Abstract
Alternative splicing is a very frequent phenomenon in the human transcriptome. There are four major types of alternative splicing: exon skipping, alternative 3′ splice site, alternative 5′ splice site, and intron retention. Here we present a large-scale analysis of intron retention in a set of 21,106 known human genes. We observed that 14.8% of these genes showed evidence of at least one intron retention event. Most of the events are located within the untranslated regions (UTRs) of human transcripts. For those retained introns interrupting the coding region, the GC content, codon usage, and the frequency of stop codons suggest that these sequences are under selection for coding potential. Furthermore, 26% of the introns within the coding region participate in the coding of a protein domain. A comparison with mouse shows that at least 22% of all informative examples of retained introns in human are also present in the mouse transcriptome. We discuss that the data we present suggest that a significant fraction of the observed events is not spurious and might reflect biological significance. The analyses also allowed us to generate a reliable set of intron retention events that can be used for the identification of splicing regulatory elements.
Keywords: Alternative splicing, intron retention, transcriptome, EST
INTRODUCTION
The past few years have witnessed the emergence of a picture showing a high frequency of alternative splicing in the human transcriptome (Hanke et al. 1999; Mironov et al. 1999; Croft et al. 2000; Modrek et al. 2001; Modrek and Lee 2002; Sakabe et al. 2003). It seems that at least half of all human genes undergo alternative splicing, but the right number probably will exceed that because the detection of splicing variants of lowly expressed genes is difficult (Kan et al. 2002; Zavolan et al. 2002). The biological significance of alternative splicing is well documented for several models. A classical example is sex determination in Drosophila, in which alternative splicing acts like a genetic trigger (Baker 1989). Another classical example is the gene slo that corresponds to a calcium-activated potassium channel (Xie and Black 2001). Splice variants of this protein form a gradient in the hair cells of the inner ear of the chicken that is involved in sound detection.
Although extremely important due to their biological significance, these examples are isolated cases reflecting particular interests of individual laboratories. Due to the genomics and bioinformatics revolution, the cDNA databases remain growing at an exponential rate. This, coupled to the availability of the human genome sequence, has allowed the use of an integrated approach for the detection of splicing variants (Modrek et al. 2001; Sakabe et al. 2003). It remains doubtful however, whether a significant fraction of the events have biological significance or whether they are spurious products from the splicing machinery (Kan et al. 2002).
There are four major types of alternative splicing: exon skipping, alternative 3′ splice site, alternative 5′ splice site, and intron retention. Exon skipping seems to be the most frequent type as shown by Modrek et al. (2001). Intron retention is certainly the least studied of all types of alternative splicing, especially because these variants are believed to be largely derived from unspliced or partially spliced pre-mRNAs. There are at least a couple of cases of intron retention events with known biological consequences. The P-element transcript in Drosophila is a transposon active in the Drosophila germ line only (Rio 1991). The blocking of the transposition in somatic cells is due to a truncation of the encoded transposase caused by the retention of the third intron in the P-element transcript. The Ret tyrosine kinase also presents a couple of intron retention events, one of them generates a truncated protein that is enriched in familial and sporadic pheochromocytomas (Le Hir et al. 2002).
Here we present a large-scale analysis of intron retention for a set of 21,106 known human genes. We evaluated the distribution of intron retention events in relation to several features such as GC content, codon usage, coding potential, and conservation in mouse among others. The observed data suggest that a significant fraction of the intron retention events is not spurious and probably reflects biological significance.
RESULTS
Identification of intron retention events
Because of the large-scale nature of their generation, EST data are enriched with artifactual sequences like genomic contamination and unprocessed or partially processed mRNAs. These issues are critical for the detection of true splicing variants. The availability of an almost complete draft of the genome sequence has allowed the integration of both genomic and cDNA data to identify splicing variants (Modrek et al. 2001). We have used a similar strategy by mapping all human cDNAs onto the draft sequence of the human genome (Sakabe et al. 2003). We have also developed a clustering strategy that groups all cDNAs mapping to the same genomic region into a single cluster (see Materials and Methods for more details). A nonredundant set of 21,106 different cDNA clusters containing at least one known human full-insert cDNA (by that we mean a fully sequenced cDNA clone) was used in the analyses described below. To validate our database, we performed an analysis of exon skipping and compared our results to data previously published by different groups. We found, for instance, that 52% of all known human genes undergo exon skipping and that 77% of the events are located within the coding region. Similar numbers were found by different groups (Hanke et al. 1999; Croft et al. 2000; Modrek et al. 2001; Kan et al. 2002).
