Abstract
During activation of the spliceosome, the U4/U6 snRNA duplex is dissociated, releasing U6 for subsequent base pairing with U2 snRNA. Proteins that directly bind the U4/U6 interaction domain potentially could mediate these structural changes. We thus investigated binding of the human U4/U6-specific proteins, 15.5K, 61K and the 20/60/90K protein complex, to U4/U6 snRNA in vitro. We demonstrate that protein 15.5K is a nucleation factor for U4/U6 snRNP assembly, mediating the interaction of 61K and 20/60/90K with U4/U6 snRNA. A similar hierarchical assembly pathway is observed for the U4atac/U6atac snRNP. In addition, we show that protein 61K directly contacts the 5′ portion of U4 snRNA via a novel RNA-binding domain. Furthermore, the 20/60/90K heteromer requires stem II but not stem I of the U4/U6 duplex for binding, and this interaction involves a direct contact between protein 90K and U6. This uneven clustering of the U4/U6 snRNP-specific proteins on U4/U6 snRNA is consistent with a sequential dissociation of the U4/U6 duplex prior to spliceosome catalysis.
Keywords: pre-mRNA splicing/protein–RNA interaction/protein 15.5K/RNA-binding domain/snRNP assembly
Introduction
Pre-mRNA splicing occurs by a two-step transesterification mechanism involving cleavage at the 5′ and 3′ splice sites and exon ligation. This reaction is catalyzed by a large protein–RNA complex termed the spliceosome. Most pre-mRNA introns are removed by the U2-dependent (major) spliceosome, which is composed of the major snRNPs U1, U2, U4/U6 and U5 and numerous non-snRNP protein splicing factors. A rare class of pre-mRNA introns is removed by the U12-dependent (minor) spliceosome, which contains a different set of snRNPs, namely U11, U12 and U4atac/U6atac, but shares U5 snRNP with the major spliceosome (reviewed by Burge et al., 1999).
Spliceosomes assemble on the pre-mRNA via an ordered pathway that involves dynamic snRNP–snRNP and snRNP–pre-mRNA interactions (reviewed by Nilsen, 1998; Staley and Guthrie, 1998). In the major spliceosome, the U1 and U2 snRNPs first recognize the 5′ splice site and branch site, respectively, forming the pre-spliceosome. Spliceosome assembly is completed by the subsequent association of the U4/U6·U5 tri-snRNP complex. Conversion of the spliceosome into a catalytically active machine requires numerous RNA rearrangements, in particular conformational changes in the RNAs of the tri-snRNP. Within the U4/U6 snRNP, the U4 and U6 snRNAs form a phylogenetically highly conserved Y-shaped U4/U6 interaction domain, consisting of two intermolecular helices (stems I and II), which are separated by the 5′ stem–loop of U4 (Figure 1). Both intermolecular helices are disrupted as the spliceosome becomes activated for catalysis; the region of U6 constituting stem II folds back on itself to form a new intramolecular stem–loop, and the region of U6 residing in stem I base-pairs with U2 snRNA to form part of the catalytic center. Concomitantly, U6 snRNA base-pairs with the 5′ end of the intron, while U1 snRNP dissociates from the 5′ splice site. In contrast to U6, U4 snRNA is released from the spliceosome, or remains only loosely attached to it. The U4atac and U6atac snRNAs of the U12-dependent spliceosome are engaged in analogous snRNA–snRNA and snRNA–pre-mRNA interactions during minor spliceosome activation and therefore represent the functional analogs of U4 and U6 snRNA (reviewed by Tarn and Steitz, 1997; Burge et al., 1999).
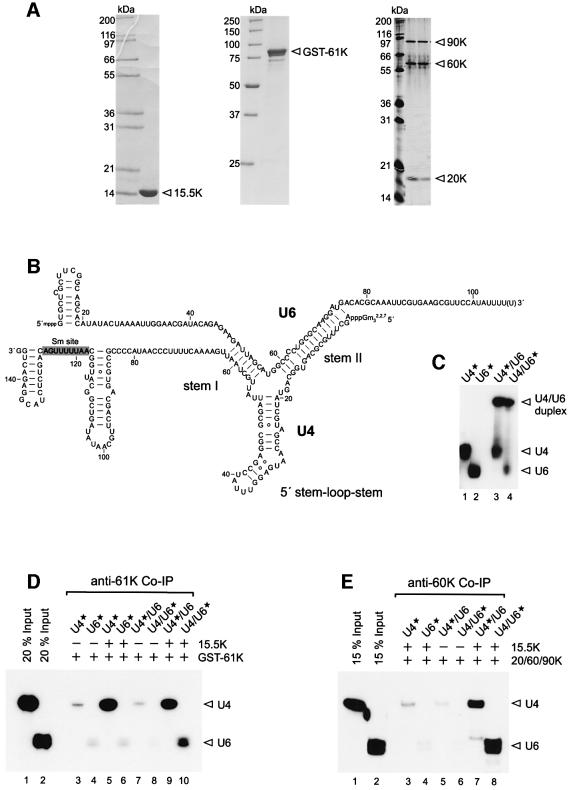
Fig. 1. Binding of protein 61K and the 20/60/90K protein complex to U4/U6 snRNA duplex requires protein 15.5K. (A) SDS–PAGE of recombinant proteins 15.5K (left panel), GST–61K (middle panel) and purified native 20/60/90K protein complex (right panel). 20/60/90K complex was visualized by silver staining, 15.5K and GST–61K were stained with Coomassie Blue. (B) Secondary structure of human U4/U6 snRNA according to Brow and Guthrie (1988). (C) U4/U6 duplex formation as analyzed by native PAGE on a 6% gel. Lanes 1 and 2, 32P-labeled human U4 and U6 snRNAs; lanes 3 and 4, 32P-labeled U4/U6 duplexes. Labeled RNA is marked by an asterisk. (D) Co-immunoprecipitation of 32P-labeled U4/U6 duplexes and U4 and U6 snRNAs with anti-61K antibody. GST–61K was incubated with each of the four RNA substrates shown in (C) in the presence (lanes 5, 6, 9 and 10) or absence (lanes 3, 4, 7 and 8) of protein 15.5K. RNA co-precipitated with anti-61K antibody was analyzed on a denaturing 10% polyacrylamide gel and visualized by autoradiography. Lanes 1 and 2: 20% of total 32P-labeled U4 and U6 snRNA input. (E) Co-immunoprecipitation of 32P-labeled U4/U6 duplexes and U4 and U6 snRNAs with anti-60K antibody. 20/60/90K protein complex was incubated with each of the four RNA substrates shown in (C) in the presence (lanes 3, 4, 7 and 8) or absence (lanes 5 and 6) of protein 15.5K. RNA co-precipitated with anti-60K antibody was analyzed as above. Lanes 1 and 2: 15% of total 32P-labeled U4 and U6 snRNA input.
The mechanism whereby the spliceosome is transformed into a catalytically active machine is only poorly understood at present. There is evidence that the yeast DExH/D-box RNA helicase Brr2p (human U5-200K) is one of the driving forces behind the disruption of the U4/U6 snRNA helices (Laggerbauer et al., 1998; Raghunathan and Guthrie, 1998), but its precise mechanism of action is still unknown. Brr2p/U5-200K could unwind the U4/U6 duplex directly. However, the in vitro RNA helicase activity of purified U5-200K is a generic one, exhibiting no specificity for naked U4/U6 duplexes (Laggerbauer et al., 1998). Alternatively, U4/U6 snRNA-binding proteins could play a key role in regulating the stabilization/destabilization of the U4/U6 RNA duplex. In this respect, information about interactions between U4/U6 snRNP proteins and the U4/U6 snRNA is of particular importance.
