Abstract
Junctional adhesion molecules (JAMs) are a family of immunoglobulin-like single-span transmembrane molecules that are expressed in endothelial cells, epithelial cells, leukocytes and myocardia. JAM has been suggested to contribute to the adhesive function of tight junctions and to regulate leukocyte trans migration. We describe the crystal structure of the recombinant extracellular part of mouse JAM (rsJAM) at 2.5 Å resolution. rsJAM consists of two immunoglobulin-like domains that are connected by a conformationally restrained short linker. Two rsJAM molecules form a U-shaped dimer with highly complementary interactions between the N-terminal domains. Two salt bridges are formed in a complementary manner by a novel dimerization motif, R(V,I,L)E, which is essential for the formation of rsJAM dimers in solution and common to the known members of the JAM family. Based on the crystal packing and studies with mutant rsJAM, we propose a model for homophilic adhesion of JAM. In this model, U-shaped JAM dimers are oriented in cis on the cell surface and form a two-dimensional network by trans-interactions of their N-terminal domains with JAM dimers from an opposite cell surface.
Keywords: homophilic adhesion/immunoglobulin superfamily/junctional adhesion molecule/protein crystallography/tight junction
Introduction
Tight junctions (TJs) are electron-dense structures connecting the lateral membranes of adjacent epithelial or endothelial cells. They exert adhesive properties and stabilize homophilic cell–cell binding. TJs serve a dual role in controlling paracellular permeability and in maintaining cell polarity. These junctional structures are particularly well developed in regions of the vascular tree where permeability has to be restricted, e.g. in the brain microvasculature and in large arteries (Mitic and Anderson, 1998; Stevenson and Keon, 1998; Dejana et al., 2000). Little is known about the molecular basis for the intercellular adhesion of TJs, despite their eminent role in organ functioning. Different transmembrane proteins have been found to be located specifically at TJs. Occludin and the claudin family belong to the class of tetra-span transmembrane proteins (Furuse et al., 1993, 1998a,b). Occludin is dispensable for TJ organization and adhesive properties (Saitou et al., 1998). In contrast, claudins promote TJ assembly when transfected in fibroblastoid cells (Furuse et al., 1998a,b), but their mode of action in intercellular adhesion remains to be elucidated. No structural information is yet available for any representative of the claudin or occludin families. Junctional adhesion molecule (JAM) was identified recently as a single-span transmembrane protein belonging to the immunoglobulin superfamily (Martìn-Padura et al., 1998) and to the CTX family of molecules that lie at the crossroads between antigen-specific receptors and adhesion molecules (Chrétien et al., 1998). Immunohistochemistry shows that JAM is located at TJs in both epithelial and endothelial cells and, to a lesser degree, on the surface of leukocytes. JAM is associated with TJ-specific cytoskeletal proteins such as cingulin and ZO-1 (Bazzoni et al., 2000a). JAM transfection induces homophilic recognition and decreases paracellular permeability in fibroblastoid cells. Antibody studies suggest that the extracellular domain of JAM is involved in the migration of leukocytes through endothelial junctions (Martìn-Padura et al., 1998). In addition, JAM has been identified as the carbohydrate-independent receptor for reovirus infection leading to viral endocytosis (Barton et al., 2001). Recently, two new members of the JAM family have been described under the names VE-JAM/JAM2/JAM-3 (Aurrand-Lions et al., 2000; Cunningham et al., 2000; Palmeri et al., 2000) and JAM-2 (Aurrand-Lions et al., 2001). Both molecules are expressed by endothelial cells and show a more restricted expression pattern than the prototype JAM. We studied the aggregation behaviour of the recombinant soluble form of JAM (rsJAM) lacking the transmembrane domain and the intracellular part. In solution, rsJAM assembles non-covalently to dimers and to a small percentage of tetramers (Bazzoni et al., 2000b). Here we describe the crystal structure of rsJAM at 2.5 Å resolution and identify a novel structural motif, R(V,I,L)E, at the dimerization interface of JAM.
Results
Structure of the monomer
The structure of rsJAM is shown in Figure 1. It consists of two immunoglobulin-like domains of the variable type (Bork et al., 1994). The c′–c′′ hairpin of the C-terminal domain is disordered. A short linker (Val127–Leu128– Val129) connects the two rsJAM domains in a rather extended conformation, leading to an estimated elbow angle of ∼125°. The transmembrane and intracellular parts of JAM, absent in rsJAM, would protrude from the C-terminus. The crystal structure of rsJAM confirms the prediction that JAM belongs to the immunoglobulin superfamily (Martìn-Padura et al., 1998). The main chain atoms of the linker residues are involved in an extensive hydrogen bond network to both domains, and the side chain of the central linker residue, Leu128, lies in a hydrophobic pocket at the interface between the two domains (Figure 2). The loops involved in domain contacts are stabilized by several vicinal proline residues (Pro130, 131, and Pro159, 160). All these interactions should stabilize the conformation of the short linker. Therefore, we expect that the elbow angle in JAM is similar to that in rsJAM.
