Abstract
The ATR protein is a member of the phosphoinositide 3-kinase-related kinase family and plays an important role in UV-induced DNA damage checkpoint response. Its role as a signal transducer in cell cycle checkpoint is well established, but it is currently unclear whether ATR functions as a damage sensor as well. Here we have purified the ATR protein and investigated its interaction with DNA by using biochemical analysis and electron microscopy. We find that ATR is a DNA-binding protein with higher affinity to UV-damaged than undamaged DNA. In addition, damaged DNA stimulates the kinase activity of ATR to a significantly higher level than undamaged DNA. Our data suggest that ATR may function as an initial sensor in the DNA damage checkpoint response.
DNA damage checkpoint is the arrest in cell cycle progression upon damage to cellular DNA. The checkpoint response, conceptually, has three biochemical components: damage sensors, signal transducers, and effector molecules (1–3). At present, the damage-sensing step of checkpoint signaling is not well understood. Thus, it is not clear whether primary lesions in the form of UV photoproducts, base adducts, single-strand nicks, double-strand breaks induced by UV, chemotherapeutic drugs, and ionizing radiation are detected by the molecular sensors that recognize one or more of these lesions or whether all these lesions eventually create a common structure on encounter with a replication fork, which then constitutes the signal for the checkpoint. Similarly, the identity of the damage sensors remains to be determined. Genetic and preliminary biochemical analyses in human cell lines and budding and fission yeasts indicate that ATM and ATR or the checkpoint Rad proteins Rad17, Rad9, Rad1, and Hus1 in mammals and their counterparts in yeasts may function as damage sensors (1, 2).
ATM and ATR belong to the phosphoinositide 3-kinase-related kinase (PIKK) family of proteins. The PIKK members are large proteins with Ser/Thr kinase activity serving important roles in DNA repair and DNA damage checkpoint (1, 4). The three PIKK proteins with repair and checkpoint functions in mammalian cells are: DNA-dependent protein kinase (DNA-PK), the kinase mutated in radiosensitive/genomic instability syndrome ataxia telangiectasia (ATM, A-T mutated), and ATR (ATM and Rad3 related) (see ref. 5). DNA-PK participates in V(D)J recombination and repair of double-strand breaks by nonhomologous end joining; it is a heterotrimer of DNA-PKcs (a PIKK member), Ku70, and Ku86, and it binds to DNA ends through the Ku70/Ku86 heterodimer. The role of DNA-PK in end joining is relatively well understood, but the physiological relevance of its kinase activity is unclear at present. In contrast, the physiological consequences of protein phosphorylation by ATM and ATR are reasonably well defined, but the DNA-binding activities of these proteins and their relevance to the ATM/ATR functions remain ill defined. Upon DNA damage (by ionizing radiation for ATM and by UV for ATR), both proteins phosphorylate p53 and the signal transduction kinases Chk1 and Chk2. However, there are no convincing data that ATM and ATR are recruited to the site of damage by functional homologs of the Ku70/Ku86 complex. It has been postulated that the mammalian checkpoint Rads (Rad17, Rad9, Rad1, and Hus1) may be the damage sensors functioning upstream of the ATM and ATR kinases (1, 5).
The checkpoint Rads are expected to form protein complexes that are the structural homologs of the Replication Factor C (RFC)/proliferating cell nuclear antigen (PCNA) clamp loader/replication clamp but with a specialized role in the DNA damage checkpoint response. Thus, it has been proposed that Rad17 forms a heteropentameric complex in which the large subunit of RFC (p140) is replaced by Rad17 (6–8). Similarly, molecular modeling studies have led to the proposal that Rad9, Rad1, and Hus1 form a heterotrimeric checkpoint sliding clamp or 9–1-1 complex similar to PCNA (9–11). In vivo (12–15) and in vitro (16) biochemical studies have supported the presence of the Rad17-RFC checkpoint clamp-loading complex and of the Rad9-Rad1-Hus1 checkpoint sliding clamp complex, and of the presence of a DNA damage checkpoint complex comprising the Rad17-RFC and the checkpoint 9–1-1 complexes (16). However, at present there is no biochemical evidence that these complexes recognize either primary or secondary DNA lesions or repair intermediates that may be the checkpoint-activating structures.