In addition to the seven Sm proteins that bind the Sm site of the U4 snRNA, and the seven LSm (‘like-Sm’) proteins (LSm 2–8) that are associated with the 3′ end of U6 snRNA, five proteins have been found by biochemical means to be associated with the human 13S U4/U6 snRNP (reviewed by Will and Lührmann, 2001). These include three proteins with mol. wts of 20, 60 and 90 kDa that form a biochemically stable, heteromeric complex (hereafter termed the 20/60/90K complex; Horowitz et al., 1997; Teigelkamp et al., 1998). U4/U6 snRNPs also contain a 15.5K protein that possesses a novel RNA-binding domain and binds directly to U4 snRNA (Nottrott et al., 1999). Interestingly, it is also present in box C/D snoRNPs, providing a link between the pre-mRNA and pre-rRNA processing machineries (Watkins et al., 2000). Finally, a 61K protein was identified recently as a fifth U4/U6 snRNP-specific protein and shown to be required for U4/U6·U5 tri-snRNP formation (Makarova et al., 2002). It shares a homologous central domain with the proteins Nop56 and Nop58, which (like protein 15.5K) are integral constituents of the box C/D snoRNPs (Gautier et al., 1997; Makarova et al., 2002). Interestingly, recent biochemical evidence indicates that these U4/U6 snRNP-specific proteins are also associated with the HeLa U4atac/U6atac snRNP (Schneider et al., 2002).
Except for the 20K protein, the U4/U6 snRNP-specific proteins are evolutionarily conserved, and orthologous proteins termed Snu13p (15.5K), Prp4p (60K), Prp3p (90K) and Prp31p (61K) are also associated with the Saccharomyces cerevisiae U4/U6 snRNP particle (Banroques and Abelson, 1989; Peterson-Bjørn et al., 1989; Anthony et al., 1997; Weidenhammer et al., 1997; Gottschalk et al., 1999; Stevens and Abelson, 1999). Like their human counterparts, Prp4p and Prp3p interact directly with each other (Ayadi et al., 1998; Gonzales-Santos et al., 2002). Genetic and biochemical studies in yeast and HeLa cells have shown that all conserved U4/U6-specific proteins are essential for cell viability and required for pre-mRNA splicing (Lustig et al., 1986; Banroques and Abelson, 1989; Peterson-Bjørn et al., 1989; Weidenhammer et al., 1997; Nottrott et al., 1999). Experimental evidence indicates a role for the 15.5K/Snu13p and Prp4p proteins in the transition of fully assembled spliceosomes toward an active form (Ayadi et al., 1997; Nottrott et al., 1999). Despite the fact that the U4/U6 snRNP-specific proteins play a crucial role in pre-mRNA splicing and are prime candidates for mediating structural changes during spliceosome activation, there is a surprising paucity of information concerning their interactions with U4 and/or U6 snRNA. The only U4/U6-specific protein shown to interact directly with U4 snRNA is the 15.5K protein, which contacts almost exclusively a purine-rich (5 + 2) internal loop within the U4 snRNA 5′ stem–loop and induces a strong bend in the RNA (Nottrott et al., 1999; Vidovic et al., 2000). In yeast, integration of Prp4p into the U4/U6 snRNP requires the 5′ portion of U4 snRNA, although it is not clear whether Prp4p binds directly to U4 snRNA or via other proteins (Bordonné et al., 1990; Xu et al., 1990).
To understand better the potential role(s) of the U4/U6 snRNP-specific proteins in spliceosome activation, we have investigated their interactions with the U4/U6 snRNAs and elucidated the assembly of U4/U6 snRNP particles in vitro.
Results
Protein 61K and the 20/60/90K complex bind U4 and U4/U6 snRNA, respectively, only in the presence of protein 15.5K
Here we have investigated the assembly of the human U4/U6 snRNP, focusing on the mode of interaction of the particle-specific proteins 15.5K, 20K, 60K, 61K and 90K with elements of the U4/U6 snRNA duplex. For this purpose, we incubated recombinant 15.5K and 61K proteins and purified 20/60/90K complexes (Figure 1A) with radiolabeled U4 and/or U6 snRNA (Figure 1B). As it was not possible to reconstitute a soluble 20/60/90K complex from recombinant proteins, it was purified biochemically from 25S U4/U6·U5 tri-snRNP particles (Figure 1A). RNA binding was analyzed by immunoprecipitation with antibodies against protein 60K or 61K, or by GST pull-down assays. We initially determined whether protein 61K and the 20/60/90K protein complex interact with U4, U6 and/or U4/U6 snRNA duplex and whether binding requires the presence of other U4/U6 snRNP proteins, in particular the 15.5K protein.
U4/U6 duplex formation, using radiolabeled U4 or U6, was confirmed on non-denaturing gels (Figure 1C). As determined by co-immunoprecipitation of the respective RNAs with anti-61K antibodies, GST–61K protein alone did not bind U4, U6 or U4/U6 duplexes to a significant extent (Figure 1D, lanes 3, 4, 7 and 8). However, in the presence of the 15.5K protein, U4 snRNA and U4/U6 duplexes, but not U6 snRNA, were precipitated (Figure 1D, lanes 5, 6, 9 and 10). Likewise, binding of the 20/60/90K complex to U4/U6 duplexes was observed only in the presence of the 15.5K protein (Figure 1E, lanes 5–8). However, 20/60/90K failed to bind U4 or U6 snRNA in either the presence or absence of 15.5K (Figure 1E, lanes 3 and 4; data not shown). We note that the failure to precipitate snRNAs in the absence of 15.5K protein was not due to RNA degradation (data not shown). These results show that protein 15.5K is required for the association of protein 61K with U4 and U4/U6 snRNAs, as well as for the association of the 20/60/90K heteromer with U4/U6 snRNA.
Protein 15.5K is required for the association of 61K and 20/60/90K with U4atac/U6atac snRNA
Minor U4atac/U6atac·U5 tri-snRNP particles can be co-immunoprecipitated specifically by anti-61K and anti-60K antibodies (Schneider et al., 2002), suggesting similar protein–protein and protein–RNA interactions in the major and minor tri-snRNPs. Thus, we next investigated whether protein 61K and the heteromeric 20/60/90K protein complex bind to the U4atac/U6atac snRNA duplex (Figure 2A) under the conditions of our in vitro assay and, if so, whether the requirements for binding are similar to those for U4/U6 snRNA. Studies analogous to those shown in Figure 1 were performed with U4atac, U6atac and U4atac/U6atac duplexes; formation of the latter was confirmed by native PAGE (Figure 2B).
Fig. 2. The 20/60/90K protein complex binds U4atac/U6atac snRNA duplex only in the presence of protein 15.5K. (A) Secondary structure of human U4atac/U6atac snRNA according to Padgett and Shukla (2002). (B) U4atac/U6atac duplex formation as analyzed by native PAGE on a 6% gel. Lanes 1 and 2, 32P-labeled human U4atac and U6atac snRNAs; lanes 3 and 4, 32P-labeled U4atac/U6atac duplexes. Labeled RNA is marked by an asterisk. (C) Co-immunoprecipitation of 32P-labeled U4atac and U6atac snRNAs and U4atac/U6atac duplexes with anti-60K antibody. The 20/60/90K complex was incubated with each of the four RNA substrates shown in (B) in the presence (lanes 3, 4, 7 and 8) or absence (lanes 5 and 6) of protein 15.5K. RNA co-precipitated with anti-60K antibody was analyzed as in Figure 1. Lanes 1 and 2: 15% of total 32P-labeled U4atac and U6atac snRNA input. (D) GST pull-down assay of [U4atac·15.5K·GST–61K] complexes. GST–61K was coupled to glutathione–Sepharose and incubated with 32P- labeled U4atac snRNA in either the absence (lane 3) or presence of protein 15.5K (lane 4). As a control, glutathione–Sepharose-coupled GST was incubated with U4atac snRNA in the presence of 15.5K (lane 2). Bound RNA was analyzed as above. Lane 1: 20% of total 32P-labeled U4atac RNA input.