Fig. 1. Schematic picture of the structure of rsJAM. The N- and C-terminal domains are coloured in cyan and orange, and the short linker between the two domains is coloured in green. Both domains have an immunoglobulin-like fold of the variable type. The β-strands are labelled according to the immunoglobulin convention. The disordered c′–c′′ hairpin in the C-terminal domain (Ala175–Asp176–Ala177–Lys178–Lys179) is indicated by a dashed line. Both disulfide bridges (Cys49–Cys108 and Cys152–C212) are shown in a yellow stick representation.
Fig. 2. Stereo view of the linker region Val127–Leu128–Val129 (green) between the N- (cyan) and C-terminal domain (orange) in a ball-and-stick representation. Oxygen and nitrogen atoms are coloured red and blue, respectively. The extensive hydrogen bond network between the main chain atoms of the linker tri-peptide and both domains is shown with black dotted lines. The side chain of Leu128 is tightly packed in a hydrophobic pocket formed by the side chains of Gln38, Pro40, Thr126, Pro159 and Tyr218. Several proline residues (Pro40, Pro130, Pro131, Pro159 and Pro160) stabilize the main chain conformation around the linker.
Structure of the dimer and the R(V,I,L)E motif
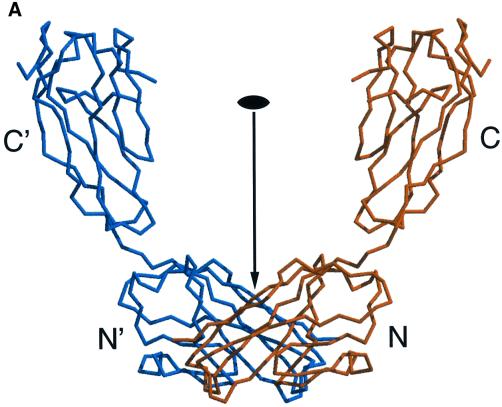
In solution, rsJAM forms ∼90% dimers and ∼10% tetramers (Bazzoni et al., 2000b). The corresponding intermolecular contacts are likely to be found in the crystal structure, in particular that of the dimeric species. We observe three extended contact areas (called interfaces I1, I2 and I3) across crystallographic 2-fold axes with contact areas of 820, 710 and 610 Å2 per monomer, respectively. In I1 and I2, contacts are formed between two C-terminal domains, i.e. between the extracellular domains of JAM that are closest to the cell membrane. In I3, contacts are formed between two N-terminal domains, i.e. between the extracellular domains of JAM that would stick out farthest from the cell membrane. When the number of atomic contacts with a distance cut-off of 3.6 Å is taken into account, I2 has significantly fewer contacts than the other two interfaces, and we regard it as a typical crystal packing interface. Although I1 is larger than I3, both include ∼250 Å2 of non-polar contact area. The larger area of I1 is due mainly to mixed atomic contacts (polar/non-polar), which could even make an unfavourable contribution to the association energy. I3 has some outstanding structural features (Figure 3). At its centre we observe two symmetry-equivalent stacked salt bridges, each formed by Arg58 from one monomer and Glu60 from the other monomer (Figure 3B). On either side of these two salt bridges is a hydrophobic stacking interaction with the aromatic side chain of Tyr74, which in turn packs against the peptide plane between Gly114 and Gly115 of the other monomer (Figure 3B). The central tri-peptide Arg58–Val59–Glu60 is located in the c-strand of the N-terminal domain (Figure 1); we call it the R(V,I,L)E motif. Tyr74 is located in the c′–c′′ hairpin, and the Gly114–Gly115 bi-peptide is located in the f–g turn. The precise stereochemical complementarity of the interactions at I3 makes us believe that it is this interface that is responsible for dimer formation in solution. The following observations support this hypothesis.
