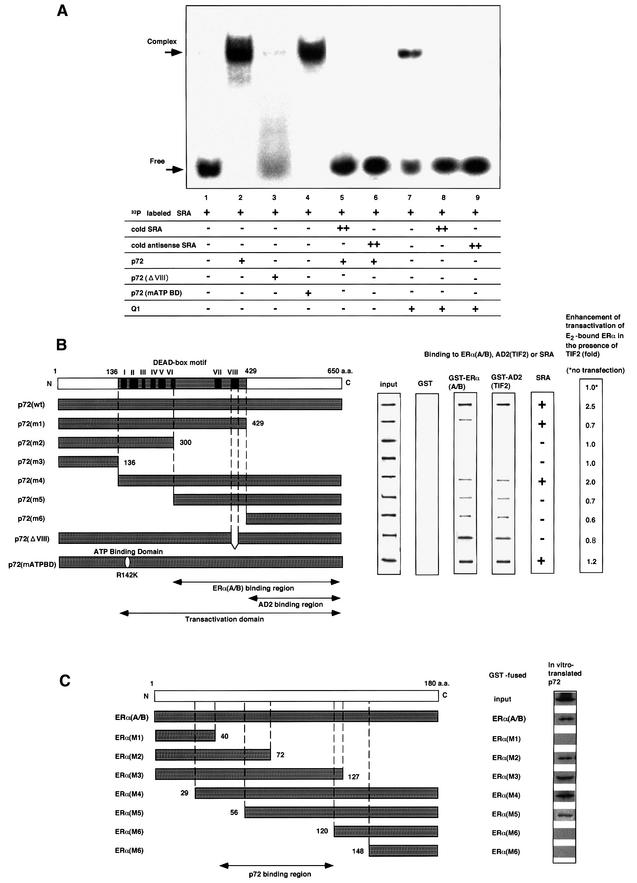
Fig. 5. p72 acts synerigistically with an RNA coactivator, SRA, through direct RNA binding. (A) Binding of p72 to an RNA coactivator, SRA. An RNA gel-shift assay was performed as described in Materials and methods. [α-32P]CTP-labeled SRA was incubated with proteins p72, p72ΔVIII, p72 mATP BD or human Q1 (10 ng) in either the presence or absence of excess non-labeled cold SRA (2 µg) (Gross and Shuman, 1998). (B) p72 functional domains for hERα, TIF2 and SRA binding. The binding regions for the SRC-1/TIF2 AD2 domains, and the hERα (A/B), and the transactivation domain in p72 are illustrated in the lower panel. The DEAD-box motif with the well-conserved I–VIII regions for putative RNA helicase activity is shown in the upper panel. The interaction domains for the hERα A/B and TIF2 AD2 domain were mapped with the deletion mutants (left panel) in the GST pull-down assay (middle panel). SRA binding to the deletion mutants was estimated by RNA gel-shift assay as shown in (A). The domain for the ligand-induced transactivation function of hERα in the presence of TIF2 was estimated by luciferase assay using the extracts from COS-1 cells expressing hERα, TIF2 and the truncated p72 mutants in the presence of E2 (10–8 M). The fold activations of the truncated p72 mutants are shown as fold induction in the right panel. (C) p72-interacting region in the A/B domain of hERα. To map the p72-interacting site in the A/B region of hERα, in vitro translated p72 was incubated with either GST–hERα(A/B) or GST–hERα(A/B) mutant (M1, M2, M3, M4, M5, M6 or M7) immobilized on glutathione–Sepharose beads (Kobayashi et al., 2000). The bound proteins were subjected to SDS–PAGE followed by autoradiography.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.