Immunoprecipitation with anti-60K antibodies demonstrated an association of the 20/60/90K complex with the U4atac/U6atac snRNA duplex, only in the presence of protein 15.5K (Figure 2C, lanes 5–8). As with the U4 and U6 snRNAs, interaction of the 20/60/90K complex could not be detected with U4atac or U6atac snRNA alone, even when protein 15.5K was included in the reaction (Figure 2C, lanes 3 and 4). In GST pull-down experiments, U4atac snRNA was bound by the GST–61K protein only when protein 15.5K was present, but not by GST alone (Figure 2D). Similarly to the U4/U6 snRNAs, 61K also bound to U4atac/U6atac snRNA duplex in the presence of 15.5K, while no interaction of 61K was detected with U6atac snRNA (data not shown). Taken together, these results show that protein 15.5K mediates the association of 20/60/90K and 61K with both U4/U6 and U4atac/U6atac snRNA, and suggest similar protein–RNA interactions in both types of particles.
The 20/60/90K complex binds to a minimal U4/U6 snRNA duplex comprising stem II and the 5′ stem–loop of U4 snRNA
The fact that the 20/60/90K protein complex bound strongly only to U4/U6 snRNA duplex suggested that characteristic features of the Y-shaped domain (Figure 1) may provide the essential RNA structural requirements for binding. These features may be stem I, stem II, the three-way junction or any combination of these. We therefore attempted to define a minimal binding site for the 20/60/90K complex on U4/U6 snRNA by progressively shortening the 3′ ends of the U4 and U6 snRNAs and testing the ability of the truncated U4/U6 snRNA duplexes to interact with the 20/60/90K complex in the presence of protein 15.5K. Surprisingly, stem I and an intact three-way junction of the duplex were not required for binding; a highly truncated U4/U6 snRNA duplex comprising the first 52 nucleotides of U4 snRNA and positions 58–87 of U6 snRNA was precipitated efficiently by anti-60K antibodies in the presence of 20/60/90K and protein 15.5K (Figure 3C). Further shortening of the U6 snRNA from the 3′ end resulted in inefficient duplex formation (data not shown), preventing a more precise investigation of the role of stem II in protein binding. Similarly, a truncated RNA duplex containing only stem I could not be generated. A U4/U6 snRNA duplex composed of U6 and U4Δ5′ stem–loop was not bound by 20/60/90K in the presence of 15.5K protein (data not shown), supporting the conclusion that 15.5K–U4 snRNA binding is a pre-requisite for 20/60/90K association. Our results thus show that the 5′ stem–loop of U4 snRNA and stem II of the U4/U6 snRNA duplex together define the RNA structural elements that are needed for specific binding of the 20/60/90K complex to U4/U6 snRNA in the presence of protein 15.5K. We therefore define the RNA substrate shown in Figure 3A as a ‘minimal’ duplex for protein binding.

Fig. 3. The 20/60/90K complex binds a truncated U4/U6 snRNA duplex. (A) Secondary structure of a ‘minimal’ U4/U6 duplex comprising nucleotides 1–52 of U4 (U4 nts 1–52) and nucleotides 58–87 of U6 snRNA (U6 nts 58–87). (B) 32P-Labeled U6 nts 58–87, alone (lane 1) and base-paired with non-labeled U4 nts 1–52 (lane 2), analyzed on a 10% native gel. (C) Co-immunoprecipitation of the ‘minimal’ U4/U6 duplex bound to the 20/60/90K complex in the presence of protein 15.5K. U6 nts 58–87 alone (lane 2) or complexed with non-labeled U4 nts 1–52 (lane 3) were incubated with protein 15.5K and the 20/60/90K complex. RNA co-immunoprecipitated with the anti-60K antibody was analyzed as in Figure 1. Lane 1: 20% of total 32P-labeled U6 RNA nts 58–87 input.
Protein 61K requires the 5′ stem–loop and the 5′ end of U4 snRNA for binding
We next investigated which region of the U4 snRNA is required for binding of protein 61K. GST pull-down experiments were performed with deletion mutants of U4 snRNA (Figure 4A), and recombinant GST–61K and protein 15.5K. Nucleotides 3′ of the 5′ stem–loop (i.e. nucleotides 53–145) could be deleted without a noticeable reduction in U4 RNA affinity for protein 61K (Figure 4B, compare lanes 1–3 with lanes 4–6). However, if the 5′-terminal region of U4 snRNA (nucleotides 1–19) was also removed, protein 61K failed to bind, even in the presence of protein 15.5K (Figure 4B, lanes 7–9). Thus, stable binding of protein 61K requires not only the 5′ stem–loop and protein 15.5K, but also nucleotides in the 5′-terminal region of U4 snRNA.
Fig. 4. Protein 61K binds the 5′ portion of U4 snRNA in the presence of protein 15.5K. (A) Secondary structure of U4 snRNA comprising nucleotide positions 1–52 and 20–52. (B) GST–61K was coupled to glutathione–Sepharose and incubated with 32P-labeled full-length U4 snRNA and the U4 snRNA fragments shown in (A), in either the presence (lanes 3, 6 and 9) or absence (lanes 2, 5 and 8) of protein 15.5K. Bound RNA was analyzed as in Figure 1. Lanes 1, 4 and 7: 20% of total input of the respective 32P-labeled RNA substrate.
Protein 61K can be cross-linked to two distinct sites on U4 snRNA in native tri-snRNP particles
The presence of protein 15.5K is an absolute require ment for the binding of protein 61K and the 20/60/90K heteromer to U4/U6 snRNA (see above). In addition, the minimal RNA requirements identified for binding of 61K and 20/60/90K strongly suggested that one or more of these proteins are directly associated with the U4 and/or U6 snRNA. We therefore performed UV cross-linking with purified native 25S tri-snRNP particles to determine whether 61K or components of the 20/60/90K complex directly contact the U4 or U6 snRNAs.
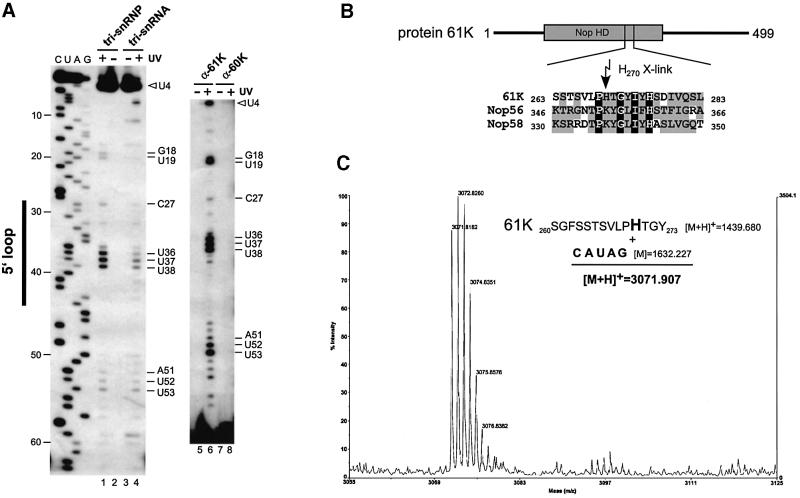
Primer extension analysis of the U4 snRNA derived from tri-snRNPs revealed strong reverse transcriptase stops at positions 37–39 (U36, U37 and U38) and weaker stops at positions 52–54 (A51, U52 and U53), 28 (C27) and 19–20 (G18 and U19) only after UV irradiation (Figure 5A, compare lanes 1 and 2). Since stops at positions 37–39, 28 and 19–20 were absent or less abundant when the snRNPs were stripped of protein before irradiation (lane 4), we infer that these stops are due to protein-specific cross-linking to these nucleotides. In contrast, the stops at positions 52–54 were undiminished in intensity, implying that they are not protein related.