In this paper, we have attempted to address two questions: whether the primary UV lesion in the form of unprocessed photoproduct can act as a signal for checkpoint activation, and whether the ATR protein can initiate the checkpoint signaling cascade by directly binding to and becoming activated by UV photoproducts. Our results indicate that ATR directly binds to UV-damaged DNA with higher affinity than undamaged DNA, and that this interaction increases its kinase activity toward p53. These results are consistent with the notion that, at least under certain circumstances, unprocessed lesions can initiate DNA damage checkpoint response and that ATR may function as damage sensor in DNA damage checkpoint response to UV.
Materials and Methods
Cell Culture and Antibodies.
The simian virus 40-transformed human embryonic kidney (HEK)293T cells were maintained in DMEM (GIBCO/BRL) supplemented with 10% FBS and 100 units of penicillin and streptomycin/ml. Mouse monoclonal anti-Flag antibody was obtained from Sigma.
Plasmid Constructs.
Two Flag-ATR constructs containing the full-length cDNA clone of human ATR and its kinase-dead version (K2327R) in pcDNA3 (Invitrogen) expression vector were kindly provided by R. T. Abraham [Duke University (17)]. By using PCR, various regions of the cDNA encoding ATR were amplified (see delineation of the regions in Fig. 1). In each case, the 5′ primer contained an ATG codon followed by a Flag-epitope in frame with the coding region that was amplified. These PCR products were digested with BamHI and XhoI restriction enzymes and ligated into pcDNA4.1 (Invitrogen) expression vector through the same enzyme sites to generate the N-terminal Flag epitope-tagged ATR fragments.
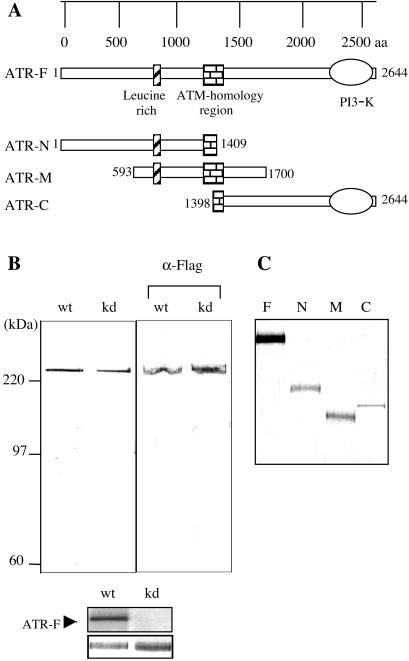
Figure 1.
Purification of full-length and various fragments of ATR from transiently transfected HEK293T cells. (A) Schematic representation of the full-length ATR (ATR-F) and its N-terminal (ATR-N), middle region (ATR-M), and C-terminal (ATR-C) fragments. (B) Recombinant wild-type (wt) and kinase-dead (kd) ATR, each containing a Flag affinity tag, purified from HEK293T cells, were separated by SDS/PAGE and visualized by silver staining shown (Left). Western blot of the ATR preparations with an antibody specific for the N-terminal FLAG tag is shown (Right). For the kinase activity of recombinant wild-type and kinase-dead ATR, purified proteins were incubated with [γ-32P]-ATP, and reaction products were electrophoresed and visualized by silver staining (Bottom) or autoradiography (Top). (C) Expression of Flag-epitope tagged and affinity-purified ATR-fragments as analyzed by silver staining.
Transfections and Cell Lysis.
Transfections were carried out by using the calcium phosphate method. For transfections, 8 × 106 293T cells were plated per 225 cm2 flask and allowed to grow for 24 h. Plasmid DNA (25 μg) was mixed with CaCl2, and this mixture was added to a 2 × HEBS (140 mM NaCl/1.5 mM Na2HPO4/50 mM Hepes, pH 7.05) solution and incubated for 8 min at room temperature. The CaCl2/DNA/HEBS precipitate was then added dropwise to the cell medium. After 16-h incubation at 37°C in 5% CO2, cells were washed twice in serum-free DMEM and fresh media added to the cells for a further 48 h of incubation. Cells were lysed in ice-cold lysis buffer [50 mM Tris, pH 7.5/150 mM NaCl/10 mM β-glycerophosphate/10% glycerol/1% Tween-20/0.1% Nonidet P-40/1 mM Na3VO4/1 mM NaF and protease inhibitors (Roche Molecular Biochemicals)] for 30 min. The cell lysates were centrifuged for 30 min at 32,000 × g or frozen in dry ice and stored at −80°C until use.