To be identified as a cDNA containing an intron retention event, a sequence was required to span the entire length of the respective retained intron. Furthermore, the same sequence had to contain at least one exon/exon boundary in order to minimize the chance of the retention being an artifact. This critical step eliminates unprocessed messages (although there is still the possibility of finding partially processed messages). The identification of an intron retention event was based on a pairwise comparison of a transcript to other sequences belonging to the same cDNA cluster. A retention event could be identified in different possible situations (Fig. 1 ▶). The prototype used to characterize the retention, spanning the respective exon/exon boundary, could be either an EST or a full-insert cDNA. The sequence containing the retained intron could also be either an EST or a full-insert cDNA. We found 3127 known genes with at least one intron retention event, which corresponds to 14.8% of the total number of known genes analyzed here. Table 1 ▶ shows the numbers of genes found for every possible case except the cases where both the prototype and the sequence containing the retention are ESTs. We decided to exclude those cases from our analysis because they involve only ESTs, which are known to be low-quality sequences. The majority of cases correspond to those pairs where the prototype is an EST and the sequence showing the retention is a full-insert cDNA. This is probably due to at least two factors: the average length of EST sequences and a bias for longer clones in the generation of full-insert cDNA sequences. A complete list of all GenBank accessions reporting an event of intron retention is available at http://www.compbio.ludwig.org.br/~pgalante/IR.
FIGURE 1.
Possible cases of intron retention. The prototype sequence and the sequence containing the retained intron can be either a full-insert cDNA or an EST. Cases in which both sequences correspond to ESTs were excluded from our data set. Exons are represented as open bars, introns as lines, and retained introns are shown as black bars (not in scale).
TABLE 1.
Number of genes containing at least one intron retention event
| Variant | |||
| Prototype | Full-insert cDNA | EST | Total |
| Full-insert cDNA | 640 | 691 (385) | 1120 |
| EST | 2594 | not analyzed | 2594 |
| Total | 2793 | 691 | 3127 |
Those cases confirmed by at least two cDNA sequences are shown in parentheses. “Total” corresponds to the nonredundant sum of the values in the corresponding row or column. The redundance is due to the fact that the same gene may have intron retention events reported by both ESTs and full-insert cDNAs (in those cases, the gene was counted only once).
Four genes reporting an intron retention event were selected for experimental validation. RT-PCR reactions were performed using primers flanking the intron being retained and primers specific to the retained sequence. Figure 2 ▶ shows the products of amplification for one of the four genes in all tissues sampled. We observed the variant containing the retained intron for all genes as well as the corresponding prototypes.
FIGURE 2.
(A) RT-PCR validation of a representative cDNA (BC004239) showing intron retention. When primers flanking the retained intron are used, two bands are visible corresponding to both variants (even numbers). If a primer specific to the intronic sequence is used, a single band corresponding to the variant with the retention is observed (odd numbers). cDNAs from colon, brain, prostate, and lung were used in the reactions. Template was excluded in the control reactions. (B) Experimental validation for all remaining genes in all tissues. (+) The retained intron was observed in the corresponding tissue. A gel containing all validation data is presented as Supplemental Figure 2, which can be found at http://www.compbio.ludwig.org.br/~pgalante/IR/
Distribution of events along transcripts
Because the presence of an intronic sequence is expected to cause a dramatic effect on the corresponding protein when it occurs within the coding sequence (CDS), we evaluated the CDS/untranslated region (UTR) distribution of all events within the data set of 640 genes where both the prototype and the sequence defining the retention event were full-insert cDNAs. For the sake of simplicity, from now on we will call these pairs of sequences the “elite group.” Because the distribution of events is primarily dependent on the frequency of introns in each part of the sequence, we first obtained the number of introns in the 5′ UTR, CDS, and 3′ UTR. This analysis was done using a set of 16,951 full-insert cDNA sequences that contained annotated 5′ and 3′ UTRs. Based on that, we calculated the number of expected intron retention events in each of the three sections of the transcript
 |
where IR is the total number of retention events, Ii is the number of introns in the respective transcript section, and I is the total number of introns in the data set, and compared that with the observed numbers of events (Table 2 ▶). We noticed a significant excess of intron retention events within both UTRs, with a corresponding decrease of observed events within the CDS (χ2 = 229, 1 degree of freedom, p < 10−49).
TABLE 2.
Number of intron retention events in the CDS and 5′ and 3′ UTRs
| Elite group | MGC | |||
| Events in | Observed | Expected | Observed | Expected |
| CDS | 287 (53%) | 502 (93%) | 87 (52%) | 155(93%) |
| 5′ UTR | 84 (15%) | 27 (5%) | 15 (9%) | 8 (5%) |
| 3′ UTR | 170 (32%) | 12 (2%) | 65 (39%) | 4 (2%) |
We considered only those cases in which both sequences correspond to full-insert cDNAs. For the statistical analysis we grouped the 5′ and 3′ UTRs. The excess of retention in both UTRs is statistically significant: elite group p = 10−49, MGC (Mammalian Gene Collection) p = 10−17. Expectation was calculated based on the density of introns in the respective transcript section (IR + Ii/I, where IR is the total number of retention events, Ii is the number of introns in the respective transcript section and I is the total number of introns in the dataset).
