Abstract
We found that heterozygous mutant alleles of E(Pc) and esc increased homologous recombination from an allelic template in somatic cells in a P-element-induced double-strand break repair assay. Flies heterozygous for mutant alleles of these genes showed increased genome stability and decreased levels of apoptosis in imaginal discs and a concomitant increase in survival following ionizing radiation. We propose that this was caused by a genomewide increase in homologous recombination in somatic cells. A double mutant of E(Pc) and esc had no additive effect, showing that these genes act in the same pathway. Finally, we found that a heterozygous deficiency for the histone deacetylase, Rpd3, masked the radiation-resistant phenotype of both esc and E(Pc) mutants. These findings provide evidence for a gene dosage-dependent interaction between the esc/E(z) complex and the Tip60 histone acetyltransferase complex. We propose that esc and E(Pc) mutants enhance homologous recombination by modulating the histone acetylation status of histone H4 at the double-strand break.
DOUBLE-STRAND breaks (DSBs) are, arguably, one of the most lethal forms of DNA damage and may lead to genomic instability or cell death in mitotically dividing cells if not repaired precisely. Eukaryotes repair DSBs through two highly conserved pathways: nonhomologous end-joining and homologous recombination. The preferred pathway seems to depend on the tissue in which the DSB has occurred (Johnson-Schlitz and Engels 1993), timing of the DSB (Hendrickson 1997; Saleh-Gohari and Helleday 2004), and histone acetylation status at the break site (Bird et al. 2002; Jazayeri et al. 2004).
When P-element excision induces a DSB in the Drosophila germ line, homologous recombination by gene conversion is the primary repair pathway (Engels et al. 1990; Gloor et al. 1991). While this repair preferentially copies information from the sister chromatid, it can also use the allelic site on the homolog (Johnson-Schlitz and Engels 1993). In somatic tissue, the predominant repair pathway is nonhomologous end-joining, but repair also occurs by gene conversion from the homolog (Gloor et al. 2000). During homologous recombination in yeast, the DSB is processed to leave 3′ overhanging ends, which are used by the homology search machinery to find and invade a homologous template in the genome (Haber 2000). DNA synthesis is primed from these ends, thereby restoring the information at the break site. Studies in higher eukaryotes have shown that repair by homologous recombination is more likely during the S and G2 phases of the cell cycle when the sister chromatid templates are available, while nonhomologous end-joining is more frequent during the G1 phase (Hendrickson 1997; Saleh-Gohari and Helleday 2004). Regardless of the tissue or timing of the DSB, for precise repair, the homology search machinery must maneuver through DNA packaged tightly into chromatin to find an appropriate template, and it is therefore reasonable to expect that proteins affecting local or global chromatin structure play a role in the repair of DNA DSBs.
The effect of chromatin structure on a genomewide homology search was examined previously by Lankenau et al. (2000). They found that gene conversion from an ectopic template was increased in the absence of the Su(Hw) chromatin insulator protein in the germ line. This protein helps in organizing the eukaryotic genome into global chromatin domains and the lack of this protein results in disorganized chromatin (Gerasimova and Corces 1998; Gerasimova et al. 2000; Byrd and Corces 2003). Thus, Lankenau et al. (2000) postulated that increased DSB repair occurred because the broken ends could more easily maneuver throughout the nucleus, making it easier to identify the template molecule.
Interestingly, many Polycomb group (PcG) and trithorax group (trxG) mutants show a loss of association between chromatin insulator sites at the nuclear periphery and a dispersal of these sites throughout the nucleus. This suggests that PcG proteins also help in promoting a structural hierarchy of chromatin within the nucleus (Gerasimova and Corces 1998). Furthermore, emerging evidence in mammalian systems implicates the PcG proteins in cellular proliferation and tumorigenesis (Bracken et al. 2003; Kirmizis et al. 2003; Raaphorst et al. 2003; Dukers et al. 2004; Leung et al. 2004; Pasini et al. 2004b; Sanchez-Beato et al. 2004; Wang et al. 2004; Attwooll et al. 2005; Gil et al. 2005; Kuzmichev et al. 2005). For these reasons, we decided to investigate the role that PcG proteins might play in DSB repair in somatic cells. Here we show that heterozygosity for mutations in either extra sex combs (esc) or Enhancer of Polycomb [E(Pc)] causes an increase in homologous recombination and enhances genome stability in somatic cells.