Fig. 5. Protein 61K can be cross-linked to U4 and U4atac snRNA. (A) Protein 61K–U4 snRNA cross-linking sites in purified native 25S tri-snRNPs. Left panel: primer extension analysis of U4 snRNA derived from UV-cross-linked tri-snRNPs (lane 1) or UV-irradiated, naked U4 snRNA (lane 4). Lanes 2 and 3: controls without UV irradiation. C, U, A and G are dideoxy sequence markers. Nucleotide positions of the reverse transcriptase stops are indicated on the right; a black bar indicates nucleotides of the U4 5′ stem–loop, and an open arrowhead marks the position of full-length U4 snRNA. Right panel: primer extension analysis of U4 snRNA after immunoprecipitation of UV-irradiated, denatured tri-snRNPs with antibodies against proteins 61K (lanes 5 and 6) or 60K (lanes 7 and 8), either with (lanes 6 and 8) or without UV irradiation (lanes 5 and 7). Nucleotide positions of the reverse transcriptase stops are indicated. (B) The cross-linked amino acid His270 is found in the central Nop56/Nop58 homology domain of protein 61K (Nop HD; residues 93–328; Makarova et al., 2002). Residues 263–283 of protein 61K were aligned with the homologous sequences of human Nop56 (amino acids 346–366; Y12065) and Nop58 (amino acids 330–350; AF123534) using the Clustal method. Identical residues are boxed in black, and conserved residues are highlighted in gray. His270 is marked by an arrow. (C) Protein 61K–U4atac snRNA cross-linking sites in reconstituted [15.5K·61K·U4atac/U6atac] complexes. MALDI-MS analysis identified the 61K peptide (260SGFSSTSVLPHTGY273) cross-linked to an RNase T1 oligonucleotide (42CAUAG46) in the U4atac 5′ stem–loop. The nucleotide composition (C1U1A2G1; [M] = 1632.227) was determined from the difference between the mono-isotopic mass of the cross-linked peptide–oligonucleotide complex ([M + H]+ = 3071.907) and that of the peptide alone ([M + H]+ = 1439.680). The cross-linked peptide moiety was determined by N-terminal sequence analysis.
To determine which tri-snRNP-specific protein was cross-linked to these nucleotides, we performed immunoprecipitations of UV-irradiated purified 25S tri-snRNPs under conditions where protein–protein interactions have been completely disrupted prior to primer extension analysis (Urlaub et al., 2002). Strong reverse transcriptase stops at nucleotides G18–U19, U36–U38 and A51–U53 were observed after immunoprecipitation with the anti-61K antibody (Figure 5A, lane 6), whereas no stops at these positions were detected after immunoprecipitation with the anti-60K antibody (lane 8), or with antibodies raised against other tri-snRNP-specific proteins (data not shown). These data demonstrate that, within native tri-snRNPs, protein 61K contacts loop nucleotides U36, U37 and U38 in the U4 5′ stem–loop and, additionally, G18 and U19 upstream of the 5′ stem–loop. Bands corresponding to nucleotides A51–U53 were also detected after immunoprecipitation (compare lanes 1 and 6), but the authenticity of these cross-links is uncertain (see above) and thus they were not analyzed further. The stop at nucleotide C27 was not enhanced, indicating that it does not arise from a U4 snRNA–61K cross-link. Identification of these distinct cross-linking sites is consistent with the fact that the 5′ stem–loop and the 5′-terminal region of U4 snRNA are required for 61K binding (see above), and demonstrate that protein 61K directly contacts the U4 snRNA.
A conserved domain of protein 61K contacts the U4atac snRNA
Protein 61K shares a homologous domain with the C/D box snoRNP-associated proteins Nop58 and Nop56 (Makarova et al., 2002; Vithana et al., 2002). As these proteins can also be cross-linked to RNA (N.Watkins, personal communication), it is likely that this common domain might be directly involved in RNA binding. We therefore set out to identify the region of protein 61K that directly contacts RNA by combining cross-linking with Edman sequencing and mass spectrometry (Urlaub et al., 2002). We chose as starting material [U4atac/U6atac·15.5K·61K] particles reconstituted in vitro, as these complexes gave the largest yield of cross-linked product (data not shown). Importantly, the requirements for binding of protein 61K to the U4atac snRNA in vitro are the same as those for its binding to U4 snRNA, since a deletion mutant of U4atac snRNA comprising nucleotides 1–57 was bound by GST–61K with the same efficiency as wild-type U4atac snRNA (data not shown). Furthermore, the overall cross-linking pattern observed with minor tri-snRNP particles is similar to that in the 25S U4/U6·U5 tri-snRNP (data not shown). Thus, data obtained from the U4atac in vitro system probably also apply to U4 and U4atac native complexes.
After digestion of reconstituted particles with chymotrypsin and RNase T1, we isolated a peptide– oligonucleotide cross-link via RP-HPLC (data not shown). N-terminal sequencing identified the peptide as a fragment of protein 61K encompassing positions 260–273 (SGFSSTSVLPHTGY). Edman sequencing failed to detect His270, demonstrating that this residue is cross-linked to the RNA (data not shown). Significantly, His270 is located within the conserved Nop58/56 homology domain (Figure 5B), confirming our hypothesis that this region directly interacts with RNA. These results thus provide the first evidence that the Nop homology domain is a conserved, novel RNA-binding domain.
Matrix-associated laser desorption ionization-mass spectrometry (MALDI-MS) analysis of the peptide– oligonucleotide showed unambiguously that the peptide is cross-linked to a T1 pentanucleotide with the composition GA2UC (Figure 5C). The human U4atac/U6atac snRNA contains only one T1 fragment with this nucleotide composition, namely positions 42–46 of the U4atac snRNA 5′ stem–loop (42CAUAG46, compare Figure 2A). These results are consistent with the cross-linking results obtained with native tri-snRNP particles, which demonstrated that the loop region of the U4 5′ stem–loop contacts protein 61K. Thus, we conclude that protein 61K is in contact with the 5′ stem–loop of both U4 and U4atac snRNA.
Protein 90K directly contacts U6 in the minimal U4/U6 snRNA duplex
To determine whether one or more proteins of the 20/60/90K complex contacts U4/U6 snRNA, we carried out UV cross-linking experiments with native tri-snRNPs as described above and probed for U6 snRNA. Although potential U6–protein cross-links were detected, these did not appear to involve proteins 61K or 60K since they were not enhanced by immunoprecipitation with the corresponding antibodies (data not shown). We therefore incubated the U4/U6 snRNA duplex composed of radiolabeled, full-length U6 and non-labeled, truncated U4 snRNA (nucleotides 1–52) with protein 15.5K and the 20/60/90K complex. The resultant particles were then UV irradiated, digested with RNases A and T1, and cross-links were analyzed by SDS–PAGE. A band migrating with an apparent molecular mass of ∼90 kDa was detected (Figure 6A, lane 4). This product was not obtained without UV irradiation (lane 1), without protein 15.5K (lane 3), if U6 snRNA alone was used instead of the U4/U6 duplex (lane 6) or upon irradiation of the U4/U6 snRNA duplex in the complete absence of proteins (lane 2). Moreover, the cross-linked product was sensitive to digestion with proteinase K (lane 5), confirming that it contained protein. We thus conclude that protein 90K, as part of the 20/60/90K heteromeric complex, directly contacts U6 snRNA within the U4/U6 snRNA duplex.