Protein Purification.
The supernatants from the spun lysates were incubated at 4°C for at least 4 h with anti-FLAG M2 affinity gel (Sigma) that had been preequilibrated in TBS buffer (50 mM Tris⋅HCl, pH 7.5/150 mM NaCl). The beads were then washed once with 1 M NaCl containing TBS buffer and three times with 150 mM NaCl containing TBS. The ATR bound to beads was either used directly for pull-down experiments or eluted with TBS buffer containing 200 μg/ml FLAG peptide (Sigma).
Human p53 protein overexpressed in SF9 cells was generated in the laboratory of J.D.G., as described (18).
DNA Substrates.
Duplex DNAs of 46 bp, containing a single T[6–4]T photoproduct and its unmodified version, were prepared from four complementary oligomers. The sequences of the oligonucleotides are:
5′-ctagcgggatccggtgcaggtaT[6-4]Tatggaattcgtagatctgcgtc-3′
3′-gatcgccctaggccacgtccataataccttaagcatctagacgcag-5′
The T[6–4]T photoproduct containing substrate was constructed by ligating the damage-containing oligomer (8-mer), which was 5′-terminally labeled, with other partially overlapping oligonucleotides to obtain the internally labeled duplex as described (19, 20). The resulting 46-bp duplex contains the (6–4) photoproduct at positions 23 and 24. The full-length duplexes were separated from the ligation mixture by gel purification through 5% nondenaturing polyacrylamide gel. These substrates were used for pull-down assays. For protein–DNA crosslinking experiments, a 50-mer substrate with an azidophenacyl photosensitizer of the following sequence was used:
5′-gcgccagctggccaccctgagaagctacgagcgagcgccaagcttgggct-3′.
These 50-mer duplex DNAs were purchased from Operon Technologies (Alameda, CA). The azidophenacyl moiety was coupled to the oligonucleotide as described (21).
For electron microscopy and kinase assays, pGEMEX1 dsDNA cooled on ice in a microtiter plate was irradiated with 254-nm light at a rate of 1 mW/cm2 by using a germicidal UV-C (254-nm) lamp (General Electric).
Electron Microscopy.
Fifty nanograms of ATR protein was incubated with 200 ng of linearized or supercoiled pGEMEX1 dsDNA in kinase buffer containing 25 mM Hepes, pH 7.9, 50 mM KCl, 10 mM MgCl2, 2 mM MnCl2, 20% glycerol, 0.1% Nonidet P-40, 1 mM DTT, and 0.5 mg/ml of BSA for 15 min at 30°C. After incubation, DNA–protein complexes were fixed with 0.6% glutaraldehyde for 10 min at 20°C. The reaction mixture was chromatographed on 2 ml of BioGel A5m (BioRad), equilibrated with TE buffer (10 mM Tris⋅HCl/1 mM EDTA, pH 8.0) to remove fixatives and unbound protein. The fractions containing DNA–protein complexes were collected and prepared for electron microscopy as described (22) and analyzed by using a Philips (Eindhoven, The Netherlands) CM12 electron microscope. The images were scanned by using a Nikon-4500 film scanner and contrast adjusted by using Adobe photoshop software (Adobe Systems, Mountain View, CA). The size of protein particles bound to DNA was estimated as described elsewhere (23).
Pull-Down Assays.
The Flag-ATR protein bound to the beads was washed first with the kinase buffer and then incubated with internally radiolabeled DNA substrates at 30°C for 30 min in a reaction buffer of 25 μl. The beads were washed three times with 0.1% Nonidet P-40 containing TBS buffer at 4°C. The DNA-bound ATR on the beads was either loaded onto 10% SDS/PAGE or eluted with TBS buffer containing 200 μg/ml of FLAG peptide (Sigma) and then visualized by silver staining. The amount of DNA bound to ATR was measured with a PhosphorImager and the imagequant system (Molecular Dynamics) by comparing the radioactivity bound to ATR-containing resin to the radioactivity in control lanes containing known amounts of DNA.
Protein–DNA Crosslinking.