MATERIALS AND METHODS
Drosophila stocks:
All stocks were cultured at 24° on standard sugar, cornmeal, and agar medium. Crosses were brooded for 4–8 days and scored until 1 day before eclosion of the F2 generation. Genetic symbols not described here can be found in Lindsley and Zimm (1992) or on FlyBase at http://www.flybase.org.
wbio; CyO/Sp; ry Sb Δ2-3/TM6 (Ubx):
This stock provided a stable transposase source, Δ2-3 (99B), on the third chromosome for P-element mobilization (Robertson et al. 1988). The white (w) gene of this stock also served as the allelic repair template for all somatic gene conversion experiments presented here. An uncharacterized mutation in the promoter region inactivates the white gene, but it retains wild-type sequence at the allelic site opposite the DSB (W. R. Engels, personal communication).
wbio; CyO amosRoi-1/esc6; ry Sb Δ2-3/TM6 (Ubx):
This stock was used to introduce a stable transposase source and the esc6 allele for somatic suppression experiments using an esc+ transgene. The CyO amosRoi-1 chromosome was obtained from W. Engels and is abbreviated as CyO Roi.
y ac whd spl and y ac sc whd-y+ spl:
These stocks each carry the 629-bp nonautonomous P-element whd80k17 (hereafter whd) insertion in exon 6 of the white gene, resulting in a bleach white-eye phenotype (Engels et al. 1990). y ac sc whd-y+ spl, a gift of William Engels, contains a 7978-bp insertion of the Drosophila yellow gene 238 bp downstream of the target break site in the 3′-UTR of the white gene. achaete (ac), scute (sc), and Notchsplit (spl) were used as markers flanking the white gene. All mutant strains were obtained from the Bloomington Stock Center, except where noted: Pcl11, Psc1, Psce22, PscvgD Su(z)2vgD, Df(2R)en-A, E(Pc)1 (obtained independently from Hugh Brock), and esc1. Chromosomes containing these mutations were balanced by either CyO or SM5 chromosomes. A stock carrying the esc6 allele as well as an esc+ transgene, P{esc+ ry+}E2, on the second chromosome balancer, CyO, hereafter CyO P{esc+}, was obtained from Erich Frei. We detected the presence of a cryptic white+ transgene on the second chromosome. This cryptic white+ transgene likely resulted when the P{esc+ ry+} construct was moved to the second chromosome from the third chromosome using the Tn70.1 P transposase source, which contains hobo ends and a w+ marker gene (Gutjahr et al. 1995), although no transposase activity remained (data not shown). L2 Pin1/CyO, P{w[+mC]=GAL4-Kr.C}DC3, P{w[+mC]=UAS-GFP.S65T}DC7 (hereafter CyO GFP) was used to create E(Pc)1 and esc6 heterozygous stocks that were balanced with CyO GFP. This stock was chosen for its GFP expression starting at early stage 9 embryogenesis through to adulthood (Casso et al. 2000). Dp(1; Y)BS; ru1 st1 e1 spn-A1 ca1/TM3, Sb1 contains a mutation for the Rad51 homolog, spindle-A (spn-A1). Df(3R)X3F, P{ry+t7.2=RP49}A3-84F e1/TM3, Sb1 is deficient for spn-A [hereafter Df(3R)X3F].
Df(3L)GN24/TM8, I(3)DTS41 is deficient for Rpd3. y1w67c23 and red1 e1 (Jim Kennison) were used as wild-type controls in the Rpd3 radiation sensitivity assays.
In(2LR)Gla, wgGla-1 Bc1/In(2LR) bwVDe2, bwVDe2 provided the Black cell (Bc1) larval marker for the second chromosome balancer [hereafter In(2LR) Gla Bc1] and Su(z)126 red1 e1/TM6B, Sb1 Tb1 (Jim Kennison) provided the Tubby (Tb1) larval marker for the third chromosome balancer to differentiate heterozygous mutants.
