Abstract
The Saccharomyces cerevisiae WD-40 repeat protein Swd2p associates with two functionally distinct multiprotein complexes: the cleavage and polyadenylation factor (CPF) that is involved in pre-mRNA and snoRNA 3′ end formation and the SET1 complex (SET1C) that methylates histone 3 lysine 4. Based on bioinformatic analysis we predict a seven-bladed β-propeller structure for Swd2p proteins. Northern, transcriptional run-on and in vitro 3′ end cleavage analyses suggest that temperature sensitive swd2 strains were defective in 3′ end formation of specific mRNAs and snoRNAs. Protein–protein interaction studies support a role for Swd2p in the assembly of 3′ end formation complexes. Furthermore, histone 3 lysine 4 di-and tri-methylation were adversely affected and telomeres were shortened in swd2 mutants. Underaccumulation of the Set1p methyltransferase accounts for the observed loss of SET1C activity and suggests a requirement for Swd2p for the stability or assembly of this complex. We also provide evidence that the roles of Swd2p as component of CPF and SET1C are functionally independent. Taken together, our results establish a dual requirement for Swd2p in 3′ end formation and histone tail modification.
Keywords: 3′ end formation, RNA polymerase II transcription termination, snoRNAs, histone methylation, WD-40 repeat
INTRODUCTION
Eukaryotic genomes encode different classes of genes, which are transcribed by RNA polymerase II (RNAP II). The major class of protein encoding genes serves as templates for the synthesis of messenger RNA precursors (pre-mRNAs). A second class of RNAP II transcripts are derived from small nucleolar RNA (snoRNA) genes and function as components of ribonucleoprotein particles in pre-rRNA processing and ribosomal RNA modification (Kiss 2002). All known primary transcripts of RNAP II require processing to be biologically functional. For pre-mRNAs this includes capping, splicing, and 3′ end formation. snoRNA transcripts generally have to be processed by exonucleolytic trimming to generate the mature RNA.
Pre-mRNA 3′ end formation requires site-specific endonucleolytic cleavage of the RNA, followed by polyadenylation of the upstream cleavage product (for review, see Zhao et al. 1999; Edmonds 2002). In Saccharomyces cerevisiae, biochemical fractionation of whole-cell extracts initially defined the cleavage factors IA and IB (CF IA and CF IB) and the cleavage factor II (CF II) as sufficient to catalyze the cleavage step (Chen and Moore 1992); polyadenylation was suggested to require CF IA, polyadenylation factor I (PF I), and poly(A) polymerase (Pap1p). Subsequently, it was found that CF II, PF I, and Pap1p are associated in vivo in the so called cleavage and polyadenylation factor CPF (Ohnacker et al. 2000). The polypeptide composition of these factors is complex (see Fig. 4D ▶ below). CF IA is a heterotetrameric protein (Kessler et al. 1996; Minvielle-Sebastia et al. 1997) and CPF consists of at least 15 different polypeptides (Dichtl et al. 2002b; Gavin et al. 2002; Dheur et al. 2003; He et al. 2003; Nedea et al. 2003). Seven CPF subunits (Pta1p, Ref2p, Pti1p, Swd2p, Ssu72p, Syc1p, and Glc7) have been suggested to form the APT subcomplex (associated with Pta1p) that together with core-CPF (previously called PF I) forms holo-CPF (Nedea et al. 2003).
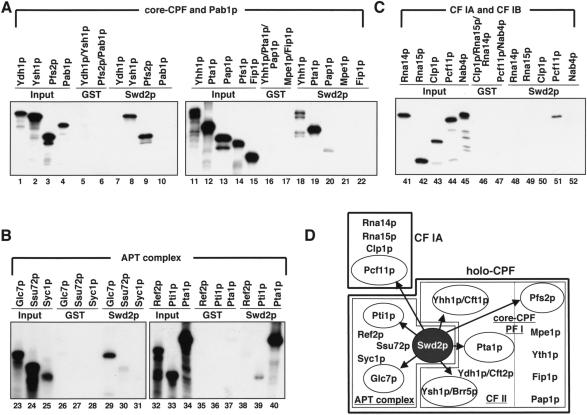
FIGURE 4.
Swd2p bridges core-CPF, APT, and CF IA subcomplexes of the 3′ end formation machinery. (A–C) Pull-down experiments with 1 μg GST or GST-Swd2p (Swd2p) and [35S]-methionine-labeled proteins as indicated. Also indicated is the association of tested subunits with core-CPF (A), the APT complex (B), or CF I (C). Input lanes show 10% of in vitro translation reactions included in binding reactions. Note that the band visible in the Pap1p lane (lane 20) does not correspond to full-length protein. (D) Schematic representation of subcomplexes and individual protein subunits of the yeast 3′ end formation machinery. Abbreviations for subcomplexes are underlined. The CF IA and holo-CPF complexes constitute most components required for 3′ end formation in vitro. Holo-CPF was suggested to contain core-CPF (previously called PF I) and APT subcomplexes (Nedea et al. 2003). Core-CPF includes the smaller CF II complex as indicated by a dashed line. Polypeptides interacting with GST-Swd2p in vitro are encircled and indicated by arrows.
Maturation of independently transcribed snoRNAs requires the production of an entry site for 3′ → 5′ exonucleases that eventually form the correct 3′ end. In one group of snoRNAs this entry site is produced by Rnt1p (yeast RNase III), which recognizes specific RNA hairpin structures (Chanfreau et al. 1998). A second group of snoRNAs, which lacks Rnt1p recognition sites, requires endonucleolytic cleavage by the 3′ end formation machinery uncoupled from the polyadenylation step (referred to as polyadenylation-independent 3′ end formation; Fatica et al. 2000). The latter genes carry specific cis-acting elements in proximity of the cleavage site (Morlando et al. 2002; Steinmetz and Brow 2003) that differ from typical polyadenylation signals found in mRNAs (Graber et al. 1999; van Helden et al. 2000). Cleavage uncoupled from polyadenylation requires components of CF IA (Fatica et al. 2000; Morlando et al. 2002) and certain subunits of CPF, which may all be contained within the APT complex (Dheur et al. 2003; Ganem et al. 2003; Nedea et al. 2003; Steinmetz and Brow 2003). Strikingly, mutations in core-CPF subunits have been found not to affect polyadenylation-independent 3′ end formation (Morlando et al. 2002). In addition, the CTD interacting protein Nrd1p and the RNA binding protein Nab3p have been shown to be involved in this process (Steinmetz et al. 2001).
Progress in the fields of RNAP II transcription and RNA processing revealed that these processes are intimately connected (for recent reviews, see Bentley 2002; Neugebauer 2002; Proudfoot et al. 2002). In vivo 3′ end formation is coupled to transcription by RNAP II (Proudfoot et al. 2002). Like other pre-mRNA processing factors, the 3′ end formation machinery is thought to associate with the carboxy terminal domain (CTD) of the largest subunit of RNAP II during transcription elongation (Proudfoot and O’Sullivan 2002). Site-specific phosphorylation and dephosphorylation of the CTD controls binding of CTD interacting proteins and integrates pre-mRNA processing with the transcription cycle. Transcription termination depends on functional polyadenylation signals and the activity of certain pre-mRNA processing factors (Proudfoot and O’Sullivan 2002). Several studies indicate that 3′ end formation factors can act during termination independent of their roles in cleavage and polyadenylation (Osheim et al. 1999; Tran et al. 2001; Orozco et al. 2002; Osheim et al. 2002; Kim and Martinson 2003; Sadowski et al. 2003).