Azidophenacetyl-derivatized DNA (2 fmol) was incubated with approximately 100 ng of ATR in 10 μl of kinase buffer at 30°C for 30 min. Reaction mixtures were transferred to a microtiter plate and irradiated with a 366-nm light from BLB lamps (General Electric) at a rate of 1 mW/cm2 at a distance of 5 cm. The samples were covered with the polystyrene lid of the microtiter plate to shield them from wavelengths below 300 nm and cooled on ice during illumination. Crosslinked proteins were resolved on 7.5% SDS/PAGE, and gels were silver stained, dried, and exposed to film.
Kinase Assays.
Kinase assays were performed in 20-μl reactions by using 50 ng of purified ATR, 50 ng of purified p53, and linearized pGEMEX1 DNA with and without UV treatment at the indicated concentrations in a kinase buffer containing 25 mM Hepes, pH 7.9, 50 mM KCl, 10 mM MgCl2, 2 mM MnCl2, 20% glycerol, 0.1% Nonidet P-40, 1 mM DTT, and 0.5 mg/ml of BSA. ATR and DNA mixtures were incubated on ice for 5 min before substrate and ATP addition. ATP was added to a final concentration of 50 μM along with 5 μCi[γ-32P]-ATP. Reaction mixtures were incubated at 30°C for 30 min. Phosphorylated proteins were subjected to 10% SDS/PAGE, visualized by silver staining, and exposed on a Molecular Dynamics PhosphorImager screen for quantitation by imagequant software.
Results
Purification of Full-Length ATR and ATR Fragments for DNA–Protein Interactions.
We wished to determine whether ATR interacts directly with DNA and if it does, whether this interaction is confined to a specific domain within the protein. Fig. 1A schematically shows the organization of the ATR protein (24). This large protein contains a limited number of sequence motifs as indicated: these include an N-terminal leucine-rich domain, an ATM homology domain of about 100 amino acids in its middle region, and a C-terminal domain with homology to PIKK family proteins, which contains the kinase active site residues. To investigate ATR-DNA interaction and the effect of this interaction on kinase activity, we used FLAG-tagged wild-type and kinase-dead ATR constructs in a mammalian expression vector as described (17, 25) as well as FLAG-tagged constructs expressing the N-terminal half (N), the middle (M), and the C-terminal half (C) of the protein as indicated in Fig. 1A. First we purified the full-length wild-type and kinase-dead ATR by affinity chromatography and tested them for purity by SDS/PAGE and silver staining and for activity by an autophosphorylation assay. As seen in Fig. 1B, the proteins are of high purity, and the kinase-dead mutant does not exhibit detectable kinase activity in our assay. Fig. 1C shows the expression of the ATR fragments on transfection of HEK293T cells with the appropriate constructs. In general, we found that the ATR full-length, ATR-N, and ATR-M proteins were expressed at comparable levels, but ATR-C was expressed at a significantly lower amount.
Binding of ATR to DNA.
Analysis of ATR-DNA binding by conventional electrophoretic mobility-shift assay in conjunction with “supershift” by anti-ATR antibodies indicated that ATR directly binds DNA (data not shown). However, because of the large size of the protein, data obtained by this method were qualitative and variable. Therefore we used a “pull-down” assay to analyze ATR–DNA interactions. HEK293T cells transfected with vectors expressing full-length ATR or ATR fragments were lysed, and the tagged ATR proteins were purified by binding to FLAG-agarose beads. Radiolabeled DNA was added to the beads and, after incubation and exhaustive washes, the proteins on the beads were released with SDS, visualized by silver staining, and the DNA was detected by autoradiography. Fig. 2 Top shows the relative levels of ATR and its derivatives on the beads. The full-length protein and the N-terminal and middle fragments are expressed at comparable levels and the C-terminal fragment is at a lower amount. Fig. 2 Middle shows the autoradiographic analysis of bound DNA and Bottom shows the quantitative analysis of the data from Middle. It appears that only full-length ATR binds DNA as the amount of DNA associated with ATR fragments is not above background. Although the use of those fragments in the pull-down experiment shows that ATR fragments alone cannot bind to DNA, the presence of ATR fragments on the beads confirms the specificity of full-length ATR interaction with DNA, indicating that the full-length ATR interaction with DNA is not a result of nonspecific binding of DNA onto resins.
Figure 2.