Double-strand break repair assay:
Males carrying the white gene repair template (wbio allele) and transposase source in the presence of a wild-type or PcG mutant allele were crossed to females bearing the target break site (recipient chromosome, whd or whd-y+) to obtain flies for gene conversion analysis (Figure 1). For each experiment, a minimum of 100 eyes were analyzed from female progeny of the genotype whd-y+ or whd/wbio; PcG mutant/+; Sb Δ2-3/+ or whd-y+ or whd/wbio; CyO/+; Sb Δ2-3/+. To analyze the trans-heterozygote, esc6/E(Pc)1, whd-y+ or whd/wbio; esc6/E(Pc)1; Sb Δ2-3/+ or whd-y+ or whd/wbio; CyO/E(Pc)1; Sb Δ2-3/+ female flies were collected in the somatic analysis. The esc+ transgene was analyzed by collecting whd-y+ or whd/wbio; CyO P{esc+}/esc6; Sb Δ2-3/+ or whd-y+ or whd/wbio; esc6/+; Sb Δ2-3/+ females. All single-mutant somatic experiments were set up such that the PcG mutant allele was transmitted paternally; all progeny retained the full maternal wild-type complement of every PcG gene tested (except when tested in trans). Consequently, we tested for the effects of decreased zygotic PcG expression on somatic gene conversion. The donor (wbio) and recipient (whd or whd-y+) chromosome was the same for each experiment so that the only variable was the presence or absence of the PcG allele.
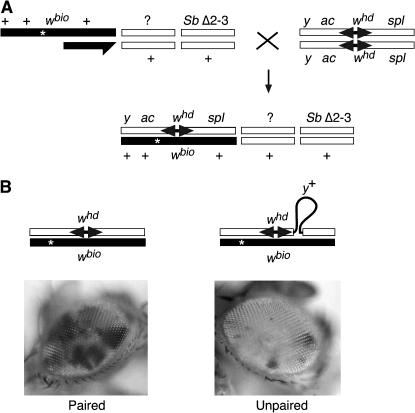
Figure 1.
Mating scheme to assess gene conversion in somatic tissue. (A) Males with a white gene repair template (wbio) on the X chromosome, a wild-type or PcG mutant allele (indicated by a question mark), and a stable transposase source (Sb Δ2-3) were crossed to females with an X chromosome bearing the target break site (whd). The break is induced by excision of the P element (double solid arrow). The template chromosome has an uncharacterized mutation in the promoter region of the white gene (asterisk) that renders it inactive. Female progeny of the genotype whd-y+ or whd/wbio; ?/+; SbΔ2-3/+ were collected for analysis of somatic gene conversion. (B) The proposed configuration of paired (left) and unpaired (right) homologous chromosomes prior to gene conversion and the typical eye phenotypes following gene conversion in an otherwise wild-type background. In the unpaired situation (right), a 7978-bp heterologous yellow gene sequence (y+) located 238 bp downstream of the P-element insertion in the whd chromosome is shown as a thin black loop.
Somatic gene conversion analysis:
Images of individual eyes were captured using a digital camera. Image analysis with pooled controls was carried out as described (Coveny et al. 2002). Photography and analysis were conducted as double-blind procedures. Internal controls were collected throughout to ensure reproducibility but were pooled across several experiments.
In addition, experiments were conducted in duplicate using wild-type sibling flies as matched controls. Following image capture, files were processed and analyzed by the less subjective method that follows. The image-processing steps were automated using a program written in Perl. JPEG images were converted to RGB split images. ImageMagick (http://www.imagemagick.org) was used to extract the red channel and to enhance the contrast to bring out areas of dark red pigment using a normalized value of 20%. These images were saved as TIFF files for analysis in Adobe Photoshop using the RGB mode. The outline of the eye was traced and a histogram of the enclosed area at level 50 gave a measure of the area covered by black pixels. For both methodologies, the data were tabulated so that the percentage of the eye covered by red tissue was associated with its image file. The proportion of the eye covered by red tissue was variable within each experiment, requiring the measurement of at least 100 eyes to achieve a representative sample.
Genome integrity assay:
The genome instability assay was adapted from Brodsky et al. (2000). esc6/CyO GFP and E(Pc)1/CyO GFP females were crossed to Canton-S wild-type males. Eggs were collected for 12 hr and allowed to develop to obtain third instar larvae. On day 5, larvae were exposed to 0 or 40 Gy of γ-irradiation in a Co60 source (Gammacell 220, Atomic Energy of Canada). Upon eclosion, 10–20 female flies of each genotype [esc6/+, E(Pc)1/+, or CyO GFP/+] were analyzed for bristle phenotypes. For the P{esc+} transgene experiment, esc6/CyO GFP females were crossed to CyO P{esc+}/esc6 males, and female flies of the reciprocal genotypes were analyzed. Wild-type flies were also tested directly as controls. A total of 34 macrochaetes were examined per fly [orbitals (6), ocellar (2), verticals (4), postvertical (2), presutural (2), notopleurals (4), supra-alars (4), dorsocentrals (4), anterior post-alars (2), and scutellars (4)] and scored as mutant if at least 50% shorter or significantly thinner than wild type (the Minute phenotype).