Posttranslational modifications of histone tails play important roles in the regulation of gene expression (Jenuwein and Allis 2001). Modifications include phosphorylation, acetylation, ubiquitination, and methylation and their combinatorial occurrence was suggested to translate into a histone code (Jenuwein and Allis 2001; Turner 2002). The S. cerevisiae genome encodes six SET domain proteins (for review, see Kouzarides 2002). Of those, Set1p is associated with seven additional proteins (Bre2p, Swd1p, Swd2p, Swd3p, Sdc1p, Spp1p, Shg1p) in SET1C and methylates H3K4 (Briggs et al. 2001; Miller et al. 2001; Roguev et al. 2001; Krogan et al. 2002; Nagy et al. 2002). H3K4 tri-methylation is associated with actively transcribed genes (Santos-Rosa et al. 2002) and was suggested to act as cellular memory for recent gene expression (Krogan et al. 2003; Ng et al. 2003).
Here, we analyzed S. cerevisiae Swd2p that is physically associated with both CPF and SET1C. We provide evidence that Swd2p is required for 3′ end formation of specific mRNAs and snoRNAs. Furthermore, the protein is necessary for SET1C methyltransferase activity on H3K4.
RESULTS
Swd2p carries seven WD-40 repeat motifs and is conserved within eukaryotes
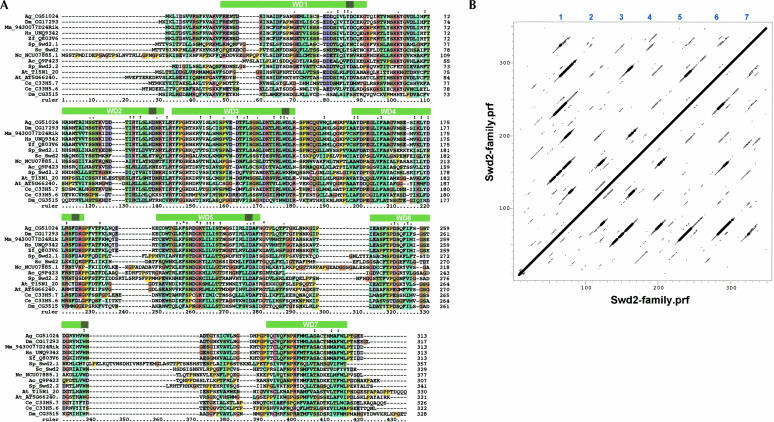
Proteomic analysis of polypeptides associated with CPF and SET1 revealed Swd2p as a common component of both complexes (Miller et al. 2001; Roguev et al. 2001; Dichtl et al. 2002b; Nagy et al. 2002; He et al. 2003). We searched databases and identified Swd2p homologs in a large number of eukaryotes (Fig. 1A ▶; data not shown; see Materials and Methods). Schizzosaccharomyces pombe, Arabidopsis thaliana, Caenorhabditis elegans, and Drosophila melanogaster have two Swd2p homologs each whereas most other species have only one. It should be noted, however, that the Swd2 family is not sharply delineated from the larger superfamily of WD-40 proteins and we cannot rule out that more distantly related proteins also belong to the Swd2 family. Standard protein motif prediction tools (SMART, PFAM; see Materials and Methods) detected up to three WD-40 repeat sequences in Swd2p (repeats 3, 5, and 6 in Fig. 1A ▶). WD-40 repeat proteins form a large protein family with diverse biological functions (Smith et al. 1999). The majority of these proteins form seven-bladed β-propeller-like structures although structures with four, five, and six blades have also been described. Because many WD-40 repeats are poorly predicted with the Pfam and SMART tools, we subjected the Swd2 family to sensitive profile–profile dot plots (Thompson et al. 1994). As shown in Figure 1B ▶, there are six distinct tiers of off-diagonal signals, strongly suggesting that the Swd2 family has a seven-bladed β-propeller structure.
FIGURE 1.
Swd2p carries seven WD repeats and is conserved within eukaryotes. (A) Multiple sequence alignment of a representative set of the Swd2 family. The sequences are: Ag_CG51024: Anopheles gambiae (tr:Q7Q1N9); Dm_CG17293 and Dm_CG3515: Drosophila melanogaster (tr:Q9VLN1 and Q9VQD1); Mm_9430077D24Rik: Mus musculus (tr:Q8BFQ4); Hs_UNQ9342: Homo sapiens (trnew:AAQ88631); Zf_Q803V6: Danio rerio (Q803V6); Sp_Swd2.1 and Sp_Swd2.2: S. pombe (tr:O60137 and tr:Q9UT39); Sc_Swd2: S. cerevisiae (sw:P36104); Nc_NCU07885.1: Neurospora crassa (tr:Q7SB08); Ac_Q9P423: Ajellomyces capsulata (tr:Q9P423); At_T15N1_20 and At_AT5G66240: Arabidopsis thaliana (tr:Q9LYK6 and tr:Q8RXD8); Ce_C33H5.7 and Ce_C33H5.6 Caenorhabditis elegans (tr:Q18403 and Q18404); the latter sequence was repredicted from the genomic sequence as described in Materials and Methods. The databases are: tr: trembl; trnew: trembl_new, and sw: SwissProt. Not included is an additional human sequence (GenBank: LOC131060), which is virtually identical to Hs_UNQ9342. It is not clear if these two sequences derive from the same gene. The first 43 residues of Ag_CG51024 and the last 72 residues of Dm_CG3515 are not shown. The approximate positions of seven predicted WD-40 repeats (see B) are indicated with green bars above the sequence. The darker green squares indicate the position of the sequence most likely to be the “WD” motif. (B) A profile–profile dot plot using a profile generated from the alignment in A shows six off-diagonal lines indicating that the family has seven WD-40 repeats. The plot was generated with a window of 17 and a dot-depth of 5. Note that the rulers on the axes are not identical to the ruler in A because gapped regions are excluded in the profile.