ATR binds DNA. HEK293T cells were transfected with vector expressing full-length ATR (ATR-F), its N-terminal (ATR-N), middle region (ATR-M), and C-terminal (ATR-C) fragments or the nontransfected (Mock) cells were lysed and the tagged proteins were purified by binding to FLAG-agarose beads. The beads were then incubated with radiolabeled DNA substrate as described in Materials and Methods. After exhaustive washing, the beads were loaded onto a 10% SDS/PAGE, and the bound proteins were visualized by silver staining (Top), and the levels of ATR-bound substrates were visualized by autoradiography (Middle). Bottom shows the quantitative analysis of data from Middle.
Our ATR preparations are quite pure (see Fig. 1), and therefore the most likely explanation of the data in Fig. 2 is that ATR directly binds DNA. However, it was conceivable that binding was mediated by small quantities of a contaminant not detectable by our analyses. Therefore, we wished to confirm direct ATR-DNA binding by photocrosslinking. DNA–protein crosslinking by UV occurs only when the two are in direct contact, and therefore this method is referred to as “zero-distance” crosslinking (26). To perform UV crosslinking, full-length ATR and ATR fragments were purified, mixed with azidophenacetyl derivatized and radiolabeled DNA, and exposed to 366-nm light. The reaction products were separated on an SDS/PAGE, which was then analyzed by silver staining (Fig. 3A), and autoradiography (Fig. 3B). We find that full-length ATR is crosslinked to DNA, whereas the ATR fragments are not (Fig. 3B). Thus, we conclude that ATR is a DNA-binding protein, and that either the DNA binding requires amino acids from the entire protein or the fragments we have expressed are not in a conformation conducive to bind DNA.
Figure 3.
UV crosslinking of full-length ATR with DNA. Purified full-length ATR (ATR-F) and its fragments, ATR-N, ATR-M, and ATR-C, were mixed with the azidophenacetyl-derivatized DNA probe, and the reaction mixtures were exposed to 366-nm light on ice. The reaction products were separated by SDS/PAGE and analyzed by (A) silver staining and (B) autoradiography.
Preferential Binding of ATR to UV (6–4) Photoproduct.
An important question in the field of DNA damage checkpoint is the identity of the signal initiating the checkpoint cascade. Although it is generally believed that lesions processed by the replication or repair systems generate a common structure that is recognized by the DNA damage checkpoint pathway, the possibility exists that under certain circumstances the damage per se is bound by the sensor. UV induces two major photoproducts in DNA: cyclobutane pyrimidine dimers, which make up 80–90% of the lesions, and (6–4) photoproducts, which constitute 10–20% of the damage. Cyclobutane pyrimidine dimers, in contrast to (6–4) photoproducts, are poorly recognized by damage sensors involved in DNA repair (27), and we reasoned that the same may be true for the damage sensors in checkpoint response as well. Therefore, we compared the binding of ATR to an oligomer of 46 bp with and without a (6–4) photoproduct by using the pull-down assay. As shown in Fig. 4, ATR has a significantly higher affinity for the oligomer with the (6–4) photoproduct. Considering that a single photoproduct increases the affinity of ATR to the 46-mer 1.6-fold, it is calculated that ATR has about 46 × 1.6≈80-fold higher affinity to a (6–4) photoproduct relative to an undamaged dinucleotide.
Figure 4.
ATR preferentially binds to UV (6–4) photoproduct. HEK293T cells transfected with a vector expressing full-length ATR or no transfected (Mock) cells were lysed and the tagged proteins were purified by binding to FLAG-agarose beads. Proteins bound to beads were incubated with either unmodified (U) 46-mer duplex or (6–4)-photoproduct containing duplex of the same sequence (M). After extensive washing, the material bound to the beads was analyzed by SDS/PAGE followed by silver staining (Top) and autoradiography (Middle). The input lanes contain 1/10th of the DNA used in the binding assay. Bottom shows the mean values and standard errors of DNA binding calculated from three independent experiments.
Analysis of ATR-DNA Complexes by Electron Microscopy.
Solution studies on ATR-DNA interaction were complemented by electron microscopy. Fig. 5A shows representative pictures of ATR-DNA complexes. The protein binds with equal affinity to linear and supercoiled DNA; under our assay conditions, 10% of supercoiled or linear DNA molecules were bound by ATR (Fig. 5A). It appears that ATR is a globular monomeric protein with no special affinity for DNA ends. However, in about half of the micrographs, the protein was seen bound to two DNA segments, indicating that ATR may have two DNA-binding sites or that alternatively two monomers bound to different regions of the DNA molecule bringing these regions into proximity by protein–protein interaction. Obviously, this point deserves further investigation by other methods.