Apoptosis assay:
Third instar larvae were collected as in the genome instability assay. On day 5, larvae were exposed to 0, 8, 20, or 40 Gy of γ-irradiation in a Co60 source and allowed to recover for 4 hr at 24°. During the recovery period, esc6/+, E(Pc)1/+, and +/+ larvae were separated from CyO GFP/+ larvae on the basis of green fluorescent protein (GFP) expression using a fluorescent dissecting microscope. Eye and wing imaginal discs from Canton-S wild-type, CyO GFP/+, esc6/+, or E(Pc)1/+ third instar larvae were dissected, stained with 1 μg/ml acridine orange (Brodsky et al. 2000) in phosphate buffered saline (PBS) for 1 min to visualize apoptotic cells, and washed twice in PBS. Discs were mounted in PBS and imaged immediately using the fluorescein channel of a fluorescence microscope. The numbers of apoptotic cells per disc were counted following extraction of wing discs from larvae irradiated with 0 or 8 Gy. At this low radiation dose, differences between wild-type and mutant discs were still evident but more easily counted. These discs were stained and allowed to flatten under a coverslip into a monolayer. The images were captured, inverted to obtain the negative in Adobe Photoshop, and printed to facilitate manual counting. Apoptotic cells in 6–12 discs were counted for each genotype.
Radiation sensitivity assays for esc6 and E(Pc)1:
Female flies with the genotypes esc6/CyO GFP, E(Pc)1/CyO GFP, or L2Pin1/CyO GFP were crossed to Canton-S wild-type males. Eggs were collected for 12 hr and allowed to develop for 4–5 days. Third instar larvae were either untreated or irradiated with 20 or 40 Gy using a Cs137 source and allowed to pupariate. Irradiated second and third instar larvae of each genotype were also collected and examined for pupariation; in these experiments, all larvae pupariated. On day 7, pupae were collected and separated on the basis of GFP expression; esc6/+, E(Pc)1/+, or L2 Pin1/+ pupae were detected by lack of GFP, while the CyO GFP/+ pupae were used as the wild-type control. The L2 Pin1/+ flies were used as a background control. The L2 allele affects the regulation of eye imaginal disc growth. Neither L2 nor Pin1 alleles were expected to have an effect on survival following irradiation. As well, the esc6/CyO GFP and E(Pc)1/CyO GFP stocks were derived from the L2 Pin1/CyO GFP stock. Eclosion of adult flies was scored until 10 days after irradiation. The number of pupae that eclosed was expressed as a percentage of the total number of pupae.
Rpd3 radiation sensitivity assay:
Chromosomes containing E(Pc)1, esc6, Df(3L)GN24, spn-A1, Df(3R)X3F, or red1 e1 were heterozygous over In(2LR) Gla Bc1 or TM6B, Sb1 Tb1. Single heterozygous males were crossed to 12–14 y1w67c23 females. For the double-mutant experiments, Df(3L)GN24/TM6B, Sb1 Tb1 males were mated to 12–14 esc6/In(2LR) Gla Bc1 or E(Pc)1/In(2LR) Gla Bc1 virgins. Matings for the y1w67c23 control stock were tested directly, without the introduction of balancer chromosomes. Bottle matings were brooded every 24–48 hr up to day 7. Progeny were identified and collected if they lacked the phenotypes associated with the balancer chromosomes. These larvae were irradiated as late third instar larvae using a Co60 source; siblings were taken as untreated controls. Eclosed adults were scored up to 10 days following treatment, and survival was expressed as a percentage of the number of flies eclosed over the total.
Statistics:
Since the somatic eye analysis data do not follow the normal distribution, we used resampling with replacement, commonly known as bootstrap analysis, to determine confidence limits (Manly 1997). This was carried out for all somatic experiments with 100,000 iterations to determine exact P-values. Standard deviation values were not determined since they are uninformative for skewed plots. For the chromosome instability, radiation sensitivity, and apoptosis experiments, weighted averages were used in a two-tailed unpaired t-test to determine statistical significance.