Swd2p is required for 3′ end formation of specific mRNAs and snoRNAs
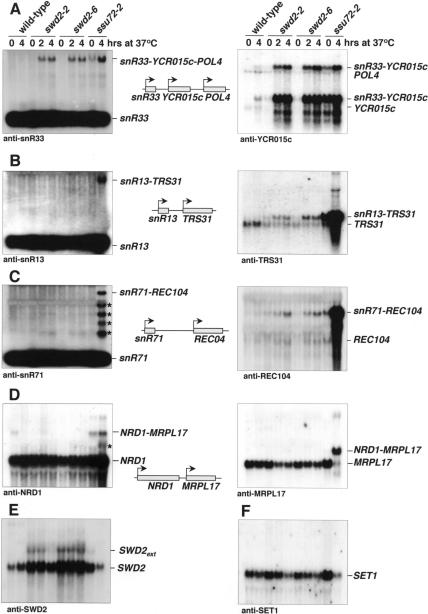
To functionally analyze SWD2 we generated temperature-sensitive alleles (see Materials and Methods). Subunits of yeast CPF have been implied in transcription termination at protein coding genes and snoRNA genes. The association of Swd2p with CPF suggested that it might also function in transcription termination. To test this, we analyzed steady-state levels of several snoRNAs and mRNAs by Northern blotting of total RNA extracted from wild-type, swd2-2 and swd2-6 strains grown at 23°C and after shift to 37°C (Fig. 2 ▶). A ssu72-2 mutant strain was analyzed in parallel. Figure 2A ▶ shows that swd2-2 and swd2-6 mutants accumulated an extended snR33 transcript following growth at 37°C. A RNA of the same length was observed in the ssu72-2 mutant, mostly at restrictive temperature. This suggested that Swd2p was required to prevent transcriptional read-through at the snR33 terminator. To prove this, we probed for the product of the YCR015c gene that lies immediately downstream of the snR33 gene. swd2 mutants strongly accumulated a snR33-YCR015c mRNA at 37°C. This mRNA was also readily detected in the ssu72-2 strain. Strikingly, levels of these transcripts were strongly increased in both swd2 and ssu72 mutants, because the endogenous YCR015c mRNA was barely detectable under these conditions in the wild type. Furthermore, we observed accumulation of an additional RNA that was previously shown to occur in ssu72-69 cells from read-through at the YCR015c terminator and to correspond to a snR33-YCR015c-POL4 transcript (Ganem et al. 2003). This RNA accumulated in swd2 mutants and ssu72-2 cells at 37°C. We could not detect read-through RNAs with oligonucleotide probes directed against snR13 and snR71 snoRNAs (Fig. 2B,C ▶); however, read-through products containing snR13 and snR71 were prominent in the ssu72-2 mutant. In contrast, the use of probes against TRS31 and REC104 mRNAs that are transcribed immediately downstream of the snR13 and snR71 genes, respectively, revealed transcriptional read-through also in the swd2 mutant strains. Strikingly, levels of these transcripts were strongly increased in the ssu72-2 mutant but not in the swd2 mutants. We therefore conclude that the failure to detect snR13 and snR71 read-through RNAs with the use of oligonucleotide probes was due to the lower abundance of these transcripts compared to the ssu72-2 mutant strain where these genes were either more heavily transcribed or more stable. Note that oligonucleotide probes used to detect snoRNAs carry only one radioactive label at the 5′ end compared to many radioactive labels in the random-primed probes that were used to detect mRNAs. We observed no read-through RNAs with the swd2 mutants when snR45 and the downstream ASN1 mRNA were tested, or when the NRD1 mRNA and the downstream MRPL17 mRNA were tested (Fig. 2D ▶; data not shown). In contrast, the ssu72-2 mutant gave strong read-through at both the snR45 and the NRD1 genes; in many cases read-through in the ssu72-2 mutant resulted in the accumulation of intermediate-sized RNAs (indicated by asterisks) that may be caused by processing at cryptic cleavage and polyadenylation sites. Figure 2E ▶ shows that SWD2 mRNA levels were increased in swd2 mutants and extended transcripts appeared. These extended SWD2 transcripts were not observed in the ssu72-2 mutant. Other mRNAs tested (SET1, CYH2, ADH1, and ACT1; Fig. 2F ▶; data not shown) did not reveal read-through transcripts in swd2 and ssu72-2 mutants; however, some reduction of these mRNAs was observed following growth at 37°C.
FIGURE 2.
Swd2 is required for 3′ end formation of specific mRNAs and snoRNAs. Northern analysis of 20 μg total RNA extracted from wild-type, swd2-2, swd2-6, and ssu72-2 cells following growth in YPD medium at 25°C or after a shift to 37°C for the indicated times. Blots were produced in duplicate. (A–F) Probed as indicated at the bottom of each panel; migration of RNAs is indicated on the right. In the middle of each panel a schematic presentation indicates the genomic arrangement of the analyzed genes and the relative direction of transcription.
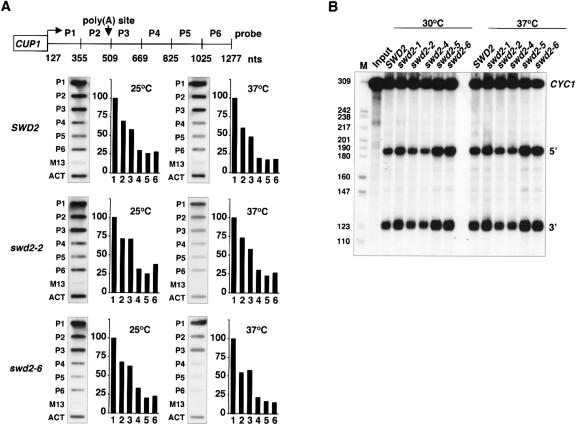
To test for transcription termination by transcriptional run-on analysis (TRO), we analyzed a plasmid-borne CYC1 gene that was under control of a minimal CUP1 promoter (pCUP-CYC1; Fig. 3A ▶). Wild-type and mutant strains carrying pCUP-CYC1 were grown in glucose-based synthetic medium containing 1 mM CuSO4. Northern analysis confirmed that expression of the pCUP1-CYC1 gene depended on the presence of CuSO4 in the medium (data not shown). TRO was performed as described (Birse et al. 1998). We found that wild-type, swd2-2, and swd2-6 strains terminated transcription efficiently following growth at 25°C and following a shift to 37°C for 1 h. These results indicated that swd2 mutant strains were not deficient in termination of the pCUP-CYC1 gene.
FIGURE 3.
Mutations in SWD2 do not interfere with 3′ end cleavage of CYC1 pre-mRNA in vitro. (A) Transcriptional run-on analysis. Slot-hybridizations and quantification of run-on transcripts obtained after transcription run-on analysis (Birse et al. 1998). Wild-type, swd2-2, and swd2-6 cells were grown in synthetic medium lacking uracil and 1 mM CuSO4 at 25°C or shifted to 37°C in a water bath for 1 h. P1 to P6 represent probes complementary to CYC1 transcripts as indicated; empty M13 and ACT1 probes served as controls. Values obtained by PhosphorImager Scanning (Storm, Molecular Dynamics) were corrected by subtraction of the M13 background signal and normalized to the value of P1 that was fixed at 100%. (B) 3′ end cleavage in vitro. Assays were performed with extracts prepared from the indicated strains. Input lanes represent mock-treated reactions. Marker bands (HpaII-digested pBR322) are indicated on the left. Internally [32P]-labeled CYC1 RNA was used as substrate. The positions of full-length RNA (CYC1), 5′ and 3′ cleavage products are indicated. Cleavage was performed either at 30°C (lanes 2–7) or at 37°C, following a 5-min preincubation of extract and reaction mixture at this temperature (lanes 8–13).
As swd2 mutants displayed transcriptional read-through at certain genes, we asked whether the protein was required for 3′ end cleavage in vitro. Processing extracts were produced from wild-type and mutant strains (swd2-1, swd2-2, swd2-4, swd2-5, swd2-6, swd2-7, swd2-8) and tested for cleavage of a synthetic CYC1 pre-mRNA substrate. In vitro cleavage occurred efficiently in wild-type and all analyzed swd2 mutant extracts (Fig. 3B ▶; data not shown). The efficiency of processing was not reduced when the extracts were preincubated at 37°C for 5 min and when the assay temperature was elevated from 30°C to 37°C. We conclude that defects in 3′ end formation of some mRNAs and snoRNAs in swd2 mutant strains did not result from a general loss of cleavage activity.
Taken together, the results in Figures 2 ▶and 3 ▶ suggest that mutations in SWD2 result in defective 3′ end formation at some genes, whereas other genes are not or are at least less strongly affected. The reasons for these apparently gene-specific effects remain unclear, but may relate to differences in 3′ end formation signals (see Discussion).
