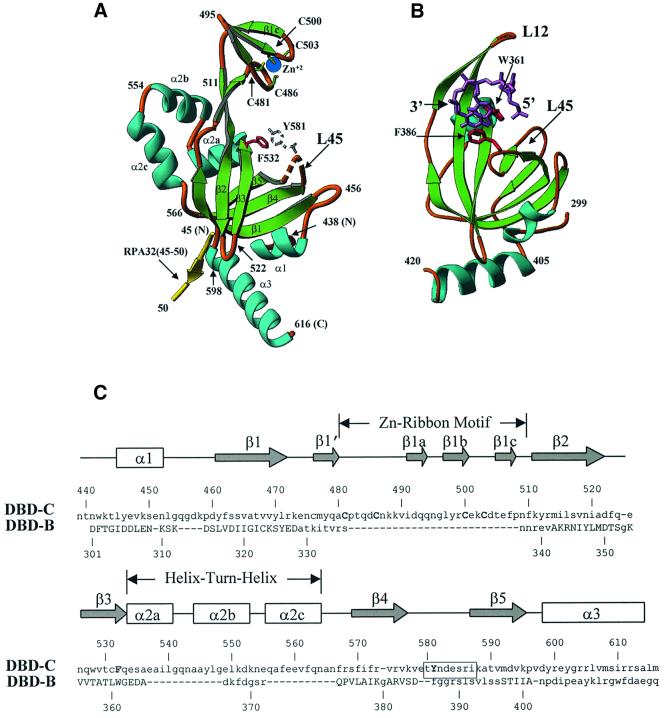
Fig. 2. Similarities and differences between DBD-C and DBD-B. (A) A detailed view of DBD-C colored in green (β-strands), sky-blue (α-helices) and gold (loops); a fragment of RPA32 (amino acids 45–50) is shown in yellow. Amino acid numbers are shown in black. A disordered loop is indicated by the dashed line. Conserved aromatic amino acids (the position of Y581 is putative) and the four cysteines that coordinate zinc ion are shown in red, colourless, green-and-yellow and blue, respectively. (B) The structure of DBD-B and the mode of its interaction with ssDNA. DBD-B is colored as DBD-C in (A) and has an orientation approximately the same as DBD-C. ssDNA is colored magenta (Protein Data Bank entry 1jmc). (C) Structure-based sequence alignment of DBD-C and DBD-B. The β-strands are indicated by arrows and the α-helices by boxes. Secondary structure elements are referred to as in agreement with the nomenclature of the OB-fold. The positions of the zinc ribbon and helix–turn–helix are outlined. The sequences of DBD-C and DBD-B were aligned using program O based on the three-dimensional structure; the residues that are capitalized in DBD-B indicate those that were included in the alignment (r.m.s.d. is 1.82 Å for 69 Cα atoms). Functionally important amino acids of DBD-C, four cysteines and two conserved aromatics, are capitalized. The boxed residues are those that are disordered in the structure.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.