Fig. 3. (A) U-shaped dimer of rsJAM shown in a Cα-representation. The two monomers are coloured in orange and blue, and the N- and C-terminal domains are labelled with N, N′, C and C′, respectively. The two monomers are related by a crystallographic dyad, which is indicated by an arrow and a dyad symbol. The two N-terminal domains contact each other via the I3 interface (see text). (B) Stereo picture of the dimer interface I3 viewed along the crystallographic dyad (indicated by a dyad symbol). For clarity, only the N-terminal domains of the two monomers are shown as a Cα-representation, using the same colour code as in (A). The N-termini are labelled with N and N′, respectively. The side chains of Arg58, Glu60 from the R(V,I,L)E motif, Tyr74 and the amino acids Gly114–Gly115 are shown in a ball-and-stick representation and are labelled accordingly. The two central salt bridges between Arg58 and Glu60′ and vice versa are indicated with black dotted lines. Each salt bridge is sandwiched by the other salt bridge on one side and by the aromatic side chain of a Tyr74 on the other side. Each Tyr74 side chain, in turn, is sandwiched by the salt bridge on one side and by the peptide bond between Gly114′ and Gly115′ on the other side.
(i) Below pH ∼5, rsJAM dissociates to monomers (Bazzoni et al., 2000b). This is consistent with the expected breakdown of the stabilizing salt bridges between Arg58 of one monomer and Glu60 of the other monomer at pH values around the pKa value for the side chain carboxylate group of Glu60.
(ii) Under non-reducing conditions, the concentration of dimers decreases slowly with time and can be restored by addition of 1 mM β-mercaptoethanol (Bazzoni et al., 2000b). The only apparent oxidation-sensitive amino acid in rsJAM is Cys73. Because Cys73 is buried in a hydrophobic environment on the back of Tyr74 and Glu60, any oxidation of its thiol group would result in a distortion of the dimer interface.
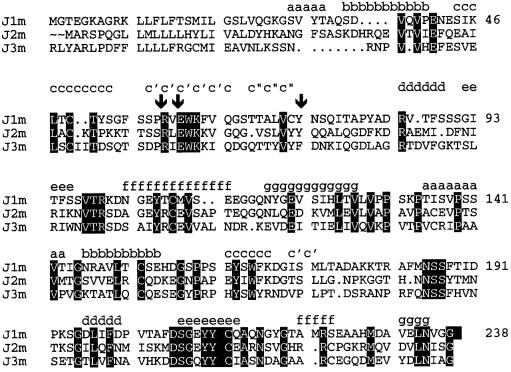
(iii) The R(V,I,L)E motif as well as the flanking residues Trp61, Lys62, Cys73 and Tyr74 are conserved in the published sequences of JAM of mouse, human and bovine origin (Ozaki et al., 1999). Moreover, the sequence R(V,I,L)E and its structurally flanking Y/F is present in two new homologues of JAM, which we call here JAM2 and JAM3 (Figure 4). JAM2 has recently been described in heart and placenta (Palmeri et al., 2000) and as VE-JAM in high endothelial venules (Cunningham et al., 2000). JAM3 has recently been described as JAM-2 in high endothelial cells in lymph node and kidney (Aurrand-Lions et al., 2001). The high sequence conservation of the R(V,I,L)E motif and of its structurally neighbouring residues at interface I3 makes it likely that the known members of the JAM family and of their homologues form homodimers via similar interfaces.
Fig. 4. Sequence alignment of rsJAM (J1m) with JAM2 (J2m) and JAM3 (J3m) (see text). The positions of Arg58, Glu60 from the R(V,I,L)E motif, and of Tyr74 are marked with arrows. The β-strands in rsJAM are indicated according to Bork (1994).
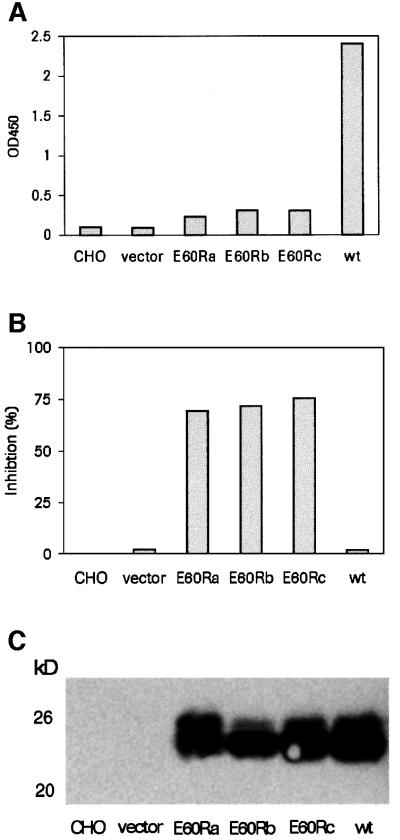
(iv) In order to prove experimentally the significance of the R(V,I,L)E sequence motif for dimer formation, we have constructed a Glu60Arg point mutant of rsJAM. In this mutant protein, we expected that dimerization should be blocked both by charge repulsion and by steric hindrance of the opposing residues: Arg58 from one monomer and Glu60Arg from the other monomer. Indeed, the Glu60Arg mutant protein did not show the presence of aggregates when the supernatant of transfected cells was analysed by a single epitope sandwich immunoassay (SESIA), which is specific for homotypic aggregates (Figure 5A). Since the lack of antibody binding could also have been the result of a misfolded Glu60Arg mutant protein, an additional SESIA experiment was performed in which the mutant protein competed with the wild-type protein for monoclonal antibody binding. The Glu60Arg mutant protein clearly inhibited binding to the wild-type protein (Figure 5B), strongly indicating a proper folding of the mutant protein. The same results were obtained with the equivalent Glu60Arg mutation in human rsJAM (data not shown). However, at a higher concentration of purified mouse Glu60Arg mutant rsJAM protein, 13% dimers were found when analysed by analytical ultracentrifugation. This indicates a second I3-independent dimerization mode, via either I1 or I2, or another site that is not obvious from the crystal packing.