Swd2p bridges the APT, core-CPF, and CF IA subcomplexes of the 3′ end formation machinery
WD-40 repeat proteins are frequently associated with mul-tiprotein complexes and are implied in mediating protein–protein contacts (Smith et al. 1999). To identify the interactions that are mediated by Swd2p we performed GST pull-down experiments with bacterially expressed GST-Swd2p and in vitro translated [35S]-methionine-labeled 3′ end formation factors. Figure 4A ▶, lanes 8, 9, 18, and 19, shows representative experiments in which GST-Swd2p bound efficiently to the core-CPF components Yhh1p/ Cft1p, Ysh1p/Brr5p, Pfs2p, and Pta1p. Other core-CPF components (Ydh1p/Cft2p, Mpe1p, Pap1p, Fip1p) did not bind. Of the APT proteins, Pta1p, Glc7p, and Pti1p showed an interaction with GST-Swd2p (Fig. 4B ▶, lanes 29,39,40); Ref2p, Ssu72p, and Syc1p did not interact significantly. Finally, the CF IA subunit Pcf11p bound to GST-Swd2p; no binding was observed with other CF IA components (Rna15p, Rna14p, Clp1p; Fig. 4C ▶, lanes 48–50), CF IB (Nab4p; Fig. 4C ▶, lane 52), or the poly(A) binding protein Pab1p (Fig. 4A ▶, lane 10). All observed interactions were specific for Swd2p because GST alone did not result in binding. Furthermore, because the binding reactions were done in the presence of RNase A, it is unlikely that RNA mediated the interactions. We propose that Swd2p contributes specific interactions between subunits of the APT, core-CPF, and CF IA subcomplexes of the 3′ end formation machinery (see Fig. 4D ▶) and may therefore function in the assembly of 3′ end formation complexes (see Discussion).
Swd2p is required for SET1C activity
Because Swd2p is a component of SET1C (Miller et al. 2001; Roguev et al. 2001; Nagy et al. 2002) we asked whether it is also required for SET1C activity, that is, methylation of H3K4. Western analysis was performed with whole-cell extracts derived from wild-type and swd2 mutant strains grown at 25°C or following growth at 37°C for 2, 4, and 8 h. For control we also analyzed extracts from strains lacking the SET1 gene (ΔSET1), from H3K4A, H3K9A mutant strains and corresponding isogenic wild-type strains. Figure 5A ▶ shows a representative experiment. Antibodies directed against di- and tri-methylated H3K4 recognized proteins in wild-type extracts that were absent in ΔSET1 and H3K4A strains and therefore corresponded to the modified forms of H3K4. swd2-2 and swd2-3 extracts showed no or only trace amounts of di- and tri-methylation in multiple experiments; these deficiencies were observed at either permissive or restrictive temperatures. Some di-methylated and tri-methylated H3K4 was detected in swd2-6 extracts at permissive temperature, which was lost upon a shift to 37°C. Cross-reacting bands that were observed served as control for loading of comparable amounts of whole-cell extract in each lane (indicated by asterisks). These data suggested a requirement for Swd2p as component of SET1C in global methylation of H3K4.
FIGURE 5.
Swd2p is required for SET1C activity. (A) Protein lysates were produced from wild-type (wt), swd2, ΔSET1, K4A, and K9A mutant strains as indicated on top of each lane. The wild-type and swd2 mutant strains were grown in YPD medium at 25°C or transferred for 4 h to a 37°C water bath. ΔSET1, K4A, and K9A strains were grown at 30°C. Following Western transfer filters were probed with polyclonal antisera directed against the indicated proteins. Asterisks mark cross-reacting bands that served as control for equal loading. (B) Southern blot of genomic DNA obtained from the indicated strains. swd2 mutants and the isogenic wild-type strain were grown in YPD medium at 25°C (lanes 1–4) or transferred to a 37°C water bath for 8 h (lanes 5–8). Ten micrograms genomic DNA were digested with XhoI and resolved on 1% agarose gels. Southern hybridization was performed with a telomere specific probe. Migration of telomere fragments is indicated on the right. (C) Western analysis of extracts from strains expressing ProtA fusions to wild-type and mutant Swd2p proteins. The indicated strains were grown at 25°C or shifted to 37°C for 4 h in YPD medium. Fifty micrograms total protein were analyzed by Western with specific antibodies. ProtA-Swd2p was detected with a polyclonal IgG. Brackets on the right indicate the association of proteins with SET1C or CPF. (D) Northern analysis as described in the legend of Figure 2 ▶. Total RNA was extracted from the strains indicated on top of each lane following growth at 30°C in YPD medium. Note that this temperature is semipermissive for swd2-2 and swd2-6 strains. Probes were as described in the legend of Figure 2 ▶ and are indicated below each panel. 18S rRNA served as loading control and was detected by a 5′ end-labeled oligonucleotide. The detected RNA species are indicated on the right of the panels.
In addition to methylation of H3K4, components of SET1C are required for the maintenance of correct telomere length, and this requirement is based at least partly on the histone methyltransferase activity of SET1C (Roguev et al. 2001). A Southern blot strategy was used to test whether mutations in SWD2 affected telomere length (Fig. 5B ▶; Roguev et al. 2001). Wild-type and swd2 mutant strains were analyzed following growth at 25°C and after an 8-h shift to 37°C. We reproducibly found that the swd2-2 mutant had shortened telomeres compared to the wild type. The swd2-3 strain appeared to be affected most severely after growth at 37°C; the swd2-6 strain displayed a weaker reduction of telomere length at both temperatures. Taken together these results suggested a requirement for Swd2p for a functional SET1C complex.
To reveal the molecular basis of loss of SET1C function we analyzed the levels of mutant Swd2p proteins. Because no specific antibody against the protein was available we used strains that expressed wild-type and mutant proteins as N-terminal ProteinA fusions. We found that the levels of protA-swd2-2p, protA-swd2-3p and protA-swd2-6p proteins were stable at permissive and nonpermissive temperatures (Fig. 5C ▶). In contrast, Set1p levels were strongly reduced in mutant extracts; Set1p levels were similarly reduced in nontagged swd2 mutant strains (data not shown). Because SET1 mRNA levels were stable in swd2 mutants at permissive temperature (see Fig. 2F ▶) underaccumulation of Set1p was likely due to a posttranscriptional defect. We suggest that SET1C might be destabilized in swd2 strains causing a reduction of Set1p levels and consequently a defect in methylation of H3K4. In contrast to Set1p, none of the tested CPF subunits (Pta1p, Ysh1p, Glc7p, Ssu72p) was significantly reduced in the swd2 mutant strains (Figs. 5C ▶, 6 ▶).
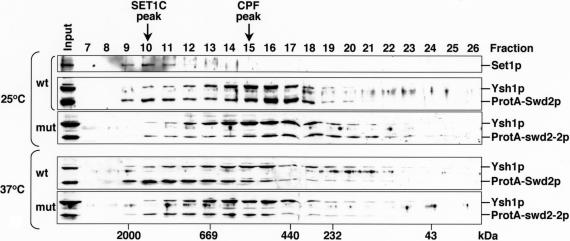
FIGURE 6.