It is well known that the presence of premature stop codons in mRNA triggers its degradation through a process of nonsense-mediated decay (NMD; Gonzalez et al. 2000). It is, therefore, possible that the observed biased distribution for retention events in the UTRs is simply a product of NMD. This would actually suggest that the rate of intron retention is higher than observed and a fraction of the events is filtered off by NMD.
Another possibility is underreporting of sequences containing a premature stop codon. To test whether this was a factor affecting the above described distribution, we selected all cases of intron retention reported by the Mammalian Gene Collection (MGC) initiative, which reports sequences to GenBank even when they contain premature stop codons (Strausberg et al. 2002). Intron retention events reported in MGC sequences are still biased for UTRs, showing that underreporting does not seem to affect the above distribution (Table 2 ▶).
Retained introns code for protein domains
One way to assess the biological significance of an intron retention event is to evaluate its contribution to the protein domains encoded by the respective full-insert cDNA. One hundred forty-seven full-insert cDNAs from the elite group containing at least one retention event totally located in the coding region were searched against Pfam (Bateman et al. 2002). See Materials and Methods for further details. Figure 3 ▶ illustrates the different types of contribution by a given retained intron to the coding of a protein domain. Only two domains were encoded entirely by two different retained introns, out of 147. Retained introns are less efficient in coding for protein domains than exons (30 exons, out of 830, encode an entire domain in the same set of full-insert sequences). There is no statistical difference between the numbers obtained for retained introns and exons (Fisher’s exact test, p = 0.21). However, retained introns are more efficient than nonretained introns present in the same set of full-insert cDNAs (no nonretained intron, out of 785, encodes an entire domain). This difference is statistically significant (Fisher’s exact test, p < 0.026).
FIGURE 3.
Contribution of retained introns to the coding of protein domains. A protein domain can be either entirely encoded by a retained intron (AF267861) or partially encoded by a retained intron (AJ005891 and AF230393). In the sequence AF267861, the retained intron encodes the entire “Elongation factor Tu” domain. In the sequence AJ005891, the entire retained intron codes for part of the “Fork head” domain. In the sequence AF230393, part of the retained intron codes for part of the SPRY domain.
Furthermore, if the retained intron was joined to the two flanking exons (exon/retained intron/exon), the number of domains entirely encoded by such segments increased to 15 (a hit was considered significant when at least 30% of the domain was encoded by the retained intron). For the 13 cases in which the intron partially encodes a domain, we tested if the exclusion of the sequence corresponding to the retained intron affected the ability to detect the correct domain. We performed a manual inspection of the protein domains encoded by a modified version of the intron retention variant (without the amino acids encoded by the retained intron). In six of these sequences, the domain encoded with the contribution of the retained intron was not found and in six others the domain was found with scores considerably lower and with e-values increased by orders of magnitude. In only one case could we not detect a substantial change in the scores after deleting the intronic sequence. This observation suggests that the amino acids encoded by the retained introns are a relevant part of a structural unit.
In addition, we found 24 other cases where the domain was longer than the “exon/retained intron/exon” unit (i.e., the domain was only partially coded by this unit) with a significant contribution of the intronic sequence (at least 30% of the domain). Thus, 39 out of 147 retained introns (26%) participated in the coding of a protein domain.
As an additional control, we tested if the retained introns located in the UTRs would participate in the coding of protein domains. None of the 762 amino acid sequences (254 UTR-retained introns translated in three phases) showed a complete or partial match against a domain in Pfam (Fisher’s exact test, p < 10−17 when compared to 39 cases in 147 retained introns in the CDS).
Selection on intron sequences involved in retention events
To evaluate whether selection for coding potential was acting on intron sequences involved in retention events, we analyzed those introns from our elite group located within the CDS (287 introns as shown in Table 2 ▶). We first compared the frequencies of all four bases in the set of introns involved in retention events to the set containing all non-retained introns. Because it has been shown that shorter introns have a higher GC content in humans (Lander et al. 2001), we divided both data sets into length categories. A significant difference (χ2 = 2276, 6 degrees of freedom, p < 10−50) is observed between retained (higher GC content) and nonretained introns (Fig. 4 ▶). Figure 4 ▶ also shows that the GC distribution for retained introns is similar to the GC distribution for their flanking exons (p = 0.23, χ2 = 7.9, 6 degrees of freedom). The distribution observed for both exons and nonretained introns is in accordance with Lander et al. (2001).