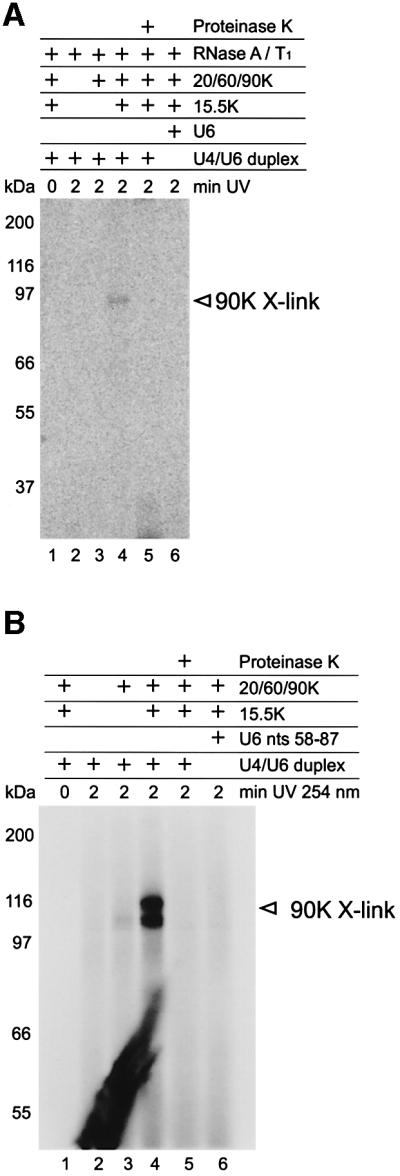
Fig. 6. Protein 90K of the 20/60/90K complex contacts U6 snRNA. (A) The 20/60/90K complex was incubated with U4/U6 duplex composed of 32P-labeled, full-length U6 snRNA and non-labeled U4 nucleotides 1–52. After UV irradiation, the products were digested with RNases A and T1, and separated by SDS–PAGE. In lane 4, U4/U6 duplex, protein 15.5K and the 20/60/90K complex were present during cross-linking. Lane 1, no UV irradiation; lane 2, all proteins omitted; lane 3, protein 15.5K omitted; lane 6, RNA duplex replaced by 32P-labeled U6 snRNA. In lane 5, the cross-linking product was digested with proteinase K. The 90K protein cross-link is marked on the right. (B) The 20/60/90K complex was incubated with the minimal U4/U6 duplex consisting of 5′ end-labeled U6 nucleotides 58–87 and non-labeled U4 nucleotides 1–52 (see Figure 3). Lane designations are as in (A) and indicated above each lane.
To map the contact site of protein 90K more precisely, we next performed cross-linking with the ‘minimal’ U4/U6 snRNA duplex, in which the U6 snRNA was 5′ labeled. Cross-linking products migrating with an apparent molecular mass of ∼100–116 kDa were detected (Figure 6B, lane 4), consistent with protein 90K contacting the U6 snRNA moiety of the minimal duplex. The exact reason why this 90K cross-link separates on the SDS–polyacrylamide gel as two closely migrating bands is not clear. One possibility is that the U6 RNA oligonucleotide becomes cross-linked to two distinct sites of the protein, resulting in a slightly different migration behavior of the two cross-linking products. In any case, the similar size of the two cross-linking products makes it unlikely that a protein other than 90K is cross-linked to the U6 snRNA. Note that RNase digestion was not performed, increasing the molecular weight of the cross-linked protein 90K. No cross-links were observed without UV irradiation (lane 1), in the absence of all proteins (lane 2) or without protein 15.5K (lane 3). Furthermore, no cross-linking product was detected when the U6 RNA oligonucleotide (nucleotides 58–87) alone was used (lane 6). This cross-link was also sensitive to digestion with proteinase K (lane 5). As the minimal U4/U6 duplex is sufficient for cross-link formation, we infer that protein 90K contacts U6 snRNA between positions 58 and 87 of the duplex. These results indicate that protein 90K may bind to stem II of the U4/U6 snRNA duplex.
Discussion
We have analyzed the assembly of spliceosomal U4/U6 snRNP particles in vitro, using purified, native and/or recombinant U4/U6 snRNP-specific proteins and in vitro transcribed U4 and U6 snRNAs. Co-immunoprecipitation/pull-down studies coupled with UV cross-linking revealed several important new aspects about the nature of the U4/U6 snRNP particle, including a conserved hierarchical assembly pathway and novel protein–RNA interactions. The possible functional role of the clustered interactions of the U4/U6 snRNP-specific proteins with the 5′ stem–loop and stem II of the U4/U6 interaction domain during activation of the spliceosome is discussed.
Conserved hierarchical assembly pathway of U4/U6 snRNP particles
We show that binding of protein 15.5K to the U4 5′ stem–loop is required for subsequent interaction of both the U4/U6 snRNP-specific 61K protein and the 20/60/90K protein complex with U4/U6 snRNA duplex (Figure 1). The association of 61K and 20/60/90K with RNA can occur independently of each other and, thus, these appear to be non-cooperative. This assembly pathway is also observed with the U4atac/U6atac snRNP, a component of the minor spliceosome (Figure 2). While a direct interaction between protein 15.5K and the 5′-terminal stem–loop of U4atac snRNA was reported in previous studies (Nottrott et al., 1999), here we demonstrate additionally the interaction of both protein 61K and the 20/60/90K complex with the U4atac/U6atac snRNAs. Thus, the network of protein–RNA interactions appears to be similar in both RNPs.
The 15.5K protein is also present in box C/D snoRNPs where it directly binds the box C/D motif. The box C/D motif shows striking similarity in primary and secondary structure to the 5′ stem–loop of the U4 snRNA and it is assumed that the protein recognizes the snoRNA motif in a similar manner (Vidovic et al., 2000; Watkins et al., 2000). Interestingly, 15.5K binding to box C/D snoRNAs has also been shown recently to be essential for the binding of the box C/D snoRNP proteins Nop56, Nop58 and fibrillarin (Watkins et al., 2002). Therefore, 15.5K binding appears to have a similar nucleation function in the assembly of both U4/U6 snRNPs and the box C/D snoRNPs.
Differential RNA structural requirements for binding of protein 61K and the 20/60/90K complex
Although both protein 61K and the 20/60/90K complex require the presence of 15.5K for interaction with U4/U6 snRNA, there are clear differences in the RNA structural requirements for their binding. Indeed, in the presence of 15.5K, protein 61K interacts with U4 and with U4/U6 snRNA duplex, but not with U6 snRNA alone, while the 20/60/90K complex exclusively interacts with U4/U6 duplexes (Figure 1). Protein 61K contacts U4 at the apical loop region of the 5′ stem–loop and at nucleotides G18/U19 (Figure 5). The latter region is also absolutely required for 61K binding (Figure 4). Thus, protein 61K bridges the 5′ half of the U4/U6 snRNA three-way junction and the apical loop of the U4 snRNA 5′ stem–loop. Interestingly, binding of protein 15.5K to the asymmetric internal loop of the U4 5′ stem–loop–stem structure results in a sharp bend between the two contiguous RNA stems, juxtaposing the extensions of the two stems (Vidovic et al., 2000). Indeed, it is precisely these extensions that protein 61K binds, i.e. the apical loop region and the 5′ half of the three-way junction. Thus, binding of 15.5K may induce the proper RNA conformation required for subsequent 61K binding.