Swd2p associates with separable high molecular weight complexes. Protein extracts were produced from wild-type (wt; ProtA-Swd2p) and mutant (mut; ProtA-swd2-2p) strains following growth at 25°C or after a shift to 37°C for 4 h as indicated. Protein was separated by SMART Superdex 200 gel filtration and analyzed by Western blotting. Fraction numbers and input material are indicated on the top of the panels and the migration of molecular weight markers separated under the same conditions is indicated at the bottom of the panels. Also indicated is the migration of peak fractions of Set1p protein (SET1C peak) and of Ysh1p protein (CPF peak). On the right of the panels the detected proteins are indicated.
To test whether the transcriptional read-through observed with swd2 mutants (Fig. 2 ▶) related to its association with SET1C, we performed Northern analysis with RNAs extracted from various strains lacking SET1C components (ΔSET1, ΔBRE2, ΔSWD1, ΔSWD3, ΔSDC1, ΔSHG1) and the H3K4A mutant strain. For better comparison all strains were grown under the same conditions (at 30°C) in this experiment; note that this temperature was semipermissive for swd2 mutants. Under these conditions the swd2-2 strain displayed strong accumulation and the swd2-6 strain weak accumulation of extended YCR015C, TRS31, and SWD2 transcripts (Fig. 5D ▶; data not shown). We did not observe extended transcripts for any strain that lacked a component of SET1C or with the H3K4A strain. Increased levels of YCR015C and SWD2-containing transcripts were observed exclusively for the swd2 mutant strains. We conclude that transcriptional read-through defects of swd2 mutants were independent of functional SET1C and H3K4.
Swd2p is independently associated with CPF and SET1C in extracts
The association of Swd2p with CPF and SET1C may indicate a physical link between both complexes. To test this we initially analyzed the size distribution of cellular Swd2p-containing complexes by gel filtration of wild-type and mutant extracts derived from strains expressing the ProteinA fusions. We used the core-CPF component Ysh1p and the Set1p methyltransferase as markers for the elution profile of both complexes (Fig. 6 ▶). SET1C displayed a higher molecular weight (peak at fraction 10) than CPF (peak at fraction 15), consistent with the previous prediction that SET1C may occur as an heterooligomeric factor in vivo (Roguev et al. 2003a). Small amounts of Ysh1p were also detected in high molecular weight fractions; however, no Set1p was detectable in CPF peak fractions. ProtA-Swd2p (from cells grown at 25°C) eluted in two major complexes, which overlapped with Set1p and with Ysh1p, consistent with its association with both complexes. Mutant ProtA-swd2-2p (from 25°C extracts) clearly shifted in its mobility to the lower molecular weight CPF complex. This is most likely due to the fact that underaccumulation of Set1p (see Fig. 5C ▶) disrupted most of SET1C. Interestingly, the distribution of wild-type ProtA-Swd2p clearly shifted to the higher molecular weight SET1C following growth at 37°C whereas the distribution of Ysh1p was the same as in extract from cells grown at 25°C. The reasons for this observation remain unclear but may indicate a cellular response to heat treatment. Mutant extracts prepared after growth at 37°C displayed reduced ProtA-swd2-2p associated with SET1C while most of the protein co-eluted with Ysh1p. These observations are consistent with the interpretation that Swd2p was associated with two separable high molecular weight complexes in extracts and that mutant ProtA-swd2-2p failed to associate efficiently with SET1C but not with CPF. The possibility remained that subpopulations of SET1C and CPF were physically associated. To further test this we probed for the presence of Set1p in CPF fractions purified from ProtA-Ydh1p- and ProtA-Pfs2p-expressing strains by Western blot. However, we could not detect Set1p above background levels (data not shown), indicating that SET1C and CPF are not stably associated with each other in extracts under nonstringent salt conditions (150 mM KCl).
DISCUSSION
Intimate connections exist between the transcription apparatus and pre-mRNA processing factors. To further understand the complex interactions between these processes we analyzed the function of the Swd2p protein, which associates with the CPF and SET1C complexes that are involved in 3′ end formation and in transcription elongation, respectively. We found that Swd2p has essential functions as component of both complexes. It is necessary to prevent transcriptional read-through at specific genes and it is required for global H3K4 methylation.
Our analysis indicated that swd2 mutants are defective in 3′ end formation of specific mRNAs and snoRNAs. Recent results on other subunits of CPF revealed similarities with the phenotypes of swd2 mutants observed here. Swd2p was suggested to associate with the APT subcomplex of CPF that includes also Pta1p, Ref2p, Pti1p, Glc7p, Ssu72p, and Syc1p (Nedea et al. 2003). A common phenotype of mutants in Ref2p, Pti1p, Ssu72p, and Swd2p is transcriptional read-through at snoRNA genes (Dheur et al. 2003; Ganem et al. 2003; Nedea et al. 2003; Steinmetz and Brow 2003). These phenotypes strongly contrast results obtained with mutants in core-CPF subunits that did not affect snoRNA termination in a snR13-based reporter assay (Morlando et al. 2002). Moreover, transcription run-on analysis revealed deficient termination on a plasmid-borne CYC1 gene in yhh1/cft1 mutants (a component of core-CPF; Dichtl et al. 2002a) but not in ssu72-2 (Dichtl et al. 2002b; He et al. 2003) and swd2 mutant strains (Fig. 3A ▶). These analyses point to distinct requirements for specific subunits of CPF for 3′ end formation at different sets of genes.
An important open question is whether distinct 3′ end formation complexes exist in vivo, which could function at specific genes. Termination depends on the cotranscrip-tional recognition of poly(A) signal sequences on pre-mRNAs. An important function of CPF lies in the recognition of poly(A) site sequences within pre-mRNAs and the RNA-binding proteins involved (Yhh1p/Cft1p, Ydh1p/Cft2p, and Yth1p) were so far exclusively assigned to core-CPF (Barabino et al. 2000; Dichtl and Keller 2001; Dichtl et al. 2002a). Poly(A) signals in pre-mRNAs and cleavage signals in snoRNAs are clearly distinct. The recognition of the respective sequences by RNA-binding components of CPF might therefore contribute to the gene-specific requirement for CPF subunits. Transcriptional read-through in swd2 and other APT mutants might reflect a deficiency in the recognition of termination signals or a defect in the transduction of the signals to RNAP II. Moreover, a deficiency in the recognition of termination signals or 3′ end formation signals, respectively, may result in failure to cleave specific transcripts at the 3′ end although the catalytic endonucleolytic activity per se may generally be unaffected. However, the phenotypes of mutants in APT subunits and core-CPF subunits cannot clearly be categorized as snoRNA and mRNA specific because APT mutants also affect termination at selected mRNA genes. Interestingly, the APT subunits Pti1p and Ref2p were suggested to uncouple cleavage from polyadenylation at 3′ ends of snoRNA genes (Dheur et al. 2003). It was proposed that the exclusion of poly(A) polymerase in Pti1p- and Ref2p-containing CPF complexes (within the so-called small nucleolar cleavage factor, snCF) may contribute to polyadenylation-independent 3′ end formation. It will be important for future analyses to test a representative set of genes (or the entire genome) for the potentially differential association of specific 3′ end formation factors, for example, APT proteins and poly(A) polymerase.