FIGURE 4.
Nucleotide composition of retained and nonretained introns and exons bordering nonretained introns, all of them located within CDS. Because nucleotide composition varies in introns of different length in humans, all comparisons were performed within specific length classes. Retained introns have a higher GC content in all length categories when compared to nonretained introns (p < 10−50). There is no difference in the distribution of retained introns and their bordering exons (p = 0.23).
The frequency of stop codons in retained introns is lower than expected. Based on the nucleotide frequencies observed for retained introns located within the CDS, the expected number of stop codons is 1064 in 26,941 codons (see Materials and Methods). The observed number of stop codons (651) was significantly smaller than expected (χ2 = 103, 1 degree of freedom, p < 10−25). We found 88 full-insert cDNAs, out of 287, in which the retention event generated a putative truncated protein due to the presence of a premature stop codon within the retained intron (available at http://www.compbio.ludwig.org.br/~pgalante/IR). The identification of 88 cases in which the retention generates a putative truncated protein allowed us to examine the GC content within the retained intron for sequences upstream of and downstream from the premature stop codon. It is expected that selection would be relaxed on intronic sequences downstream from the premature stop codon. The GC content for sequences upstream of and downstream from the premature stop codon is 58% and 49%, respectively.
Codon usage in retained introns, exons, and nonretained introns
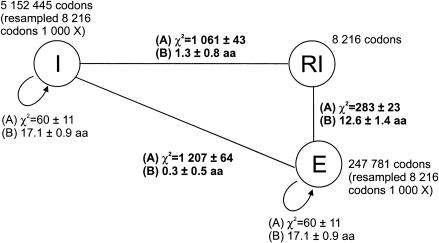
Codon usage in retained introns, exons, and nonretained introns was evaluated as described in Materials and Methods. Two strategies were used to compare the different data sets. The first strategy compares the whole codon usage table (61 codons) of the three data sets (results are labeled A in Fig. 5 ▶). The second strategy is based on the calculation of the average number of amino acids using the same codons in the three different data sets (results are labeled B in Fig. 5 ▶). Both approaches show that retained introns are more similar to exons than to nonretained introns.
FIGURE 5.
Similarity of codon usage in introns (I), exons (E), and retained introns (RI). Codon usage of retained introns is considerably more similar to exons than to that of nonretained introns as evaluated by two different measurements: (A) the mean χ2 value for the comparison of all 61 codons at once in 1000 simulations; (B) the average number of amino acids with similar codon usage frequency. Curved arrows represent internal variations within each subset.
Conservation of intron retention in mouse cDNA sequences
To assess if events of intron retention found in the human transcriptome are conserved in the mouse transcriptome we aligned human full-insert cDNAs presenting intron retention (elite group with at least one mouse cDNA hit) to mouse full-insert cDNAs and ESTs using BLAST 2.0 (Altschul et al. 1997) with an e-value threshold of e−10. We found that 38%–57% of all retained introns (depending on the UTR/CDS localization) present a mouse hit (Table 3 ▶, first line). The sequence identity of orthologous retained introns is 84%, significantly higher than the average identity found for orthologous nonretained introns (60%) and similar to the average degree of identity found for orthologous exons (87%). These numbers are similar to those observed by Waterston et al. (2002). Interestingly, we found only three cases of retained intron in mouse with a premature stop codon, suggesting that the putative retention in mouse preserved the open reading frame.
TABLE 3.
Intron retention events in human (detected in the elite group with annotated CDS) that also occur in mouse
| 5′ UTR | CDS | 3′ UTR | |
| Intron retention in human with mouse cDNA hits | 9/21 (43%) | 74/130 (57%) | 15/39 (38%) |
| Mouse clusters with more than one cDNA in the region | 1/9 | 40/74 | 5/15 |
| Conserved intron retention event | 0/1 | 9/40 | 1/5 |
We performed the analysis separately for CDS and UTR. The first row corresponds to the results of searching the retained introns in human against mouse cDNAs (ESTs and full-insert cDNA). The second row defines the cases that have more than one mouse cDNA in the region of the retained intron (these cases are informative for seeking an intron at the genome level). The third row corresponds to those cases defined in the second row where an intron is observed at the genome level. See text for more details.
To verify if the mouse cDNA also corresponds to an intron retention variant, we searched for at least one other cDNA that could define the corresponding intron at the genome level. For many examples, the region containing the retained intron was not covered by any other cDNA sequence, thus not allowing the identification of an intron in that region (second line of Table 3 ▶). An intron retention in human is deemed “conserved” in mouse if a mouse hit covers the retained intron in human and if there is also evidence that the retained sequence is also an intron in the mouse genome. We found 10 cases, out of 46, of conserved intron retention. This number is certainly an underestimate because the prototype may not have been sequenced and all cDNA sequences covering that region may correspond to the variant showing the retention, as also pointed out by Thanaraj et al. (2003).