Binding of the 20/60/90K complex also requires the U4 5′ stem–loop, and is observed only when stem II of the U4/U6 duplex is formed (Figure 3). Stem I of the U4/U6 duplex can be deleted without loss of binding, implying that an intact three-way junction is not required. Our data suggest that the 20/60/90K complex may contact simultaneously both the U4 5′ stem–loop/15.5K protein and part of stem II, thus bridging these two parts of the U4/U6 duplex. A direct contact of the 20/60/90K heteromer with stem II is supported by the fact that the 90K protein could be cross-linked to an oligonucleotide encompassing nucleotides 58–87 of U6 only when it was base-paired to the 5′ region of U4 snRNA, i.e. when stem II was formed (Figure 6). This strongly suggests that part of stem II is recognized by 90K, irrespective of whether the cross-linked nucleotide(s) is located within the portion of U6 RNA base-paired to U4 RNA or in the short single-stranded U6 overhang (Figure 3). Finally, we note that the association of the 20/60/90K protein complex with U4/U6 snRNAs occurs in the absence of Lsm proteins in vitro. This is not in contradiction to the previously reported contribution of the Lsm 2–8 proteins in U4/U6 di-snRNP formation (Achsel et al., 1999), since we assembled the 20/60/90K proteins to pre-formed U4/U6 snRNA duplexes.
61K and 90K are novel RNA-binding proteins
The 61K and 90K proteins could be UV cross-linked to U4 or U6 snRNA, respectively (Figures 5 and 6), indicating that they are bona fide RNA-binding proteins. Interestingly, protein 61K shares significant homology with the box C/D snoRNP proteins Nop56 and Nop58 within a conserved central domain spanning residues 97–328, referred to as the Nop homology domain (Vithana et al., 2001; Makarova et al., 2002). As the U4 snRNP and box C/D snoRNPs share a similar core structure, with protein 15.5K bound to the respective RNAs (Watkins et al., 2000), it is highly interesting that one of the 61K–RNA cross-links mapped to an amino acid within the Nop homology domain (Figure 5). These results are consistent with the idea that these proteins belong to a family of RNA-binding proteins with a hitherto unidentified RNA-binding motif. Cross-linking of protein 90K to U6 snRNA requires formation of stem II of the U4/U6 duplex, suggesting that 90K might be a double-stranded (ds) RNA-binding protein. Indeed, sequence homology has been detected previously between 90K and the known dsRNA-binding proteins, RNase III of Escherichia coli and the Drosophila melanogaster Staufen protein (Lauber et al., 1997). Recently, mutations in the gene encoding the 61K protein were found to be responsible for the autosomal form of retinitis pigmentosa (Vithana et al., 2001). Most of these mutations lie within the Nop homology domain. Significantly, analysis of the 61K cross-linking site demonstrated that it directly contacts the apical loop region of the U4atac 5′ stem–loop via this conserved domain (Figure 5). Similarly, mutations in protein 90K are also linked to this genetic disease (Chakarova et al., 2002) and are clustered adjacent to 90K’s putative dsRNA-binding motif. These mutations thus may have profound effects on protein–RNA and/or protein–protein interactions that may be crucial for the dynamics of tri-snRNP and spliceosome assembly.
What determines the RNA-binding specificity of these proteins?
The finding that protein 61K and the 20/60/90K complex interact with U4/U6 and U4atac/U6atac snRNAs strongly suggests that the common structural features which they recognize are not conserved bases, but rather conserved elements of secondary structure. This idea is supported by the following results. (i) Protein 61K cross-links to the apical loop of the 5′ stem–loop of both U4 and U4atac snRNA. This cannot be due to similarity of the RNA sequences, as the 5′ apical loop sequences of U4 snRNA and U4atac snRNA are 36UUUAU40 and 41GCAUA45, respectively. Furthermore, the 5′-terminal sequences of both the U4 and U4atac snRNA share little sequence identity, but both are required for 61K protein binding. (ii) The 20/60/90K complex binds efficiently a minimal U4/U6 duplex containing nucleotides 58–87 of U6 snRNA (Figure 3). As there is only a very low degree of sequence identity between this region of U6 and the corresponding region of U6atac snRNA, we infer that the 20/60/90K complex recognizes RNA secondary structure rather than primary sequence. Our data support a model in which the binding of protein 15.5K to U4 or U4atac snRNA generates particular secondary structure elements in the U4/U6 snRNA that are recognized by protein 61K and the 20/60/90K complex. This mode of RNA interaction is not uncommon for RNP complexes. In the E.coli ribosome, for example, secondary 16S rRNA-binding sites for the protein heterodimer S6–S18 are generated only upon prior binding of protein S15 to the 16S rRNA (Agalarov et al., 2000). It is also possible that the conformation of protein 15.5K is changed likewise by its binding to U4 snRNA via a mutually induced fit mechanism (Williamson, 2000), allowing 15.5K to bind directly to protein 61K or the 20/60/90K complex.
Functional implications for the activation of the spliceosome
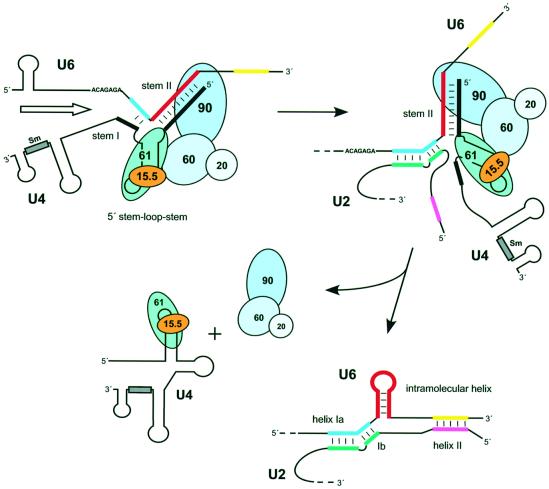
A striking feature of the 15.5K, 61K and 90K protein–RNA contacts is that they are clustered on the RNA within the 5′ stem–loop of U4 and stem II of the U4/U6 duplex, suggesting that the U4/U6-specific proteins are found predominantly in these regions (see Figure 7). This agrees very well with chemical modification experiments on native tri-snRNP particles in which the 5′ stem–loop of U4 and stem II of U4/U6, but neither the 3′ half of the three-way junction nor stem I of U4/U6 duplex appears to be strongly protected (Mougin et al., 2002). This particular distribution of the U4/U6 protein components has potential functional implications for the activation of the spliceosome prior to catalysis, during which unwinding of the U4/U6 duplex occurs.
Fig. 7. Proposed mechanism for disruption of U4/U6 snRNA during activation of the spliceosome. See text for details.
Spliceosome activation is accompanied by the dissociation of U4 snRNA from the U4/U6 duplex, allowing U6 snRNA to pair with U2 snRNA. At present, it is not clear whether U4/U6 stems I and II dissociate simultaneously, or in separate, regulated steps. In light of our data, it is attractive to suggest that the 20/60/90K complex might stabilize stem II, enabling stem I to be opened first (Figure 7). Consistent with the idea of sequential dissociation, an intermediate in the catalytic core of the U12-dependent spliceosome has been detected that has an intact U4atac/U6atac stem II, but in which parts of stem I are already base-paired with U12 snRNA (Frilander and Steitz, 2001). This, together with the absence of any detectable protein–RNA interaction on stem I and at the 3′ half of the three-way junction, could allow reversible opening/closing of stem I, thus introducing the possibility of a proofreading step during the initial formation of the novel U2/U6 snRNA duplex.