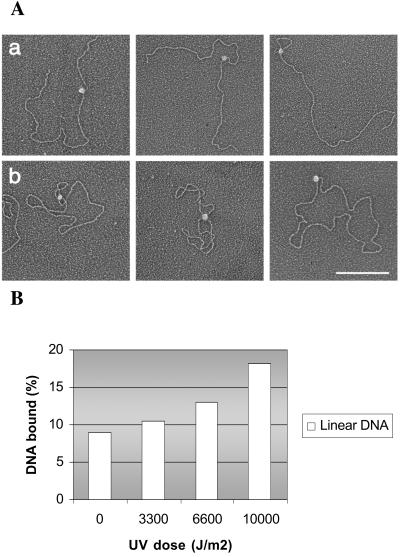
Figure 5.
ATR-DNA binding analyzed by electron microscopy. (A) Visualization of the binding of ATR protein to (a) linearized or (b) supercoiled pGEMEX1 dsDNA. The samples were directly mounted onto thin carbon-coated foils and rotary shadowed with tungsten. The complexes shown in a and b are representatives of the total population of protein-bound DNA molecules observed. (Bar = 250 nm.) (B) Quantitative comparison of ATR DNA binding to unirradiated or UV-irradiated plasmid DNA. The number of protein–DNA complex is given in relation to the number of DNA molecules counted in each reaction.
In addition to providing information regarding the size and the shape of ATR, electron microscopy also enabled us to measure the affinity of ATR to damaged DNA by an independent method. Unirradiated or UV-irradiated plasmid DNA was mixed with ATR, and the fractions of DNA molecules with bound ATR were scored. As seen in Fig. 5B, gradually more ATR is bound to DNA with increasing UV dose. Thus, this experiment, combined with the pull-down assay shown in Fig. 4, strongly suggests that this binding may enable ATR to act as a sensor in the DNA damage checkpoint cascade.
Stimulation of ATR Kinase Activity by DNA Damage.
If specific binding of ATR to DNA damage is an early step in the UV-induced DNA damage checkpoint response, DNA damage might be expected to modulate the kinase activity of ATR. To test this prediction, ATR was incubated with one of its better known substrates, p53, in the presence of undamaged and UV-damaged DNA (Fig. 6A). There is a high basal level of p53 phosphorylation in the absence of DNA, which is stimulated by DNA (28) and to a significantly higher level by UV-damaged DNA (Fig. 6B). Thus, our data are consistent with the notion that ATR is a damage sensor and is directly activated by the primary DNA lesion.
Figure 6.
Phosphorylation of p53 by purified ATR is stimulated by damaged DNA. (A) Kinase reactions were performed with p53 alone (lane 1) or p53 and ATR in the absence of DNA (lane 2), in the presence of untreated DNA (lanes 3–5), or in the presence of UV-treated DNA (lanes 6–8), as indicated. Proteins were separated by SDS/PAGE and visualized by silver staining (Bottom), and phosphorylated p53 was detected by PhosphorImager (Top). (B) Quantitative analysis of ATR kinase activity. The mean values and standard errors were calculated from three independent experiments. The relative kinase activity of ATR is normalized against the highest kinase activity obtained, which was at UV irradiated DNA concentration of 750 nM.
Discussion
In this study, we have investigated the role of ATR as a damage sensor in DNA damage checkpoint response. Our results show that ATR directly recognizes UV damage and, as a result of binding to UV lesions, its kinase activity is stimulated about 2-fold. The discrimination by ATR between damaged and undamaged DNA is modest, and hence it may be asked whether such low selectivity is sufficient to confer a damage sensor function to ATR. Although this is a legitimate concern, it must be pointed out that well characterized damage recognition proteins XPA, RPA, and XPC, which are necessary and sufficient to confer high specificity to human excision nuclease, display a similar preference for DNA as that of ATR (29). It has been suggested that the modest discriminatory power of the damage recognition proteins of the nucleotide excision repair system is amplified by cooperative action of these proteins and by the kinetic proofreading mechanism (30), which results from the differential stability of the excision nuclease subassemblies that form on damaged compared with undamaged DNA. The same kinetic discrimination is known to occur by the damage recognition proteins of Escherichia coli excision nuclease. In fact, modest discriminatory power between target and nontarget sequences is a common theme in eukaryotic DNA acting proteins, and high specificity is achieved by either combinatorial action of multiple factors as often occurs in transcriptional regulation (31), or by a kinetic proofreading mechanism (30) such as the one operative in nucleotide excision repair, where low-specificity ATP-independent damage recognition is amplified by subsequent recruitment of ATPase/helicases that aid in the formation of high-specificity complexes (29). Currently it is unclear whether checkpoint sensors achieve high specificity by a combinatorial mechanism or by a kinetic proofreading mechanism. Regardless of the specifics, our data strongly suggest that ATR can directly bind to DNA and, by virtue of its higher affinity for damaged DNA, it may function as a sensor that recognizes the primary DNA lesion.