DISCUSSION
One could expect that the splicing process has an intrinsic error rate that is difficult to estimate. It is reasonable to suppose that at least a fraction of all splicing variants found in the cDNA databases correspond to artifacts caused by such errors. This is even more pronounced nowadays as databases are reaching a critical in-depth sampling. Thus, an important question refers to the amount of artifactual variants identified by the use of cDNA databases. Although valid for all types of alternative splicing, this seems to be more critical for intron retention, because unprocessed or partially processed messages would be easily detected as variants.
Kan et al. (2002) found an intron retention rate of around 35%. They then used some statistics to infer the reliability of a given splicing variant. They found, for instance, that less than 5% of all genes exhibited intron retention at a 95% confidence interval (p < 0.05). We have found that 14.8% of all genes sampled here exhibited putative events of intron retention. This discrepancy may be due to the fact that we have not applied any statistics in our data set and/or to differences in the data sets used. On the other hand, we considered only sequences showing evidence of splicing to exclude totally unprocessed transcripts, and most of our analyses were performed on an elite group. Interestingly, if we consider the elite group only, we find evidence of intron retention in 4.6% of the whole data set used (640 genes with intron retention divided by 13,841 genes with at least two full-insert cDNAs).
An important aspect of our analysis is the apparent non-random distribution of intron retention events regarding some specific features. The rate of intron retention is more pronounced in the UTRs as clearly shown in Table 2 ▶. There is a threefold and 14-fold excess (above the expectation) of events in the 5′ and 3′ UTRs, respectively. Much of the bias is probably due to NMD that would trigger the degradation of transcripts bearing premature stop codons. Although this would explain the lower number of events within the CDS, it does not explain the different rates of intron retention between the 5′ and 3′ UTRs. This bias toward the 3′ UTR could be explained by the direction of RNA synthesis, because splicing seems to be coupled to transcription (for review, see Black 2003) and therefore the last intron would have less time available to be spliced out. However, for 90% of the retained introns, we were able to find at least one exon/exon boundary downstream from the retained intron, suggesting that the last intron was spliced as efficiently as the first introns in the transcript. One possible explanation implying biological significance is an effect of the retained intron on mRNA stability. Cis-acting elements present in the 3′ UTR are known to alter the stability of mRNAs (Bashirulla et al. 2001), making them either more or less stable. The presence of such elements in the retained introns would affect the stability of that variant transcript.
Stamm et al. (2000) observed that retained introns are significantly shorter than nonretained introns. We observed the same trend in our data set (see Supplemental Fig. 1 ▶ at http://www.compbio.ludwig.org.br/~pgalante/IR/). However, the interpretation that this feature is related to biological significance is not straightforward because the retention of very long introns is unlikely to occur. Furthermore, other factors like cloning efficiency could contribute to the biased length distribution observed for retained introns.
Some analyzed features show that the retained introns are considerably more similar to exons than to other introns. The GC content of such sequences is higher than that of nonretained introns in the same size range and is similar to exons. Introns with a higher GC content might have a lower excision rate as demonstrated by Goodall and Filipowics (1991) in dicots. This phenomenon, however, does not seem to exist for other organisms, including vertebrates (Goodall and Filipowics 1989). The codon usage follows the same trend and is overall much more similar to that of exons than to introns. Likewise, retained introns encode more domains than introns but less than exons, showing that the intronic sequence is a relevant part of a larger functional unit, even when it is not coding the entire domain.
These aspects show that the pattern emerging from the data set of variants is not random and therefore can be interpreted as having some biological significance. We cannot rule out, however, that unknown properties of the splicing process would generate such a bias in the pattern of intron retention events.
We have also found evidence that retained introns within the CDS are under selective pressure for coding potential. Not only is the GC content different from that of introns, as discussed above, but also the frequency of stop codons is considerably lower than random expectation. It should be noted, however, that the reduced frequency of stop codons can also be explained by NMD. Other evidence of selective pressure is that a fraction of introns being retained in human is also retained in mouse, presenting an average sequence identity comparable to orthologous exons (83%). As observed by Thanaraj et al. (2003) the presence of a same variant in two species is strongly affected by the cDNA representation of that gene in both transcriptomes. Therefore, it is reasonable to assume that the observed rate of cooccurrence is an underestimate.