How does Swd2p function as a component of CPF and SET1C? Our bioinformatic analysis of Swd2p proteins suggests a seven-bladed β-propeller structure. Many members of this large protein family have been found to associate with multiprotein complexes, including numerous factors involved in pre-mRNA processing (Smith et al. 1999). Our protein interaction studies indicate that Swd2p can engage in multiple interactions with other 3′ end formation components. Interestingly, we identified interaction partners within the core-CPF, the APT, and the CF IA subcomplexes of the 3′ end formation machinery. Thus, it is tempting to speculate that Swd2p may contribute to the assembly of 3′ end formation complexes, which may be required primarily at selected genes. Alternatively, Swd2p may play solely a structural role within CPF. However, mutant Swd2p proteins did not appear to destabilize the complex significantly, as no reduction of any of the tested CPF components could be observed. In contrast, mutations within Swd2p resulted in strong underaccumulation of Set1p, most likely due to a posttranscriptional defect. This may reflect a crucial structural role for Swd2p in SET1C. Alternatively, SET1C assembly may be affected. We do not know if the same part of Swd2p is responsible for the interaction with the two complexes. Because no obvious common domains have been found in SET1C and CPF, it is possible that the Swd2p interaction depends on a short motif found in two different types of proteins in the two complexes.
Despite its bifunctional nature, the roles of Swd2p as a component of CPF and SET1C appear to occur independently of each other. This conclusion is based on three important observations: First, CPF and SET1C do not detectably interact in extracts; however, the possibility remains that a putative interaction of both complexes may be very labile or even transient. Second, phenotypes of mutations in Swd2p in 3′ end formation are duplicated by other mutations in CPF but not by mutations in SET1C or H3K4. Third, the bifunctional nature of Swd2p is not conserved in evolution, as two different Swd2p proteins associate with CPF and SET1C in S. pombe (Swd2.1 and Swd2.2, respectively; Roguev et al. 2003b). Thus, the functions that are played by a single Swd2p protein in S. cerevisiae have been separated in S. pombe. It will be very interesting to analyze the proteomic distribution of human Swd2p. Although there may be more than one Swd2p protein encoded in the human genome, this requires experimental verification. However, none of the known components of the mammalian 3′ end formation machinery displayed clear homology to S. cerevisiae Swd2p in our database searches. The association of Swd2p with both CPF and SET1C remains intriguing, as both of these complexes interact with elongating RNAP II. SET1C was suggested to associate with genes primarily at 5′ regions (Krogan et al. 2003; Ng et al. 2003), whereas CPF associates with genes increasingly toward their 3′ end (Komarnitsky et al. 2000; Licatalosi et al. 2002; Nedea et al. 2003). Moreover, recent reports suggested a requirement for SET1C in 3′ end formation/termination on certain genes that may depend on the interaction of the chromatin remodeling ATPase Isw1p with di- and tri-methylated H3K4 (Santos-Rosa et al. 2003). Isw1p itself was previously implied in RNAP II termination and may function to establish chromatin boundaries at 3′ ends of genes (Alen et al. 2002). However, Isw1p has crucial functions already at the beginning of genes, where it controls the association of the transcription machinery by nucleosome remodeling and influences the phosphorylation status of the RNAP II CTD during distinct phases of the transcription cycle (Morillon et al. 2003). The common denominator for CPF and SET1C function may therefore be the interaction with the elongating RNAP II complex, which ultimately controls termination of transcription and the formation of 3′ ends.
MATERIALS AND METHODS
Yeast growth, strain construction, and plasmids
Strains used in this study are listed in Table 1 ▶. SET1C deletion strains (ΔSET1, ΔBRE2, ΔSWD1, ΔSDC1, and ΔSHG1) were generously provided by A. Roguev and F. Stewart (Technical University of Dresden, Germany) and will be described elsewhere. Yeast were grown in rich YPD medium (2% glucose, 2% bactotryptone, 1% yeast extract) or synthetic drop-out medium (2% glucose, 0.67% yeast nitrogen base, 1× amino acids) as indicated in the figure legends. Temperature-sensitive SWD2 alleles were isolated by in vivo gap repair (Muhlrad et al. 1992). The SWD2 gene (nt -817–1932 relative to the A of the translational start codon, which corresponds to +1) was cloned as a SalI/BamHI fragment into pRS416 (pBD236). A 2.3-kb SalI/PstI fragment from pBD236 carrying SWD2 was cloned into pRS415ΔSpeI (generated by SpeI digest, Klenow treatment, and religation) to generate pBD238. The SWD2 gene was amplified by Taq Polymerase under error-prone conditions (Ohnacker et al. 2000) with primers swd2mut2 (TTG GTATGATACGCAAGGTGATG) and swd2mut3 (TGCTGTATT GAGATTGATCTCTTC). PCR products and linear SpeI-digested pBD238 (removing nt -100–1207 of SWD2 sequence) were co-transformed into YWK164. LEU+ transformants were first replica-plated on 5-FOA; after subsequent replica-plating on YPD growth was tested at 15°C, 23°C, 30°C, 34°C, and 37°C. Plasmid isolation from candidate mutant strains and reintroduction into a swd2 deletion background identified nine strains, which were nonviable at 37°C. Detailed analyses were performed with swd2-2, swd2-3, and swd2-6 strains. Amino acid changes in the mutant alleles were inferred by DNA sequencing of plasmids carrying mutant alleles. The following mutations were identified: swd2-2, F14P, C27G, F41I, K185E, S253L, C257R; swd2-3, G185E, S253E, C257R; swd2-6, L12P, K120E, F324E. Plasmids encoding proteinA-tagged fusions of wild-type and mutant SWD2 alleles were generated by PCR amplification of the respective open reading frame and cloning into pNOP1 (Senger et al. 1998) using primer-encoded NcoI and BamHI restriction sites: pBD239 (ProtA-SWD2-LEU2); pBD377 (ProtA-swd2-2-LEU2); pBD378 (ProtA-swd2-2-LEU2); pBD379 (ProtA-swd2-2-LEU2). Plasmids were transformed into strain YWK164, followed by plasmid shuffling on 5-FOA.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
| YWK164 | Matahis3Δ ura3Δ leu2Δ swd2::KANMX [SWD2-URA3-CEN] | This work |
| YWK168 | Matahis3Δ ura3Δ leu2Δ swd2::KANMX [SWD2-LEU2-CEN] | This work |
| YWK171 | Matahis3Δ ura3Δ leu2Δ swd2::KANMX [swd2-LEU2-CEN] | This work |
| YWK172 | Matahis3Δ ura3Δ leu2Δ swd2::KANMX [swd2-3-LEU2-CEN] | This work |
| YWK176 | Matahis3Δ ura3Δ leu2Δ swd2::KANMX [swd2-6-LEU2-CEN] | This work |
| YWK186 | Mataura3-1 trp1-1 ade2-1, leu2-3,112 his3-11,15 ssu72-2 | Dichtl et al. (2002b) |
| YWK167 | Matahis3Δ ura3Δ leu2Δ swd2::KANMX pBD239 [ProtA-SWD2-LEU2-CEN] | This work |
| YWK287 | Matahis3Δ ura3Δ leu2Δ swd2::KANMX pBD377 [ProtA-swd2-2-LEU2-CEN] | This work |
| YWK288 | Matahis3Δ ura3Δ leu2Δ swd2::KANMX pBD378 [ProtA-swd2-3-LEU2-CEN] | This work |
| YWK289 | Matahis3Δ ura3Δ leu2Δ swd2::KANMX pBD379 [ProtA-swd2-6-LEU2-CEN] | This work |
| MX1-4C | Mat α ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ (hht1-hhf1)3(hh2-hhf2) [CEN ARS URA3 HHT-1-HHF1] | Hsu et al. (2000) |
| YWK247 | Mat α ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ (hht1-hhf1)3(hht2-hhf2) pJH18 [CEN ARS TRP1 HHT1-HHF1] | This work |
| YWK245 | Mat α ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ (hht1-hhf1)3(hht2-hhf2) pBD364 [CEN ARS TRP1 HHT1-(K4A)-HHF1] | This work |
| YWK246 | Mat α ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ (hht1-hhf1)3(hht2-hhf2) pBD364 [CEN ARS TRP1 HHT1(K9A)-HHF1] | This work |
pCUP-CYC1 (pBD352) used for TRO was produced by replacing the EcoRI/BglII fragment containing the GAL1/10 promoter from pGAL-CYC1 (Birse et al. 1998) with a PCR fragment spanning bp −400 to 37 (relative to the A of the translational start codon) of the CUP1 promoter using primer-encoded EcoRI (CATAGGGAATTCTTAAAACACTTTTGTATTATTTTT) and BglII (TAAATGAGATCTGACCTTCATTTTGGAAGTTAAT TA) restriction sites. GST-Swd2p (pBD241) was produced by PCR amplification of the SWD2 open reading frame with primers encoding NcoI (CATGATCCATGGGTATGACCACCGTGTCCATCAAT) and BamHI (ATGCGGGATCCTCATTCATCGTACACGTAAAAATC) restriction sites and cloning into pGDV1 (Dichtl and Keller 2001). H3K4 and H3K9 mutations were produced by site-directed mutagenesis on plasmid pJH18 (CEN ARS TRP1 HHT1-HHF1; details available upon request; Hsu et al. 2000). The resulting plasmids pBD364 (H3K4A) and pBD365 (H3K9A) were transformed into the shuffling strain MX1-4C (Hsu et al. 2000). Following selection for transformants, URA3-marked H3K4 plasmids were shuffled out by plating on 5-FOA medium.