Activation of the spliceosome prior to catalysis is finally achieved by the complete unwinding of U4/U6 stem II and the subsequent formation of an intramolecular U6 snRNA helix. Thus, the 20/60/90K complex may play a crucial role in the formation of catalytically active RNA elements in the spliceosome (i.e. U2/U6 duplex and the intramolecular U6 helix). Our model predicts that unwinding of stem II would be accompanied by the dissociation of the 20/60/90K complex from the U4/U6 duplex (Figure 7).
What triggers the destabilization of the U4/U6 snRNA interaction domain prior to catalysis? The most prominent candidate is the RNA-dependent helicase U5-200K (Brr2p in S.cerevisiae), which has been demonstrated to affect U4/U6 unwinding (Laggerbauer et al., 1998; Raghunathan and Guthrie, 1998), very probably in cooperation with additional U4/U6·U5 tri-snRNP proteins such as Prp8 (U5-220K), Snu114p (U5-116K) or Prp38p (Xie et al., 1998; Kuhn et al., 1999; Bartels et al., 2002). Whether they act directly on U4/U6 snRNA, or on protein–RNA complexes stabilizing the U4/U6 duplex presently is not clear. Another possibility is that a switch is induced in the conformation of one or more of the U4/U6 snRNA-binding proteins, which in turn leads to destabilization of the duplex. Clearly, further experiments are needed to specify the role of the U4/U6 snRNP-specific proteins in the regulation of the spliceosomal activation step.
Materials and methods
Purification of recombinant proteins
Protein 15.5K was purified according to Nottrott et al. (1999). The 61K cDNA was subcloned into pGEX-6-P1 (Amersham Pharmacia Biotech), and recombinant GST–61K protein was purified by affinity chromatography on glutathione–Sepharose according to Makarova et al. (2002).
Purification of the native 20/60/90K protein complex
Peak glycerol gradient fractions of purified native 25S HeLa tri-snRNPs were concentrated by ultracentrifugation (Laggerbauer et al., 1998), resuspended in buffer T [50 mM Tris–HCl pH 8.0, 1 mM dithiothreitol (DTT), 0.5 mM EDTA] containing 400 mM NaCl and loaded onto a 1 ml heparin HiTrap column (Amersham Pharmacia Biotech). Proteins were eluted by a 0.4–0.9 M NaCl gradient (4°C). Fractions containing the 20/60/90K complex subsequently were centrifuged on a 5–20% glycerol gradient (Sorvall TST 41.14 rotor, 35 000 r.p.m., 21 h, 4°C) containing buffer G (20 mM HEPES/KOH pH 7.9, 700 mM NaCl, 1.5 mM MgCl2). Fractions containing 20/60/90K complexes were pooled, diluted 1:2 in buffer T and loaded again onto the heparin column. Bound proteins were eluted by a linear 0.4–0.9 M NaCl gradient (4°C).
RNA synthesis and duplex formation
RNA transcripts were generated by run-off transcription of linearized plasmids encoding human U4, U6, U4atac or U6atac snRNA. The DNA template encoding U4 nts 1–52 was generated from the U4 plasmid by PCR. Transcription of 32P-labeled RNA was performed in the presence of either 0.66 µM [α-32P]UTP or [α-32P]CTP (3000 Ci/mmol). RNA oligonucleotides (U4 nts 20–52; U6 nts 58–87) were chemically synthesized and 5′ end-labeled using [γ-32P]ATP (5000 Ci/mmol).
RNA duplexes were formed by annealing 20 nM 32P-labeled RNA with 200 nM non-labeled RNA in buffer A (Brow and Vidaver, 1995). For UV cross-linking studies shown in Figure 6A, RNA duplexes were generated by annealing 0.14 µM [α-32P]CTP-labeled, full-length U6 snRNA with 2 µM non-labeled U4 nts 1–52 in buffer A. All samples were incubated at 80°C for 1 min, cooled slowly to 30°C and placed on ice. U4/U6 duplexes containing full-length U6 snRNA were formed in the presence of 20 µM oligonucleotide hU6cen2 (Brow and Vidaver, 1995). Duplex formation was monitored by native PAGE on either 6 or 10% (80:1) gels.
Binding reactions, immunoprecipitations and GST pull-downs
A 40 pmol concentration of GST–61K protein or 20/60/90K complex was incubated with 5 µl of RNA substrate in the presence or absence of 35 pmol of 15.5K protein in 15–30 µl of buffer A for 20 min at 30°C. Binding reactions were performed in the presence of 10 µg of E.coli tRNA unless used for in vitro cross-linking. Coupling of anti-61K and anti-60K antibodies to protein A–Sepharose and immunoprecipitation were performed as described by Nottrott et al. (1999). Co-precipitated RNA was separated on a 10% polyacrylamide–7 M urea gel and visualized by autoradiography.
For GST pull-downs, 40 pmol of either GST or GST–61K protein were coupled to glutathione–Sepharose according to Nottrott et al. (1999) and incubated with pre-formed U4atac–15.5K complex in buffer D150 (20 mM HEPES/KOH pH 7.9, 150 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100) for 2 h at 4°C. Pre-formed complex was obtained by incubating 20 pmol of recombinant 15.5K protein with 32P-labeled RNA substrate (0.2–0.4 pmol) in a final volume of 10 µl of buffer D150 for 45 min at 4°C. Beads were washed four times with buffer D150 and once with buffer IPP500 (50 mM Tris–HCl pH 8.0, 500 mM NaCl, 0.05% NP-40). Co-precipitated RNAs were recovered and analyzed as above.
UV cross-linking studies
For UV cross-linking of reconstituted protein–RNA complexes, RNA duplexes containing 0.7 pmol labeled RNA were generated in a final volume of 30 µl as described above. UV irradiation at 254 nm and subsequent RNase T1/A or proteinase K digestion were performed as described by Urlaub et al. (2002). Samples were separated by SDS–PAGE on 10% gels and cross-linked proteins were visualized by autoradiography. UV cross-linking of purified native 25S tri-snRNP particles, immunoprecipitation under denaturing conditions and primer extension analysis were carried out according to Urlaub et al. (2002). Antibodies against the following tri-snRNP proteins were used for immunoprecipitation: 15K, 15.5K, 27K, 40K, 60K, 61K, 65K, 100K, 102K, 110K, 116K, 200K and 220K. Oligo B1 was used for primer extension analysis (Nottrott et al., 1999).
For large-scale cross-linking experiments, U4atac/U6atac duplex was formed by incubating 10 nmol of U4atac snRNA with 25 nmol of U6atac snRNA in 500 µl of buffer A as described above. The sample volume was adjusted to 13 ml with buffer P (250 mM NaCl, 50 mM Tris–HCl pH 7.5). Duplex was incubated for 20 min at 4°C with 10 nmol of 15.5K protein and 5 nmol of GST–61K protein in the presence of 400 U of RNasin and 20 mM DTT, and UV irradiated as described by Urlaub et al. (2002). Isolation and identification of cross-linked 61K peptide–oligonucleotides by Edmann degradation and MALDI-MS was performed as described by Urlaub et al. (2002).