As noted originally by Gilbert (1978), and recently stressed by Black (2003), new variants are expected to correspond to a small fraction of all transcripts from a given gene. Mutations that affect splicing can allow the production of new proteins without the loss of the original one. If these new splicing variants are spurious products of the splicing reaction, filtering selection would be relaxed on the machinery that generates them, because they correspond to a small fraction of all transcripts and the original version is maintained. On the other hand, if the new variants have some biological significance, it is expected that selection will act on them to maintain the new biological function. Thus, the observed signals of selection support a biological role for the retained introns.
We have, however, identified 88 cases in which the intron retention event generates a putative truncated protein. For 40% of these cases the premature stop codon was still located in the 3′-most exon, which can explain why they were not filtered out by NMD. Among these 88 cases there are genes that seem to be involved in several types of syndromes such as Williams Beuren syndrome, chromosome 22 region, and the Betten, Spielmeyer-Vogt disease gene. Furthermore, there are several genes with retained introns clearly related to the tumorigenic process like p19A, kallikrein 3 and 4, TNF receptor, BCL2-like 11, and CDC2-like 10, among others. Further analyses are needed to explore the possibility of an association between the splicing variants and any feature of these diseases.
For those cases showing a putative premature stop codon, the GC content is higher in the intronic sequence upstream of the stop codon (58%) than in the downstream sequences (49%). This reinforces the notion that selection is acting on the retained introns.
It is important to stress that our results do not exclude the possibility that a fraction of intron retention events is indeed artifactual. The data indicate, however, that this fraction is not sufficiently large to eliminate signals that can be interpreted as products of events with biological significance. We make available subsets of retained introns that are more likely to be free of artifacts (www.compbio.ludwig.org.br/~pgalante/IR). These include, for instance, a set of 385 cases of intron retention that are confirmed by at least two cDNA sequences. These sets might be useful for the identification of putative regulatory elements.
The definition of introns and exons may become fuzzy when we take into account intron retention. One could establish two requirements for a genomic sequence to be considered an exon: (1) It should be present in the mature mRNA, and (2) it should be flanked by sequences excised at the RNA level. Because the retained introns identified in this study (which seem to be under selection) violate the second requirement, they should be considered introns subjected to regulated alternative splicing. This raises the question of whether retained introns are portions of exons becoming introns or introns being incorporated into exons. The analysis performed here cannot answer this question. Expression data coupled to more complex interorganism comparisons could help to elucidate this issue.
Finally, regarding the mechanistic nature of the observed retention events, it is possible that the flanking exons or the retained introns themselves bear weak splice sites and/or do not contain proper splicing regulatory elements. An analysis of splice sites and regulatory elements currently underway will hopefully shed some light on this subject.
MATERIALS AND METHODS
cDNA mapping and clustering
All human cDNAs available in dbEST (July 2002, Boguski et al. 1993) and mRNA sequences from known human genes from Uni-Gene release 153 (Schuler et al. 1996) were aligned to the masked human genome sequence (build 29, obtained from NCBI) by using pp-Blast (Osorio et al. 2003), an implementation of MEGA-BLAST (Zhang et al. 2000) for a parallel cluster. The parameters used in MEGABLAST were: -f T -J F -F F -W 24. The MEGA-BLAST output was parsed and a MySQL database was loaded with the mapping information. Spurious hits were excluded from the mapping database by using an additional set of alignment criteria. These include a minimum degree of identity for a cDNA/genome alignment set to 93% over at least 45% of the total EST length or 55% of the total length of the full-insert sequence. Furthermore, sequences mapping to more than one location on the genome were given a score for alignment quality. Higher score was associated with a higher identity over a longer alignment. Only the sequences with the highest scores were used in the analyses reported here. Clustering of cDNA sequences was based on their genomic coordinates as described by Sakabe et al. (2003). Briefly, if two sequences shared at least partially the same gene structure they were joined into the same cluster. If no exon/intron boundary was defined, a sequence had to have at least a 100-bp overlap with another sequence at the genome level to be added to the respective cluster.
Identification of intron retention events
A set of 21,106 distinct cDNA clusters containing at least one known full-insert cDNA sequence was used in the present analysis. Intron retention events were characterized by the presence of at least one cDNA sequence in which the corresponding intron was not spliced out. We considered valid only those cases in which the sequence characterizing the retention also defined at least one more intron that was not retained. This excludes from our analysis artifacts derived from unprocessed messages.
Distribution of events along transcripts
We selected from the 21,106 full-insert cDNAs those sequences (16,951) containing an annotated CDS. We then mapped all retention events to these sequences. For this analysis, we have only used those cases in which both the prototype and the sequence containing the retained intron were full-insert cDNAs to avoid any positional bias that could be introduced by using 3′ and 5′ ESTs. A retention was considered to be located within a given transcript section if the 5′ end of the intron was mapped to that section. In a few cases in which the presence of the intron generated a stop codon, the 3′ end of the intron would be located in the 3′ UTR whereas the 5′ end would be located in the CDS.