RESULTS
Somatic DSB repair assays:
We investigated the effect of several PcG genes on somatic cell DSB repair by measuring the proportion of the eye covered by red pigmentation (Figure 1 and materials and methods). Red patches are produced in the eye when a DSB, caused by excision of the P-element from the whd allele, is repaired by homologous recombination using the allelic white gene as a template. Since every cell in the adult head has undergone P-element excision and subsequent repair, white areas indicate repair by nonhomologous end-joining. Thus, we can measure the proportion of DSBs repaired by each pathway (Gloor et al. 2000). We initially tested the effect of a number of PcG genes using the pooled control assay in both the paired and unpaired configurations explained in Figure 1. Plots of the cumulative proportion of eyes covered by at least the indicated percentage of red pigment for a variety of conditions are shown in Figure 2A. In these plots, a rightward shift of the experimental line relative to the control line indicates that more eyes are covered by more red pigment; this represents an increase in homologous recombination and a corresponding decrease in nonhomologous end-joining. At least 100 eyes were analyzed for each experiment. We used resampling with replacement (bootstrapping) (Manly 1997) to determine the likelihood that an experimental distribution as extreme as that observed could be chosen by chance from a joint distribution composed of the control and the experimental distributions.
Figure 2.
The effect of mutant PcG genes on DSB repair using pooled controls. The fraction of the eye covered with red pigment was measured for at least 100 eyes in flies heterozygous for the mutations in the indicated genes. Shown here is the cumulative proportion of eyes covered by at least the indicated percentage of pigment. (A) Measurements from the unpaired and paired chromosomal configurations are solid and dashed lines, respectively. The controls for this analysis were pooled across several experiments and are shown as solid boxes associated with thick lines. For convenience, the pooled control distributions were derived from sibling CyO/+ flies. Control distributions from flies lacking any balancer chromosomes were indistinguishable from the CyO/+ distribution (not shown). Lines associated with each genotype are indicated in the key. (B) The pooled control for the paired configuration (solid box), esc6/+ (open box), and esc6/P{esc+} (open circle).
The number of eyes analyzed and the likelihood that the experimental and control distributions are the same are given in Table 1. We found that flies heterozygous for either single mutant, E(Pc)1 or esc6, showed a significant shift to the right compared to the controls in both the paired and the unpaired arrangements. Flies heterozygous for the Pcl11 mutation also had a strong shift to the right in the unpaired configuration, but this mutation was not tested in the paired configuration. Flies heterozygous for a chromosome containing both the Pscvg-D and Su(z)2vg-D mutant alleles showed a slight shift to the right in both the paired and the unpaired assays. In contrast, flies heterozygous for single mutants in Psc (either Psc1 or Psce22) had distributions indistinguishable from the control in the unpaired configuration. The P-values reported in Table 1 allowed us to infer that flies heterozygous for the E(Pc)1, Pcl11, or esc6 mutations had significantly more red pigment in their eyes than the control flies following the repair of somatic DSBs. The pooled control analysis was relatively rapid, but was potentially susceptible to the effects of genetic background. In addition, only one allele of the esc and E(Pc) genes was examined. Therefore, we repeated the analysis for esc and E(Pc) using a second allele with the matched-control method. Eyes from sibling wild-type flies were collected as controls to reduce the effect of genetic background in this analysis. Figure 3A shows the results for the paired configuration with matched controls. Flies heterozygous for the E(Pc)1 mutant allele showed a moderate but significant shift to the right in both the paired and the unpaired chromosomal arrangements (P-values and sample sizes given in Table 2). In addition, we used the unpaired assay and tested the effect of flies heterozygous for Df(2R)en-A, a large deficiency that includes the E(Pc) gene. In this experiment a greater proportion of eyes contained more red pigment than the controls, although this difference was not quite significant (Table 2). Note that the unpaired E(Pc)1 and Df(2R)en-A distributions are virtually superimposable, but that the two control distributions are somewhat different. Therefore, the differences in significance between E(Pc)1 or Df(2R)en-A and their control distributions is due to the different control distributions. Figure 3B shows the results of a similar analysis on the esc gene. Here we tested flies heterozygous for either the null esc6 allele or the hypomorphic esc1 allele and found that the esc6 allele resulted in significantly more red pigment in the eyes compared to the controls in both the paired and the unpaired assays (Table 2). However, we found that flies heterozygous for the esc1 allele showed a significant increase in pigmentation only in the paired configuration.