The prototype of the PIKK family, the DNA–PKcs, is recruited to DNA ends by the Ku heterodimer, and it has been speculated that other members of this family may also possess Ku-like small subunits with the potential of binding to DNA and recruiting the large cognate PIKK. Indeed, after this work was completed, a paper was published showing that ATR is in association with a 90-kDa protein named ATRIP (32). It appears that ATRIP is important for the stability of ATR in vivo. However, whether ATRIP affects any of the ATR functions is not known. In the ATR preparations used in the present study, we do not see the ATRIP subunit, and hence we can state with relative confidence that the binding properties we observe are those of the ATR polypeptide itself. Furthermore, that UV irradiation crosslinked ATR to DNA strongly indicates that whether or not it is aided by another protein such as ATRIP, ATR is in direct contact with DNA. The effect of ATRIP on the DNA-binding and kinase activities of ATR is currently under investigation.
UV-induced checkpoint response depends on ATR. However, in contrast to ATM, whose kinase activity increases after ionizing radiation, when ATR is purified from UV- and IR-treated cells, it does not exhibit increased kinase activity (1). Nevertheless, it has been found that Xenopus ATR can be separated by DNA affinity chromatography (33), and that DNA stimulates the kinase activity of human ATR (28, 34), suggesting that this protein as well may bind to DNA and function as a sensor. However, these experiments do not give a clue why ATR specifically functions in the UV-induced cell cycle checkpoint. In fact, studies with Xenopus extracts show that ATR associates with chromatin on initiation of replication and dissociates on completion of replication (35), and that the amount of ATR bound to chromatin increases on inhibiting DNA (but not RNA primer) synthesis by aphidicolin. These findings have led to the proposal that ATR binds to the RNA primer or the primer-associated structure to activate the intrinsic S-phase checkpoint as well as the G2/M checkpoint. However, two recent studies in yeast have shown that the counterparts of the ATR–ATRIP complex can also be recruited to an HO-induced double-strand break (36, 37), suggesting that ATR may recognize multiple DNA structures and may not be strictly specific to cell cycle phase or DNA damage to activate the cell cycle checkpoint. In addition to ATR and ATM, the checkpoint Rad proteins have also been implicated in damage sensing. It has been proposed that either the primary or processed DNA lesions are recognized by Rad17-RFC, which loads the 9–1-1 complex onto DNA and in doing so initiates the DNA damage checkpoint response (2, 11) by recruiting and/or activating the ATR kinase to initiate phosphorylation of downstream effectors. One fission yeast study raises some doubts about this model (38), where it was found that the counterpart of ATRIP is phosphorylated by ATR after DNA damage independently of other checkpoint components, including Hus1, Rad1, and Rad9, suggesting that ATR can be directly activated by DNA damage, which might place the 9–1-1 complex downstream from ATR. Surprisingly, recently two studies in yeast and one study in humans have shown that the checkpoint 9–1-1 and ATR-ATRIP complexes or their yeast counterparts can be recruited to chromatin independently (36, 37, 39). It has also been shown that Rad17 is required for the recruitment of Rad9 onto chromatin (39). However, at present there is no direct evidence that Rad17-RFC loads the 9–1-1 complex onto DNA, damaged or undamaged, and interestingly, the S. cerevisiae homologs of human Rad17 and Rad9 are not required for cell cycle checkpoint in S-phase.
In summary, currently available evidence from in vivo studies is insufficient to allow a firm conclusion regarding the roles of various “upstream” checkpoint proteins in damage sensing and whether these proteins sense the primary lesion or a common intermediate resulting from the processing of the various primary lesions. Within the context of these uncertainties, we believe that the data presented in this paper constitute the most compelling evidence to date that in UV-induced checkpoint response in mammalian cells, at least part of the checkpoint activation is initiated by binding of ATR to the UV photoproducts and stimulation of its kinase activity as a consequence of this binding.