RNA analyses
Northern analyses were done as described (Dichtl et al. 2002a). PCR products covering the following sequences (relative to the A of the translational start codon) served as templates for probes: PGK1 (nt 1–1251), CYH2 (nt 1–450), ADH1 (nt 1–1049), CUP1 (nt 1–186), ACT1 (nt 1–677), anti-TRS31 (nt 34–849), anti-YCR015c (nt 60–654), anti-MRPL15 (nt 28–838), anti-NRD1 (nt 3–706), anti-SWD2 (nt 1–990) and anti-SET1 (nt 1–772). Oligonucleotide probes were labeled with γ[32P]ATP and T4 polynucleotide kinase: anti-18S (CAGACAAATCACTCCA), anti-snR13 (GGTCAGATAAAAGTAAAAAAAGGTAGC), anti-snR33 (CCGGTGTAAAAGCTAGGCTTCAATC), anti-snR45 (ATCGCT CCGAGAAGAATTGTTCGAT), and anti-snR71 (TCTGAGTGAG CTGAGAAGGTTATCA). Transcriptional run-on analysis was performed according to Birse et al. (1998).
Protein analyses and in vitro 3′ end processing
Protein extracts for analysis of histone proteins were prepared from 50-ml cultures of exponentially growing yeast by glass-bead homogenization [50% (v/v) glass beads; 800 μL total volume] in homogenization buffer A (20 mM HEPES-KOH at pH 8.0, 350 mM KCl, 10% glycerol, 0.1% Tween 20, 1 mM PMSF) at 4°C. Crude extracts were cleared by three consecutive 30-min centrifugation steps at maximum speed in a table-top centrifuge at 4°C. Concentration of total protein was determined by Bradford analysis and 50 μg total protein were analyzed from each sample by SDS-PAGE on 15% gels. Antibodies for detection of di- and tri-methylated H3K4 were obtained from Abcam. Protein extracts for gel-filtration analysis were prepared as described for histone extracts described above, but in homogenization buffer B (150 mM KCl, 20 mM Tris-HCl at pH 7.9, 10% glycerol, 0.01% NP-40, 1 mM DTT, 1 mM PMSF). Three hundred fifty micrograms total protein were applied to a SMART Superdex 200 column, equilibrated in homogenization buffer B. Eighty-milliliter fractions were collected and 20 μL were used for SDS-PAGE on 12% gels. Molecular weight standards (Blue dextran: 2000 kD; thyroglobin: 669 kD; ferritin: 440 kD; catalase: 232 kD; ovalbumin 43 kD) were resolved under identical conditions. Production of GST-fusion proteins, in vitro translation, and GST pull-down assays were performed as described (Dichtl et al. 2002b). Preparation of extracts and in vitro 3′ end processing was done as described (Ohnacker et al. 2000).
Database searches and sequence analysis
Database searches in the UniProt database (Apweiler et al. 2004) were performed with Blastp (Altschul et al. 1997) as implemented at ExPASy (URL: http://au.expasy.org/tools/blast/). Searches were performed with S. cerevisiae Swd2 and S. pombe Swd2.1 and Swd2.2 as queries using default parameters. Top scoring sequences from a representative set of organisms were collected and multiply aligned using Clustal_X (Thompson et al. 1997) and manually edited to optimize minor poorly aligned regions. The C. elegans sequence Ce_C33H5.6 (trembl:Q18404) was obviously mispredicted and was corrected aided by profile alignment to the genomic sequence (embl:CEC33H5) using PairWise (Birney et al. 1996) and the Swd2 family profile, which led to the correction of exon 4 to position 3292–3582 and a new last exon at 4016–4252. The profile was generated from the multiple alignment using PairWise and used to generate the profile–profile dotplot using Pro-Plot (Thompson et al. 1997).
Acknowledgments
We thank D. Blank for technical help in the initial phase of this work. We are grateful to A. Roguev, F. Stewart, N. Proudfoot, A. Hsu, E. Hurt, S. Röck, C. Moore, and P. Nagy for generous gifts of yeast strains, plasmids, M13 phages, and antibodies. This work was supported by the University of Basel, the Swiss National Science Fund, the European Community (see www.eurnomics.org) via the Bundesamt für Bildung und Wissenschaft, Bern (grant 01.0123), the Louis-Jeantet-Foundation for Medicine and the Norwegian Research Council (grant 146652/431 to RA).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7090104.