Fig. 5. (A) SESIA results with culture media from transiently transfected CHO cells. Three independent transfections with the rsJAM Glu60Arg mutant (E60Ra, E60Rb and E60Rc) were compared with wild-type rsJAM (wt) and control cell medium. Only the wild-type rsJAM protein indicates the presence of homotypic aggregates. (B) Competitive SESIA experiment in which the same culture media were tested for the ability to inhibit antibody binding to the wild-type protein when added to wild-type rsJAM culture medium. The three E60R mutant proteins inhibit antibody binding to wild-type rsJAM. (C) Western blot detection of rsJAM with monoclonal antibody BV12 in transfected cell supernatant.
Discussion
A model for homophilic adhesion
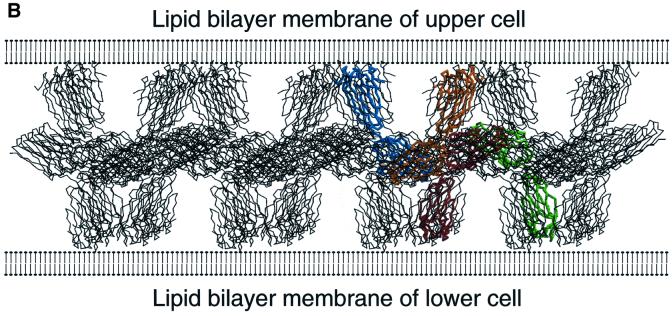
We examined whether the crystal packing contains networks of molecules that could serve as plausible models for homophilic adhesion of JAM. The presumed cooperative nature of such a network makes it likely that weak molecular contacts (as they occur in crystals) would be sufficient for their formation. We are aware that any such model is rather speculative at this point. In the case of the cadherins, involved in homophilic cell–cell recognition and adhesion (Takeichi, 1990), a molecular model for homophilic adhesion (‘cell-adhesion zipper’) was proposed based on crystal contacts in three different crystal forms of the N-terminal domain of N-cadherin (Shapiro et al., 1995). However, subsequent crystallographic studies on N-terminal two-domain fragments of N-cadherin (Tamura et al., 1998) and E-cadherin (Nagar et al., 1996; Pertz et al., 1999) could not confirm this model. On the other hand, such a model makes specific predictions that can be tested experimentally. In selecting a plausible network for homophilic adhesion, we made the assumption that the U-shaped dimer shown in Figure 3A is a cis-dimer (both subunits anchored in the same membrane) with the R(V,I,L)E motif at the dimer interface. In the crystal packing, there is a pair of cis-dimers contacting each other at their N-terminal domains such that their C-terminal domains point pair-wise in opposite directions (Figure 6A). This would then represent a trans-interaction (both subunits anchored in opposite membranes). If we repeat this structural motif of two dimers over several neighbouring unit cells, we obtain a two-dimensional molecular network in which the N-terminal domains contact each other in a common central plane. The C-terminal domains stick out almost perpendicular on either side of that plane, as if they were emanating from opposing cell surfaces (Figure 6B). In the full-length JAM, each C-terminal domain of rsJAM would then be connected to the transmembrane domain located in the cell surface. Parallel channels with diameters of ∼30–35 Å run between the C-terminal domains of adjacent cis-dimers.