Frequency of stop codons
The number of observed stop codons in the set of retained introns (287 sequences with 26,941 codons from the elite group) was counted considering codons in frame with the upstream exon. The number of expected stop codons is ((f(T) × f(G) × f(A) + f(T) × f(A) × f(A) + f(T) × f(A) × f(G)) × number of codons in the data set) where f(T) is the frequency of T in the data set and so on.
Codon usage in nonretained introns, exons, and retained introns
Codon usage was determined for those cases belonging to the elite group. The exons and nonretained introns used as a control came also from the elite group to avoid any possible bias in the selection of genes. In the case of both retained and nonretained introns, the frame was defined by the frame of the upstream exon. To evaluate the similarity in codon usage in the three sets we made two distinct measurements. The first was a comparison of the entire codon usage table (61 codons, 60 degrees of freedom) among the three sets. We performed 1000 pairwise comparisons of the random distributions of 8216 codons (the number of codons in the set of retained introns) along the 61 possible codons to calculate the average χ2 values (labeled A in Fig. 5 ▶) and standard deviation. Internal variations within each set (curved arrows in Fig. 5 ▶) were also calculated as an internal control. The second measurement refers to the number of amino acids that presented similar codon usage (χ2 tests at p ≤ 0.05; labeled B in Fig. 5 ▶). As methionine and tryptophan have only one codon, they were excluded from this last analysis. Again 1000 random sets were built from each set and pairwise compared, grouped by amino acid. The numbers of amino acids with p values <0.05 in χ2 tests were counted and presented in Figure 5 ▶.
Analysis of protein domains encoded by retained introns
Amino acid sequences of full-insert cDNAs, belonging to the elite group, with a retained intron located entirely in the CDS were submitted to domain search in Pfam 9.0 (Bateman et al. 2002) using the program hmm in a GeneMatcher hardware (Paracel Inc). Retained introns smaller than 99 bp and longer than 990 bp were excluded from this analysis. The final number of sequences submitted to Pfam was 147. To be considered a significant hit, the retained intron had to encode at least 30% of the protein domain and have an e-value <10−1. The coordinates of the domains were then converted to their cDNA positions. The cDNA coordinates of retained introns or exons (control set) entirely in the CDS were then compared to domain coordinates and grouped in those matching a whole domain or part of it. Domains were considered different in a given protein sequence when they presented distinct annotation and coordinates. For the comparison of exons, introns, and retained introns, domain search was performed with each exon or intron (of the full-insert cDNAs with intron retention) being a separate query. Only sequences with similar lengths were analyzed (<330 and >33 amino acids).
Experimental validation
The four genes tested by RT-PCR are: (1) BC015569—similar to SRp25 nuclear protein, (2) BC004239—jumping translocation breakpoint, (3) BC025673—similar to transcription elongation factor B (SIII), and (4) BC011903—similar to the small subunit of calpain 4. Reverse transcription was carried out using the Superscript First Strand Synthesis Kit, according to the manufacturer’s instructions (Life Technologies). The cDNA was synthesized by incubating 2 μg of either brain, lung, prostate, or breast total RNA in 20 μL of 1× reverse transcriptase buffer containing 0.5 μg oligo dT primer, 0.5 mM dNTP, 5.0 mM MgCl2, 10 μM DTT, 80 U ribonuclease inhibitor, and 200 U Superscript II reverse transcriptase at 42°C for 1 h. The quality of total RNA was tested by PCR using MLH1 primers located at intronic sequences flanking exon 12 (forward: 5′-TGGTGTCTCTAGTTCTGG-3′, and reverse: 5′-CATTGTTGTAGTAGCTCTGC-3′), as an indicator of possible genomic DNA contamination. Three specific primers were designed for each intron retention event: two primers annealing to the flanking exons of the retained intron and one additional primer annealing to the retained intron itself. RT-PCR reactions were carried out in a 25-μL reaction mixture containing 1 μL of cDNA, 1× Taq DNA polymerase buffer, 1 μL of dNTP 2.5 mM, 6 pmoles of primers, 0.5 μL of MgCl2 50 mM, and 1 unit Taq DNA polymerase (GIBCO/BRL). Standard PCR conditions were: 4 min at 94°C (initial denaturation), 45 sec at 94°C, 45 sec at 58°C, and 1 min at 72°C for 40 cycles and a final extension step of 10 min at 72°C. PCR products were analyzed both on 8% silver-stained polyacrylamide gels and on ethidium bromide agarose gel.