TABLE 1.
Sample sizes and P-values for the pooled control analysis
| Paired
|
Unpaired
|
|||
|---|---|---|---|---|
| Genotype | No.a | P-value | No. | P-value |
| CyO/+ | 322 | ND | 570 | ND |
| esc6/+ | 150 | <1 × 10−5 | 351 | <1 × 10−5 |
| E(Pc)1/+ | 114 | <1 × 10−5 | 491 | 0.0063 |
| Pcl11/+ | ND | ND | 334 | 4.3 × 10−4 |
| Psc Su(z)2/+ | 314 | 0.0062 | 328 | 0.052 |
| Psce22/+ | 101 | 0.13 | ND | ND |
| Psc1/+ | 238 | 0.21 | ND | ND |
ND, not done.
Number of eyes measured.
Figure 3.
The effect of heterozygous mutations of E(Pc) and esc on DSB repair using matched controls. Measurements from unpaired and paired chromosomal arrangements are displayed as solid or dashed lines, respectively. Each mutant allele was heterozygous for a wild-type chromosome; the controls in this case were sibling flies with a CyO/+ genotype. (A) Results for the E(Pc) alleles. (B) The effect of mutant alleles of esc on DSB repair. The paired controls for esc1 and esc6 were indistinguishable and were merged into one joint distribution. This is indicated as the pooled control line. (C) The results for the E(Pc)1, esc6 double mutant. Flies with the second chromosome genotypes indicated in the key were generated and the pigmentation coverage was measured in both the paired (dashed lines) and the unpaired chromosomal arrangements (solid lines).
TABLE 2.
Sample sizes and P-values for the matched control analysis
| Experimental genotype
|
Paired
|
Unpaired
|
||||
|---|---|---|---|---|---|---|
| Controla | Experimentala | P-value | Control | Experimental | P-value | |
| Df(2R)en-A | ND | ND | NDb | 333 | 419 | 0.061 |
| E(Pc)1 | 211 | 261 | 0.016 | 309 | 305 | 0.0063 |
| esc1 | 317c | 253 | <1 × 10−5 | 207 | 359 | 0.24 |
| esc6 | 317 | 342 | <1 × 10−5 | 296 | 355 | 0.0063 |
Number of eyes measured.
P-value not determined.
Matched controls for esc1 and esc6 were indistinguishable (P = 0.21) and were pooled.
Genomic integrity in esc6 and E(Pc)1 mutants:
Homologous recombination reconstitutes the information at the break site and generally stabilizes the genome. In contrast, nonhomologous end-joining is imprecise and the end-rejoining may be promiscuous, leading to the production of chromosome rearrangements and fragments (Guirouilh-Barbat et al. 2004). Therefore, if the effects of the esc or E(Pc) mutations on DSB repair seen at the white locus were genomewide, we predicted that flies heterozygous for either the esc6 or the E(Pc)1 mutation should have fewer genome rearrangements following exposure to ionizing radiation than wild-type flies. Genome integrity can be measured by observing the production of clonal populations of cells lacking one or more of the ∼50 Minute loci (which encode ribosomal proteins) after exposure to ionizing radiation (Brodsky et al. 2000). One manifestation of such clonal populations is the production of macrochaete bristles that appear shorter and thinner or are lacking altogether (Minute phenotype).
There was no significant difference in macrochaete appearance of unirradiated esc6/+ and E(Pc)1/+ animals compared to control flies. Interestingly, no bristle defects were observed in any of the 20 CyO GFP/+ control flies examined, but loss of 1 bristle was observed in 2 of the 16 E(Pc)1/+ flies and in 1 of the 16 esc6/+ flies. Following exposure to 40 Gy of ionizing radiation, an average of 5.6 ± 1.1 bristles were defective from CyO GFP/+ control flies compared to only 2.9 ± 0.62 for E(Pc)1/+ (P = 0.028) and 1.9 ± 0.41 for esc6/+ flies (P = 0.0051). We conclude that the genome is significantly more stable following exposure to ionizing radiation in E(Pc)1/+ and esc6/+ mutants than in the control flies.