Fig. 6. (A) Crystallographic complex of two U-shaped dimers, shown in a Cα-representation. One dimer is coloured in orange and blue in the same orientation as shown in Figure 3A. Its N-terminal domains are labelled N1 and N1′. The other dimer is coloured in red and green. Its N-terminal domains are labelled N2 and N2′. (B) Schematic picture of a model for homophilic adhesion of JAM. The two-dimensional network is constructed by repeating the structural motif of the two dimers shown in (A) over several neighbouring unit cells. The assumed surfaces of two opposing cells are drawn schematically as lipid bilayers. In this model, the N-terminal domains of the U-shaped dimers lie almost parallel to the cell surfaces and contact each other in a common central plane. The C-terminal domains stick out almost perpendicular from the cell surfaces. The predicted distance between two assumed opposing cell surfaces is ∼105–110 Å, which is comparable with the estimated distance of ∼100 Å from electron micrographs of TJs.
This model for homophilic adhesion of JAM shows some attractive features. (i) The highly complementary dimer interface at the N-terminal domains makes it likely that formation of cis-dimers occurs prior to any adhesive trans-interaction. This assumption is supported by our finding that rsJAM, but not mutant rsJAM, assembles during or shortly after the passage through the secretory pathway of Chinese hamster ovary (CHO) cells (Figure 5A). This would be analogous to the ECADCOMP model system for homophilic adhesion of E-cadherins (Pertz et al., 1999), in which cis-dimers are precursors for trans-interactions.
(ii) The C-terminal domains of rsJAM are positioned close to the assumed cell membrane and thus to the transmembrane domain of JAM. They protrude almost perpendicular from the assumed cell surfaces, optimally presenting the contact areas in the N-terminal domains for trans-interactions. (iii) The predicted distance between two assumed cell surfaces would be ∼105–110 Å. This is of the same order of magnitude as our estimated distance of ∼100 Å between opposing cell surfaces observed in electron micrographs of TJs (see Figure 5A in Bolton et al., 1998).
Clearly, more data are needed to define the role of JAM in the context of the adhesion complex and of the dynamic behaviour of TJs. The possibility of heterodimerization of JAM homologues via the R(V,I,L)E motif adds to the expected complexity of the adhesive JAM interactions.
Materials and methods
Protein production, purification and crystallization
The rsJAM form was expressed in insect cells and purified as described (Bazzoni et al., 2000b). The homogeneous protein was analysed using dynamic light scattering to determine its suitability for crystallization (Zulauf and D’Arcy, 1992). At pH 7.0, the protein was monodisperse, and the estimated mol. wt was 51 kDa, corresponding to a dimer. The protein was concentrated to 10–15 mg/ml for crystallization trials with a sparse matrix screen (Jancarik and Kim, 1991) using the vapour diffusion method (McPherson, 1982). We found crystals in 16 out of 48 conditions. The most promising crystals were observed in drops mixed from 3 µl of protein solution and 3 µl of reservoir solution containing 25% polyethylene glycol (PEG) 3350, 200 mM MgCl2 and 100 mM Tris pH 8.5. Although the crystals were large and well formed, they diffracted only to ∼8 Å resolution. Transfer of crystals into the same buffer, but with an increased PEG concentration of 40%, improved the diffraction to 2.5 Å resolution. A heavy atom derivative was obtained by soaking crystals overnight in this solution additionally containing 30 mM K2PtCl4.
Data collection
The native data set was collected on a HISTAR multi-wire area detector using CuKα radiation produced by an Enraf-Nonius FR571 rotating anode generator. Rotation images were taken every 0.2° in two runs with orthogonal orientations of the capillary to increase the data completeness. The K2PtCl4 heavy atom derivative data set was collected on a 30 cm MAR imaging plate using CuKα radiation produced by an Enraf-Nonius FR591 rotating anode generator. Rotation images were taken every 1.0°. The crystals were cryo-cooled at 120 K using the Oxford Cryosystem. The high concentration of PEG was sufficient to act as a cryo-protectant. The diffraction data were processed with the XDS suite (Kabsch, 1988). Data statistics are given in Table I.