Acknowledgments
We are grateful to Robert T. Abraham for gifts of plasmids expressing full-length wild-type and mutant ATR. We thank Thomas Petes and Jerard Hurwitz for critical comments on the manuscript. This work was supported by National Institutes of Health Grants GM32833 (to A.S.) and GM31819 and CA16086 (to J.D.G.).
Abbreviations
- PIKK
phosphoinositide 3-kinase-related kinase
- DNA-PK
DNA-dependent protein kinase
- RFC
Replication Factor C
- HEK
human embryonic kidney
References
- 1.Abraham R T. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 2.Lowndes N F, Murguia J R. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 3.Zhou B B, Elledge S J. Nature (London) 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 4.Shiloh Y. Curr Opin Genet Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 5.Durocher D, Jackson S P. Curr Opin Cell Biol. 2001;13:225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 6.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai K K, Shimoda C, Nojima H. Mol Biol Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green C M, Erdjument-Bromage H, Tempst P, Lowndes N F. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 9.Thelen M P, Venclovas C, Fidelis K. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 10.Caspari T, Dahlen M, Kanter-Smoler G, Lindsay H D, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr A M. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venclovas C, Thelen M P. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volkmer E, Karnitz L M. J Biol Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- 13.St. Onge R P, Udell C M, Casselman R, Davey S. Mol Biol Cell. 1999;10:1985–1995. doi: 10.1091/mbc.10.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hang H, Lieberman H B. Genomics. 2000;65:24–33. doi: 10.1006/geno.2000.6142. [DOI] [PubMed] [Google Scholar]
- 15.Burtelow M A, Roos-Mattjus P M, Rauen M, Babendure J R, Karnitz L M. J Biol Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 16.Lindsey-Boltz L A, Bermudez V P, Hurwitz J, Sancar A. Proc Natl Acad Sci USA. 2001;98:11236–11241. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tibbetts R S, Brumbaugh K M, Williams J M, Sarkaria J N, Cliby W A, Shieh S Y, Taya Y, Prives C, Abraham R T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degtyareva N, Subramanian D, Griffith J D. J Biol Chem. 2001;276:8778–8784. doi: 10.1074/jbc.M006795200. [DOI] [PubMed] [Google Scholar]
- 19.Huang J C, Svoboda D L, Reardon J T, Sancar A. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsunaga T, Park C H, Bessho T, Mu D, Sancar A. J Biol Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- 21.Yang S W, Nash H A. Proc Natl Acad Sci USA. 1994;91:12183–12187. doi: 10.1073/pnas.91.25.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffith J D, Christiansen G. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- 23.Griffith J D, Makhov A, Zawel L, Reinberg D. J Mol Biol. 1995;246:576–584. doi: 10.1016/s0022-2836(05)80107-x. [DOI] [PubMed] [Google Scholar]
- 24.Cimprich K A, Shin T B, Keith C T, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hockensmith J W, Kubasek W L, Vorachek W R, Evertsz E M, von Hippel P H. Methods Enzymol. 1991;208:211–236. doi: 10.1016/0076-6879(91)08015-a. [DOI] [PubMed] [Google Scholar]
- 27.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 28.Hall-Jackson C A, Cross D A, Morrice N, Smythe C. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- 29.Wakasugi M, Sancar A. J Biol Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 30.Hopfield J J. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolberger C. Annu Rev Biophys Biomol Struct. 1999;28:29–56. doi: 10.1146/annurev.biophys.28.1.29. [DOI] [PubMed] [Google Scholar]
- 32.Cortez D, Guntuku S, Qin J, Elledge S J. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 33.Guo Z, Kumagai A, Wang S X, Dunphy W G. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lakin N D, Hann B C, Jackson S P. Oncogene. 1999;18:3989–3995. doi: 10.1038/sj.onc.1202973. [DOI] [PubMed] [Google Scholar]
- 35.Hekmat-Nejad M, You Z, Yee M C, Newport J W, Cimprich K A. Curr Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- 36.Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- 37.Melo J A, Cohen J, Toczyski D P. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards R J, Bentley N J, Carr A M. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 39.Zou L, Cortez D, Elledge S J. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]