mishkin M, Gadian DG, Passingham RE (1998) Neural basis of an inherited speech and language disorder. Proc Natl Acad Sci USA 95:12695–12700 10.1073/pnas.95.21.12695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley AC, Metsala JL (1990) The growth of lexical contraints on spoken word recognition. Perceptual Psychophysiology 47:267–280 [DOI] [PubMed] [Google Scholar]
- Wechsler D (1989) Wechsler preschool and primary scale of intelligence–revised (WPPSI-R). The Psychological Corporation, San Antonio, TX [Google Scholar]
- ——— (1991) Wechsler intelligence scale for children–third edition. The Psychological Coporation, San Antonio, TX [Google Scholar]
- ——— (1992) Wechsler individual achievement test. The Psychological Corporation, San Antonio, TX [Google Scholar]
- Woodcock R (1987) Woodcock reading mastery tests–revised. American Guidance Service, Circle Pines, MN [Google Scholar]