Table I. Crystallographic data and refinement statistics.
| Native data set | K2PtCl4 derivative data setg | |
|---|---|---|
| Space group | I222 (No. 23) | I222 (No. 23) |
| Cell axes a, b, c | 42.6Å, 83.3 Å, 130.7 Å | 42.8Å, 83.6 Å, 131.0 Å |
| Resolution range | 25.0–2.5 Å | 35.0–2.65 Å |
| No. of observed reflections | 18 966 | 18 004 |
| No. of unique reflections | 7497 | 6930 |
| Rsym overall/outer shella | 5.4%/13.0% (2.6–2.5 Å) | 7.6%/28.0% (2.7–2.65 Å) |
| I/σ(I) overall/outer shell | 14.4/3.2 | 12.3/3.2 |
| Completeness overall/outer shell | 89.5%/68.3% | 95.9%/92.6% |
| Phasing power (centric/acentric)b | 1.29/1.85 | |
| RCullis (centric/acentric)c | 0.73/0.67 | |
| Figure-of-merit (centric/acentric)d | 0.35/0.39 | |
| No. of refined non-hydrogen atoms | ||
| protein | 1566 | |
| Mg2+ | 1 | |
| water | 70 | |
| R-factor/free R-factore | 0.15/0.21 | |
| R.m.s.d. bond lengths/bond anglesf | 0.005 Å/1.3° |
aRsym = Σh Σi|Ii(h) – <I(h)>|/Σh Σi Ii(h), where Ii(h) and <I(h)> are the ith and mean measurement of the intensity of reflection h.
bPhasing power =ΣFH/Σ|FPH – |FP + FH||, where FP + FH is the vector sum of the protein structure factor FP and the heavy atom structure factor FH; FPH is the heavy atom derivative structure factor amplitude.
cRCullis = Σ|FPH – |FP + FH||/Σ|FPH – FP|, where FP + FH is the vector sum of the protein structure factor FP and the heavy atom structure factor FH; FPH is the heavy atom derivative structure factor amplitude; FP is the protein structure factor amplitude.
dFigure-of-merit = |∫>α P(α)exp(iα)dα/∫α P(α)dα|, with α ranging from 0 to 2π.
eR = Σ|FC – FP|/Σ FP, where FC is the calculated structure factor amplitude for the refined model; FP is the observed structure factor amplitude; the free R-factor is the R-factor for a 5% test data set that was excluded from the refinement; neither a resolution cut-off nor an amplitude cut-off was applied to the data.
fR.m.s.d., root-mean-square deviation from the Engh and Huber parameter set.
gPlatinum-binding sites were at Met109:Sδ, His124:Nε2, Met172:Sδ and Met222:Sδ.
Structure determination
All computer programs used are part of the CCP4 program suite (CCP4, 1994), except where indicated. The structure was solved by single isomorphous replacement with anomalous scattering (SIRAS). The ambiguity in the choice of the correct space group, either I222 (No. 23) or I212121 (No. 24), was resolved by a single clear platinum position explaining the strongest difference Patterson function peaks using the program SHELXS (Sheldrick et al., 1993) for space group I222, but not for space group I212121. Heavy atom refinement and phasing were performed with the program SHARP (De la Fortelle and Bricogne, 1997). During heavy atom refinement, three additional platinum sites were identified. The resulting electron density was modified with the program SOLOMON (Abrahams and Leslie, 1996). An initial 83% complete protein model could be built and real-space refined using the computer graphics modelling program MOLOC (Gerber, 1992).
Protein refinement
Initial protein refinement was performed with the maximum-likelihood refinement program REFMAC (Murshodov et al., 1997) using the phase probability distribution from the heavy atom phasing. Final refinement was performed with the maximum-likelihood refinement program X-PLOR 98.0 (Brünger et al., 1987) using the Engh and Huber (1991) parameters for ideal stereochemistry and an appropriate bulk solvent correction (Jiang and Bruenger, 1994; Kostrewa, 1997). Refinement statistics are given in Table I. The refined model consists of amino acid residues 27–174 and 180–238, one Mg2+ and 70 water molecules. The Mg2+ has bound at the protein surface to His155:Nε2 and to five water molecules involved in hydrogen bonds to a neighbouring protein molecule in the crystal. Presumably, the bound Mg2+ results from the crystallization condition rather than from a structural or functional role. The missing residues Ala175–Asp176–Ala177–Lys178–Lys179 are part of a surface loop. The N-terminal residue Lys27 is the first amino acid after the main signal peptide cleavage site. The stereochemical quality of the refined structure was checked with the programs PROCHECK (Laskowski et al., 1993) and WHAT_CHECK (Hooft et al., 1996). There are no amino acids in disallowed regions of the Ramachandran plot (Ramachandran et al., 1963). All figures were prepared with the programs MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Bacon, 1997).