Acknowledgments
Acknowledgements
We thank M.Raabe for excellent technical assistance, C.Lenz for support in mass spectrometry, and Cindy L.Will for critical and helpful comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
References
- Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalarov S.C., Prasad,G.S., Funke,P.M., Stout,C.D. and Williamson,J.R. (2000) Structure of the S15, S6, S18–rRNA complex: assembly of the 30S ribosome central domain. Science, 288, 107–112. [DOI] [PubMed] [Google Scholar]
- Anthony J.G., Weidenhammer,E.M. and Woolford,J.L.,Jr (1997) The yeast Prp3 protein is a U4/U6 snRNP protein necessary for integrity of the U4/U6 snRNP and the U4/U6·U5 tri-snRNP. RNA, 3, 1142–1152. [PMC free article] [PubMed] [Google Scholar]
- Ayadi L., Miller,M. and Banroques,J. (1997) Mutations within the yeast U4/U6 snRNP protein Prp4 affect a late stage of spliceosome assembly. RNA, 3, 197–209. [PMC free article] [PubMed] [Google Scholar]
- Ayadi L., Callebaut,I., Saguez,C., Villa,T., Mornon,J.P. and Banroques,J. (1998) Functional and structural characterization of the Prp3 binding domain of the yeast Prp4 splicing factor. J. Mol. Biol., 284, 673–687. [DOI] [PubMed] [Google Scholar]
- Banroques J. and Abelson,J.N. (1989) PRP4: a protein of the yeast U4/U6 small nuclear ribonucleoprotein particle. Mol. Cell. Biol., 9, 3710–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C., Klatt,C., Lührmann,R. and Fabrizio,P. (2002) The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO rep., 3, 875–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordonné R., Banroques,J., Abelson,J. and Guthrie,C. (1990) Domains of yeast U4 spliceosomal RNA required for PRP4 protein binding, snRNP–snRNP interactions and pre-mRNA splicing in vivo. Genes Dev., 4, 1185–1196. [DOI] [PubMed] [Google Scholar]
- Brow D.A. and Guthrie,C. (1988) Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature, 334, 213–218. [DOI] [PubMed] [Google Scholar]
- Brow D.A. and Vidaver,R.M. (1995) An element in human U6 RNA destabilizes the U4/U6 spliceosomal RNA complex. RNA, 1, 122–131. [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosome. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Chakarova C.F., et al. (2002) Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum. Mol. Genet, 11, 87–92. [DOI] [PubMed] [Google Scholar]
- Frilander M.J. and Steitz,J.A. (2001) Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol. Cell, 7, 217–226. [DOI] [PubMed] [Google Scholar]
- Gautier T., Berges,T., Tollervey,D. and Hurt,E. (1997) Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-Santos J.M., Wang,A., Jones J., Ushida C., Liu,J. and Hu,J. (2002) Central region of the human splicing factor Hprp3p interacts with Hprp4p. J. Biol. Chem., 277, 23764–23772. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Neubauer,G., Banroques,J., Mann,M., Lührmann,R. and Fabrizio,P. (1999) Identification by mass spectrometry and functional analysis of novel proteins of the yeast U4/U6·U5 tri-snRNP. EMBO J., 18, 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz D.S., Kobayashi,R. and Krainer,A.R. (1997) A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA, 3, 1374–1387. [PMC free article] [PubMed] [Google Scholar]
- Kuhn A.N., Li,Z. and Brow,D.A. (1999) Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell, 3, 65–75. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B., Achsel,T. and Lührmann,R. (1998) The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl Acad. Sci. USA, 95, 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber J., Plessel,G., Prehn,S., Will,C.L., Fabrizio,P., Gröning,K., Lane,W.S. and Lührmann,R. (1997) The human U4/U6 snRNP contains 60 and 90kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA, 3, 926–941. [PMC free article] [PubMed] [Google Scholar]
- Lustig A.J., Lin,R.J. and Abelson,J. (1986) The yeast RNA gene products are essential for mRNA splicing in vitro. Cell, 47, 953–963. [DOI] [PubMed] [Google Scholar]
- Makarova O.V., Makarov,E.M., Liu,S., Vornlocher,H.P. and Lührmann,R. (2002) Protein 61K, encoded by a gene (PRPRF31) linked to autosomal dominant retinitis pigmentosa, is required for [U4/U6·U5] tri-snRNP formation and pre-mRNA splicing. EMBO J., 21, 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougin A., Gottschalk,A., Fabrizio,P., Lührmann,R. and Branlant,C. (2002) Direct probing of RNA structure and RNA–protein interactions in purified HeLa cells and yeast spliceosomal U4/U6·U5 tri-snRNP particles. J. Mol. Biol., 317, 631–649. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. (1998) RNA–RNA interactions in nuclear pre-mRNA splicing. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 279–307.
- Nottrott S., Hartmuth,K., Fabrizio,P., Urlaub,H., Vidovic,I., Ficner,R. and Lührmann,R. (1999) Functional interaction of a novel 15.5kD [U4/U6·U5] tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J., 18, 6119–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R.A. and Shukla,G.C. (2002) A revised model for U4atac/U6atac snRNA base pairing. RNA, 8, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson-Bjørn S.P., Soltyk,A., Beggs,J.D. and Friesen,J.D. (1989) PRP4 (RNA4) from Saccharomyces cerevisiae: its gene product is associated with the U4/U6 small nuclear ribonucleoprotein particle. Mol. Cell. Biol., 9, 3698–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan P.L. and Guthrie,C. (1998) RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol., 8, 847–855. [DOI] [PubMed] [Google Scholar]
- Schneider C., Will,C.L., Makarova,O.V., Makarov,E.M. and Lührmann,R. (2002) Human U4/U6·U5 and U4atac/U6atac·U5 tri-snRNPs exhibit similar protein compositions. Mol. Cell. Biol., 22, 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Stevens S.W. and Abelson,J. (1999) Purification of the yeast U4/U6·U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl Acad. Sci. USA, 96, 7226–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W.Y. and Steitz,J.A. (1997) Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci., 22, 132–137. [DOI] [PubMed] [Google Scholar]
- Teigelkamp S., Achsel,T., Mundt,C., Gothel,S.F., Cronshagen,U., Lane,W.S., Marahiel,M. and Lührmann,R. (1998) The 20kD protein of human [U4/U6·U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins. RNA, 4, 127–141. [PMC free article] [PubMed] [Google Scholar]
- Urlaub H., Hartmuth,K. and Lührmann,R. (2002) A two-tracked approach to analyze RNA–protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods, 26, 170–181. [DOI] [PubMed] [Google Scholar]
- Vidovic I., Nottrott,S., Hartmuth,K., Lührmann,R. and Ficner,R. (2000) Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell, 6, 1331–1342. [DOI] [PubMed] [Google Scholar]
- Vithana E.N. et al. (2001) A human homolog of yeast pre-mRNA splicing gene PRP31 underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol. Cell, 8, 375–381. [DOI] [PubMed] [Google Scholar]
- Watkins N.J. et al. (2000) A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- Watkins N.J., Dickmanns,A. and Lührmann,R. (2002) Conserved stem II of the box C/D motif is essential for nucleolar localisation and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenhammer E.M., Ruiz-Noriega,M. and Woolford,J.L.,Jr (1997) Prp31p promotes the association of the U4/U6·U5 tri-snRNP with prespliceosomes to form spliceosomes in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 3580–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- Williamson J.R. (2000) Induced fit in RNA–protein recognition. Nat. Struct. Biol., 7, 834–837. [DOI] [PubMed] [Google Scholar]
- Xie J., Beickman,K., Otte,E. and Rymond,B.C. (1998) Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J., 17, 2938–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Petersen-Bjørn,S. and Friesen,J.D. (1990) The PRP4 (RNA4) protein of Saccharomyces cerevisiae is associated with the 5′ portion of the U4 small nuclear RNA. Mol. Cell. Biol., 10, 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]