Decreased apoptosis in esc6 and E(Pc)1 mutants:
Not surprisingly, following irradiation, we noted that the eyes were often slightly smaller in size and showed disordered ommatidia in the wild-type and in the E(Pc)1/+ and esc6/+ flies. This rough-eye phenotype is typical of radiation-induced apoptosis and is commonly seen, in part, because the products of promiscuous nonhomologous end-joining include acentric and dicentric chromosomes (Zhivotovsky and Kroemer 2004). This phenotype increased in severity with increased radiation dose in both wild-type and mutant flies and appeared qualitatively similar, although it was not completely penetrant (not shown).
We predicted that because of increased genome integrity, fewer cells in E(Pc)1 and esc6 heterozygotes than in the controls would undergo radiation-induced apoptosis. Imaginal discs were removed from third instar larvae following exposure to 0 or 8 Gy of ionizing radiation and stained for apoptotic cells using the vital dye acridine orange (Ciapponi et al. 2004). Within each genotype, wing and eye discs showed similar levels of fluorescent nuclei (not shown); therefore wing discs were used in the analysis. Unirradiated discs from wild-type, esc6, or E(Pc)1 mutant discs had very few apoptotic cells, although both the esc6 and E(Pc)1 mutant wing discs showed a slightly higher basal level of apoptosis than control discs (Figure 4). This was an interesting observation given that the unirradiated esc6/+ and E(Pc)1/+ flies showed slightly more defective macrochaetae than wild-type flies. However, following treatment with 8 Gy of radiation, there were significantly fewer apoptotic cells from both esc6 and E(Pc)1 heterozygous mutants (779 ± 90 and 676 ± 106) than from wild-type (1419 ± 110) discs (P < 0.0005 for both mutations; Figure 4, A and B). Qualitatively similar results were observed at doses of 20 or 40 Gy, but there were too many apoptotic cells to derive an accurate count. We concluded that esc6 and E(Pc)1 reduced radiation-induced apoptosis by about twofold in wing imaginal discs.
Figure 4.
Reduced radiation-induced apoptosis in E(Pc)1 and esc6 mutants. Wild-type and E(Pc)1 and esc6 heterozygous mutant third instar larvae were exposed to 0 or 8 Gy of γ-irradiation. Wing discs were dissected, stained with acridine orange, and flattened under a coverslip. (A) Typical images for a wild-type and an esc6/+ wing disc. (B) The numbers of apoptotic cells were counted in each disc treated with 0 or 8 Gy. The average number of apoptotic cells for each genotype was determined by subtracting the average number of apoptotic cells in untreated discs for each genotype from the average number of apoptotic cells following treatment with 8 Gy. For each genotype and treatment, 8–12 discs were examined. Error bars represent the standard error of the mean.
Flies carrying the esc6 and E(Pc)1 mutants are radiation resistant:
Finally, we reasoned that the mutant animals should become resistant to ionizing radiation because of the decrease in apoptosis and the increase in genome stability. Third instar larvae were treated with 0, 20, or 40 Gy of ionizing radiation and allowed to pupariate before being separated on the basis of GFP expression. We tested the following genotypes: esc6/+, E(Pc)1/+, L2Pin1/+ as a background control, and a wild-type control CyO GFP/+. Table 3 shows the number of pupae that eclosed to adulthood expressed as a percentage of the total number of pupae at each dose. We found that survival following irradiation was strongly enhanced in animals heterozygous for a single-mutant copy of either the esc6 or the E(Pc)1 mutation and that the greatest difference between the wild-type and mutant animals was observed at the highest radiation dose. At 40 Gy, 77.9% of esc6/+ and 80.0% of E(Pc)1 flies eclosed compared to 54.0 and 53.5% of their controls. As expected, there was no significant difference in survival of the background control flies, L2 Pin1/+, over the same CyO GFP/+ control regardless of radiation dose. We conclude that the esc6 and E(Pc)1 mutations confer radiation resistance in the heterozygous state, possibly by increasing genomewide homologous recombination.
TABLE 3.