Cloning and analysis of the mutant rsJAM
The Glu60Arg mutation was introduced into rsJAM-pcDNA3 and expressed by transient transfection into CHO cells. Culture medium of three independent transfections (E60Ra, E60Rb and E60Rc) was compared with supernatant of non-transfected cells, of cells transfected with the empty vector, and of wild-type rsJAM. An SESIA, recognizing only aggregated rsJAM, was performed as described (Bazzoni et al., 2000b). Briefly, the anti-JAM monoclonal antibody BV12 was used in a sandwich immunoassay for antigen capture and simultaneously for detection of rsJAM in cell supernatant (Figure 5A). Expression of mutant rsJAM in cell supernatant was confirmed by the inhibition of the SESIA signal (Figure 5B) by western blot detection with BV12 (Figure 5C). For large-scale production, the mutant rsJAM was expressed in insect cells and purified as described for rsJAM (Bazzoni et al., 2000b). Equilibrium sedimentation centrifugation was performed and analysed as described (Bazzoni et al., 2000b).
Coordinates
The atomic coordinates of the refined structure of rsJAM have been deposited with the Protein Data Bank (Bernstein et al., 1977) with the entry code 1F97.
Acknowledgments
Acknowledgements
We thank Louis Du Pasquier for his helpful comments, Christine Kocyba for constructing the mutant protein, and Emil Kuznir for operating the analytical centrifuge.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Aurrand-Lions M., Duncan,L., DuPasqier,L. and Imhof,B.A. (2000) Cloning of JAM-2 and JAM-3: an emerging junctional adhesion molecular family? Curr. Top. Microbiol. Immunol., 251, 91–98. [DOI] [PubMed] [Google Scholar]
- Aurrand-Lions M., Duncan,L., Ballestrem,C. and Imhof,B.A. (2001) JAM-2, a novel immunoglobulin superfamily molecule, expressed by endothelial and lymphatic cells. J. Biol. Chem., 276, 2733–2741. [DOI] [PubMed] [Google Scholar]
- Barton E.S., Forrest,J.C., Connolly,J.L., Chappell,J.D., Yuan,L., Schnell, F.J., Nusrat,A., Parkos,C.A. and Dermody,T.S. (2001) Junction adhesion molecule is a receptor for reovirus. Cell, 104, 441–451. [DOI] [PubMed] [Google Scholar]
- Bazzoni G., Martinez-Estrada,O.M., Orsenigo,F., Cordenonsi,M., Citi,S. and Dejana,E. (2000a) Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin and occludin. J. Biol. Chem., 275, 20520–20526. [DOI] [PubMed] [Google Scholar]
- Bazzoni G., Martinez-Estrada,O.M., Mueller,F., Nelboeck,P., Schmid, G., Bartfai,T., Dejana,E. and Brockhaus,M. (2000b) Homophilic interaction of junctional adhesion molecule. J. Biol. Chem., 275, 30970–30976. [DOI] [PubMed] [Google Scholar]
- Bernstein F.C., Koetzle,T.F., Williams,G.J.B., Meyers,E.F., Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The Protein Data Bank: a computer based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- Bolton S.J., Anthony,D.C. and Perry,V.H. (1998) Loss of the tight junction proteins occludin and zonula occludens-1 from cerebral vascular endothelium during neutrophil-induced blood–brain barrier breakdown in vivo. Neuroscience, 86, 1245–1257. [DOI] [PubMed] [Google Scholar]
- Bork P., Holm,L. and Sander,C. (1994) The immunoglobulin fold: structural classification, sequence patterns and common core. J. Mol. Biol., 242, 309–320. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Kuriyan,J. and Karplus,M. (1987) Crystallographic R factor refinement by molecular dynamics. Science, 235, 458–460. [DOI] [PubMed] [Google Scholar]
- Chrétien I., Marcuz,A., Courtet,M., Katevuo,K., Vainio,O., Heath,J.K., White,S.J. and Du Pasquier,L. (1998) CTX, a Xenopus thymocyte receptor, defines a molecular family conserved throughout vertebrates. Eur. J. Immunol., 28, 4094–4104. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cunningham S.A., Arrate,M.P., Rodriguez,J.M., Bjercke,R.J., Vanderslice,P., Morris,A.P. and Brock,T.A. (2000) A novel protein with homology to the junctional adhesion molecule. J. Biol. Chem., 275, 34750–34756. [DOI] [PubMed] [Google Scholar]
- Dejana E., Lampugnani,M.G., Martinez-Estrada,O. and Bazzoni,G. (2000) The molecular organization of endothelial junctions and their functional role in vascular morphogenesis and permeability. Int. J. Dev. Biol., 44, 743–748. [PubMed] [Google Scholar]
- De la Fortelle E. and Bricogne,G. (1997) Maximum likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Engh R.A. and Huber,R. (1991) Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A, 47, 392–400. [Google Scholar]