REFERENCES
- Alen, C., Kent, N.A., Jones, H.S., O’Sullivan, J., Aranda, A., and Proudfoot, N.J. 2002. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell 10: 1441–1452. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Bairoch, A., Wu, C.H., Barker, W.C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. 2004. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 32: 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino, S.L.M., Ohnacker, M., and Keller, W. 2000. Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 19: 3778–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, D. 2002. The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell. Biol. 14: 336–342. [DOI] [PubMed] [Google Scholar]
- Birney, E., Thompson, J.D., and Gibson, T.J. 1996. PairWise and SearchWise: Finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res. 24: 2730–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse, C.E., Minvielle-Sebastia, L., Lee, B.A., Keller, W., and Proudfoot, N.J. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301. [DOI] [PubMed] [Google Scholar]
- Briggs, S.D., Bryk, M., Strahl, B.D., Cheung, W.L., Davie, J.K., Dent, S.Y., Winston, F., and Allis, C.D. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 15: 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Legrain, P., and Jacquier, A. 1998. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284: 975–988. [DOI] [PubMed] [Google Scholar]
- Chen, J. and Moore, C. 1992. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol. 12: 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheur, S., Vo, L.T.A., Voisinet-Hakil, F., Minet, M., Schmitter, J.M., Lacroute, F., Wyers, F., and Minvielle-Sebastia, L. 2003. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 22: 2831–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl, B. and Keller, W. 2001. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 20: 3197–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl, B., Blank, D., Sadowski, M., Hübner, W., Weiser, S., and Keller, W. 2002a. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 21: 4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl, B., Blank, D., Ohnacker, M., Friedlein, A., Roeder, D., Langen, H., and Keller, W. 2002b. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell. 10: 1139–1150. [DOI] [PubMed] [Google Scholar]
- Edmonds, M. 2002. A history of poly A sequences: From formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 71: 285–389. [DOI] [PubMed] [Google Scholar]
- Fatica, A., Morlando, M., and Bozzoni, I. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19: 6218–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, C., Devaux, F., Torchet, C., Jacq, C., Quevillon-Cheruel, S., Labesse, G., Facca, C., and Faye, G. 2003. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 22: 1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Graber, J.H., Cantor, C.R., Mohr, S.C., and Smith, T.F. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. 96: 14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Khan, A.U., Cheng, H., Pappas Jr., D.L., Hampsey, M., and Moore, C.L. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes & Dev. 17: 1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, J.Y., Sun, Z.W., Li, X., Reuben, M., Tatchell, K., Bishop, D.K., Grushcow, J.M., Brame, C.J., Caldwell, J.A., Hunt, D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7 phosphatase in budding yeast and nematodes. Cell 102: 279–291. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T. and Allis, C.D. 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kessler, M.M., Zhao, J., and Moore, C.L. 1996. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. J. Biol. Chem. 271: 27167–27175. [DOI] [PubMed] [Google Scholar]
- Kim, S.J., and Martinson, H.G. 2003. Poly(A)-dependent transcription termination: Continued communication of the poly(A) signal with the polymerase is required long after extrusion in vivo. J. Biol. Chem. 278: 41691–41701. [DOI] [PubMed] [Google Scholar]
- Kiss, T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Komarnitsky, P., Cho, E.J., and Buratowski, S. 2000. Different phos-phorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 14: 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12: 198–209. [DOI] [PubMed] [Google Scholar]
- Krogan, N.J., Dover, J., Khorrami, S., Greenblatt, J.F., Schneider, J., Johnston, M., and Shilatifard, A. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277: 10753–10755. [DOI] [PubMed] [Google Scholar]
- Krogan, N.J., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Ryan, O.W., Golshani, A., Johnston, M., et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell 11: 721–729. [DOI] [PubMed] [Google Scholar]
- Licatalosi, D.D., Geiger, G., Minet, M., Schroeder, S., Cilli, K., McNeil, J.B., and Bentley, D.L. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9: 1101–1111. [DOI] [PubMed] [Google Scholar]
- Miller, T., Krogan, N.J., Dover, J., Erdjument-Bromage, H., Tempst, P., Johnston, M., Greenblatt, J.F., and Shilatifard, A. 2001. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. 98: 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Preker, P.J., Wiederkehr, T., Strahm, Y., and Keller, W. 1997. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in pre-messenger RNA 3′-end formation. Proc. Natl. Acad. Sci. 94: 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon, A., Karabetsou, N., O’Sullivan, J., Kent, N., Proudfoot, N., and Mellor, J. 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymer-ase II. Cell 115: 425–435. [DOI] [PubMed] [Google Scholar]
- Morlando, M., Greco, P., Dichtl, B., Fatica, A., Keller, W., and Bozzoni, I. 2002. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 22: 1379–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., Hunter, R., and Parker, R. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8: 79–82. [DOI] [PubMed] [Google Scholar]
- Nagy, P.L., Griesenbeck, J., Kornberg, R.D., and Cleary, M.L. 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. 99: 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea, E., He, X., Kim, M., Pootoolal, J., Zhong, G., Canadien, V., Hughes, T., Buratowski, S., Moore, C.L., and Greenblatt, J. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278: 33000–33010. [DOI] [PubMed] [Google Scholar]
- Neugebauer, K.M. 2002. On the importance of being co-transcriptional. J. Cell. Sci. 115: 3865–3871. [DOI] [PubMed] [Google Scholar]
- Ng, H.H., Robert, F., Young, R.A., and Struhl, K. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709–719. [DOI] [PubMed] [Google Scholar]
- Ohnacker, M., Barabino, S.M., Preker, P.J., and Keller, W. 2000. The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 19: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco, I.J., Kim, S.J., and Martinson, H.G. 2002. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J. Biol. Chem. 277: 42899–42911. [DOI] [PubMed] [Google Scholar]
- Osheim, Y.N., Proudfoot, N.J., and Beyer, A.L. 1999. EM visualization of transcription by RNA polymerase II: Downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell 3: 379–387. [DOI] [PubMed] [Google Scholar]
- Osheim, Y.N., Sikes, M.L., and Beyer, A.L. 2002. EM visualization of pol II genes in Drosophila: Most genes terminate without prior 3′ end cleavage of nascent transcripts. Chromosoma 111: 1–12. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. and O’Sullivan, J. 2002. Polyadenylation: A tail of two complexes. Curr. Biol. 12: 855–857. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 502–512. [DOI] [PubMed] [Google Scholar]
- Roguev, A., Schaft, D., Shevchenko, A., Pijnappel, W.W., Wilm, M., Aasland, R., and Stewart, A.F. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20: 7137–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev, A., Schaft, D., Shevchenko, A., Aasland, R., and Stewart, A.F. 2003a. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J. Biol. Chem. 278: 8487–8493. [DOI] [PubMed] [Google Scholar]
- Roguev, A., Shevchenko, A., Schaft, D., Thomas, H., and Stewart, A.F. 2003b. A comparative analysis of an orthologous proteomic environment in the yeasts S. cerevisiae and S. pombe. Mol. Cell. Proteomics. 3: 125–132. [DOI] [PubMed] [Google Scholar]
- Sadowski, M., Dichtl, B., Hübner, W., and Keller, W. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 22: 2167–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Schneider, R., Bannister, A.J., Sherriff, J., Bernstein, B.E., Emre, N.C., Schreiber, S.L., Mellor, J., and Kouzarides, T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., Schneider, R., Bernstein, B.E., Karabetsou, N., Morillon, A., Weise, C., Schreiber, S.L., Mellor, J., and Kouzarides, T. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12: 1325–1332. [DOI] [PubMed] [Google Scholar]
- Senger, B., Simos, G., Bischoff, F.R., Podtelejnikov, A., Mann, M., and Hurt, E. 1998. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 17: 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.F., Gaitatzes, C., Saxena, K., and Neer, E.J. 1999. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185. [DOI] [PubMed] [Google Scholar]
- Steinmetz, E.J. and Brow, D.A. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 23: 6339–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, E.J., Conrad, N.K., Brow, D.A., and Corden, J.L. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput. Appl. Biosci. 10: 19–29. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, D.P., Kim, S.J., Park, N.J., Jew, T.M., and Martinson, H.G. 2001. Mechanism of poly(A) signal transduction to RNA polymerase II in vitro. Mol. Cell. Biol. 21: 7495–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B.M. 2002. Cellular memory and the histone code. Cell 111: 285–291. [DOI] [PubMed] [Google Scholar]
- van Helden, J., del Olmo, M., and Perez-Ortin, J.E. 2000. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 28: 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]