Acknowledgments
We thank Maria D. Vibranovski, Anamaria A. Camargo, and Ricardo R. Brentani for critical reading of the manuscript. N.J.S. is supported by a Ph.D. fellowship from Fapesp. N.K.S. is supported by a Ph.D. fellowship from Capes. The authors are also indebted to two anonymous reviewers for excellent comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5123504.
REFERENCES
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, B.S. 1989. Sex in flies: The splice of life. Nature 340: 521–524. [DOI] [PubMed] [Google Scholar]
- Bashirullah, A., Cooperstock, R.L., and Lipshitz, H.D. 2001. Spatial and temporal control of RNA stability. Proc. Natl. Acad. Sci. 98: 7025–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291–336. [DOI] [PubMed] [Google Scholar]
- Boguski, M.S., Lowe, T.M., and Tolstoshev, C.M. 1993. dbEST—Database for “expressed sequence tags.” Nat. Genet. 4: 332–333. [DOI] [PubMed] [Google Scholar]
- Croft, L., Schandorff, S., Clark, F., Burrage, K., Arctander, P., and Mattick, J.S. 2000. ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nat. Genet. 24: 340–341. [DOI] [PubMed] [Google Scholar]
- Gilbert, W. 1978. Why genes in pieces? Nature 271: 501. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C.I., Ruiz-Echevarria, M.J., Vasudevan, S., Henry, M.F., and Peiltz, S.W. 2000. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 5: 489–499. [DOI] [PubMed] [Google Scholar]
- Goodall, G.J. and Filipowics, W. 1989. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58: 473–483. [DOI] [PubMed] [Google Scholar]
- ———. 1991. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J. 10: 2635–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke, J., Brett, D., Zastrow, I., Aydin, A., Delbruck, S., Lehmann, G., Luft, F., Reich, J., and Bork, P. 1999. Alternative splicing of human genes: More the rule than the exception. Trends Genet. 15: 389–390. [DOI] [PubMed] [Google Scholar]
- Kan, Z., States, D., and Gish, W. 2002. Selecting for functional alternative splices in ESTs. Genome Res. 12: 1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Bald-win, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Charlet-Berguerand, N., de Franciscis, V., and Thermes, C. 2002. 5′ end Ret splicing: Absence of variants in normal tissues and intron retention in pheocromocytomas. Oncology 63: 84–91. [DOI] [PubMed] [Google Scholar]
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9: 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic view of of alternative splicing. Nat. Genet. 30: 13–19. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29: 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio, E., de Souza, J.E., Zaiats, A.C., de Oliveira, P.S.L., and de Souza, S.J. 2003. pp-Blast: A “pseudo-parallel” Blast. Braz. J. Med. Biol. Res. 36: 463–464. [DOI] [PubMed] [Google Scholar]
- Rio, D.C. 1991. Regulation of Drosophila P-element transposition. Trends Genet. 7: 282–287. [DOI] [PubMed] [Google Scholar]
- Sakabe, N.J., de Souza, J.E., Galante, P.F.A., de Oliveira, P.S.L., Passetti, F., Brentani, H., Osorio, E.C., Zaiats, A.C., Leerkes, M.R., Kitajima, J.P., et al. 2003. ORESTES are enriched in rare exon usage variants affecting the encoded protein. C. R. Biol. 326: 979–985. [DOI] [PubMed] [Google Scholar]
- Schuler, G.D., Boguski, M.S., Stewart, E.A., Stein, L.D., Gyapay, G., Rice, K., White, R.E., Rodriguez-Tome, P., Aggarwal, A., Bajorek, E., et al. 1996. A gene map of the human genome. Science 274: 540–546. [PubMed] [Google Scholar]
- Stamm, S., Zhu, J., Nakai, K., Stoilov, P., Stoss, O., and Zhang, M.Q. 2000. An alternativeexon database and its statistical analysis. DNA Cell Biol. 19: 739–756. [DOI] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Grouse, L.H., Derge, J.G., Klausner, R.D., Collins, F.S., Wagner, L., Shenmen, C.M., Schuler, G.D., Altschul, S.F., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 99: 16899–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj, T.A., Clark, F., and Muilu, J. 2003. Conservation of human alternative splice events in mouse. Nucleic Acids Res. 31: 2544–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Xie, J. and Black, D.L. 2001. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature 410: 936–939. [DOI] [PubMed] [Google Scholar]
- Zavolan, M., van Nimwegen, E., and Gaasterland, T. 2002. Splice variation in mouse full-length cDNAs identified by mapping to the mouse genome. Genome Res. 12: 1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. 2000. A greedy algorithm for aligning DNA sequences. J. Comp. Biol. 7: 203–214. [DOI] [PubMed] [Google Scholar]