- Furuse M., Hirase,T., Itoh,M., Nagafuchi,A., Yonemura,S., Tsukita,S. and Tsukita,S. (1993) Occludin: a novel integral membrane protein localizing at tight junctions. J. Cell Biol., 123, 1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Fujita,K., Hiiragi,T., Fujimoto,K. and Tsukita,S. (1998a) Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol., 141, 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Sasaki,H., Fujimoto,K. and Tsukita,S. (1998b) A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol., 143, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber P.R. (1992) Peptide mechanics: a force field for peptides and proteins working with entire residues as small units. Biopolymers, 32, 1003–1017. [Google Scholar]
- Hooft R.W.W., Vriend,G., Sander,C. and Abola,E.E. (1996) Errors in protein structures. Nature, 381, 272. [DOI] [PubMed] [Google Scholar]
- Jancarik J. and Kim,S.-H. (1991) Sparse matrix sampling: a screening method for crystallization of proteins. J. Appl. Crystallogr., 24, 409–411. [Google Scholar]
- Jiang J.-S. and Bruenger,A.T. (1994) Protein hydration observed by X-ray diffraction. J. Mol. Biol., 243, 100–115. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1988) Evaluation of single crystal X-ray diffraction data from a position sensitive detector. J. Appl. Crystallogr., 21, 916–924. [Google Scholar]
- Kostrewa D. (1997) Bulk solvent correction: practical application and effects in real and reciprocal space. Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr., 34, 9–22. [Google Scholar]
- Kraulis, P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Martìn-Padura I. et al. (1998) Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol., 142, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A. (1982) Preparation and Analysis of Protein Crystals. John Wiley & Sons, New York, NY.
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505. [DOI] [PubMed] [Google Scholar]
- Mitic L.L. and Anderson,J.M. (1998) Molecular architecture of tight junctions. Annu. Rev. Physiol., 60, 121–142. [DOI] [PubMed] [Google Scholar]
- Murshodov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Nagar B., Overduin,M., Ikura,M. and Rini,J.M. (1996) Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature, 380, 360–364. [DOI] [PubMed] [Google Scholar]
- Ozaki H., Ishii,K., Horiuchi,H., Arai,H., Kawamoto,T., Okawa,K., Iwamatsu,A. and Kita,T. (1999) Cutting edge: combined treatment of TNF-α and IFN-γ causes redistribution of junctional adhesion molecule in human endothelial cells. J. Immunol., 163, 553–557. [PubMed] [Google Scholar]
- Palmeri D., van Zante,A., Chiao-Chain,H., Hemmerich,S. and Rosen, S.D. (2000) Vascular endothelial junction-associated molecule, a novel member of the immunoglobulin superfamily, is localized to intercellular boundaries of endothelial cells. J. Biol. Chem., 275, 19139–19145. [DOI] [PubMed] [Google Scholar]
- Pertz O., Bozic,D., Koch,A.W., Fauser,C., Brancaccio,A. and Engel,J. (1999) A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J., 18, 1738–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran G.N., Ramakrishnan,C. and Sasisekharan,V. (1963) Stereochemistry of polypeptide chain configurations. J. Mol. Biol., 7, 95–99. [DOI] [PubMed] [Google Scholar]
- Saitou M., Fujimoto,K., Doi,Y., Itoh,M., Fujimoto,T., Furuse,M., Takano,H., Noda,T. and Tsukita,S. (1998) Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions J. Cell Biol., 141, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L. et al. (1995) Structural basis of cell–cell adhesion by cadherins. Nature, 374, 327–337. [DOI] [PubMed] [Google Scholar]
- Sheldrick G.M., Dauter,Z., Wilson,K.S., Hope,H. and Sieker,L.C. (1993) The application of direct methods and Patterson interpretation to high resolution native protein data. Acta Crystallogr. D, 49, 18–23. [DOI] [PubMed] [Google Scholar]
- Stevenson B.R. and Keon,B.H. (1998) The tight junction: morphology to molecules. Annu. Rev. Cell Dev. Biol., 14, 89–109. [DOI] [PubMed] [Google Scholar]
- Takeichi M. (1990) Cadherins: a molecular family important in selective cell–cell adhesion. Annu. Rev. Biochem., 59, 237–252. [DOI] [PubMed] [Google Scholar]
- Tamura K., Shan,W.S., Hendrickson,W.A., Colman,D.R. and Shapiro,L. (1998) Structure–function analysis of cell adhesion by neural (N-) cadherin. Neuron, 20, 1153–1163. [DOI] [PubMed] [Google Scholar]
- Zulauf M. and D’Arcy,A. (1992) Light scattering of proteins as a criterion for crystallisation. J. Crystal Growth, 122, 102–106. [Google Scholar]