Survival of esc and E(Pc) heterozygous flies following γ-irradiation
| % survival (N,a SD,bP-valuec)
|
|||
|---|---|---|---|
| Genotype | 0 Gy | 20 Gy | 40 Gy |
| Wild typed | 77.0 (1849, 7.4) | 58.5 (2270, 7.8) | 54.0 (1557, 4.1) |
| esc6/+ | 88.0 (1629, 5.4, NS) | 82.1 (1848, 3.7, ***) | 77.9 (1570, 2.8, ***) |
| Wild type | 79.0 (3897, 5.0) | 62.1 (3774, 2.2) | 53.5 (1749, 6.5) |
| E(Pc)1/+ | 85.9 (3188, 1.6, *) | 74.7 (3100, 3.3, **) | 80.0 (1465, 7.1, ***) |
| Wild type | 81.9 (2080, 4.1) | 67.8 (2023, 7.6) | 56.5 (1360, 7.5) |
| L2Pin1/+ | 83.5 (1854, 2.3, NS) | 74.8 (2151, 4.4, NS) | 53.5 (1983, 7.8, NS) |
Total number of flies irradiated.
Standard deviation of the mean.
Compared to wild type: NS, not significant; * <0.05; ** <0.01; *** <0.005.
CyO GFP/+ sibling wild-type controls for next experimental row.
Effects suppressed by an esc+ transgene:
The effects that we observed on the repair of the P-element-induced DSB were modest, but consistent between the pooled and matched control experiments and were found with two different alleles of each gene. Corresponding effects were observed in the genome integrity, apoptosis, and ionizing radiation sensitivity assays. However, it was still formally possible, although unlikely, that an uncharacterized mutation on one or more of the chromosomes was responsible for the observed effects. Therefore, we used a wild-type esc transgene that could rescue both the maternal and zygotic esc6 phenotypes (Gutjahr et al. 1995) to see if it would suppress the increase in pigmentation observed following P-element excision and suppress the increase in genome integrity associated with the esc6 mutation.
We first used the pooled control method to measure suppression in the DSB repair assay at the white locus. The results, shown in Figure 2B, demonstrate that flies heterozygous for the esc6 allele and the P{esc+} transgene (N = 268) showed a significant decrease in pigment coverage compared to flies heterozygous for only the esc6 allele (P = <1 × 10−5). This suppression was not quite complete, as the distribution of pigment coverage for flies with both the esc6 allele and the P{esc+} transgene was also different from the pooled control (P = 0.001). Further investigation showed that the CyO P{esc+} chromosome also carried a cryptic P{w+} transgene that contributed some red pigment by gene conversion from the ectopic white gene (not shown). This would tend to increase the amount of pigment in the eyes, but we observed a strong decrease in pigmentation. It is therefore likely that the extra pigment contributed by gene conversion from the ectopic P{w+} transgene prevented complete suppression in this experiment.
We also measured the ability of the P{esc+} transgene to suppress the ionizing-radiation-dependent increase in genome stability associated with the heterozygous esc6 mutation. We observed that adults derived from esc6/CyO GFP mutant third instar larvae exposed to 40 Gy of ionizing radiation had about one-third the number of mutant bristles (3.8 ± 0.4, N = 25) than did control wild-type animals (12.1 ± 1.4, N = 8). This effect in the esc6/CyO GFP animals was completely suppressed by the addition of the P{esc+} transgene; esc6/CyO P{esc+} animals had an average of 12.35 ± 1.1 mutant bristles (N = 26). The numbers of defective bristles in the wild-type control and the transgene suppression groups were indistinguishable, but both were significantly different from the esc6/CyO GFP group (P < 0.0001). Together with the results from the DSB repair assay, we concluded that the effect on DSB repair, genome integrity, apoptosis, and ionizing radiation was caused by the mutation in extra sex combs.
Mutants in esc and E(Pc) genes are not additive:
We next focused on the mechanism by which mutants in esc and E(Pc) increased homologous recombination in somatic cells. It was possible that these genes affected the transcription of DNA repair genes. Expression of mRNA corresponding to several DNA repair genes, spnA (Rad51), okra (Rad54), and Rad50, was similar in an E(Pc)1/+ and a wild-type genetic background when compared by Northern blots (not shown).
As a further test of the transcriptional model, we used the somatic DSB repair assay to analyze the well-known genetic interaction between E(Pc)1 and esc6. Strong alleles of E(Pc) are recessive lethal, have no phenotype as heterozygotes, and enhance the phenotypes of a large number of PcG genes, including esc (Lindsley and Zimm 1992).
In the pooled control assay, we found that flies heterozygous for both the esc6 and the E(Pc)1 alleles had a distribution of pigmentation that was indistinguishable from either single mutant in either the paired (N = 216, P = 0.25 vs. esc6 alone, P = 0.16 vs. E(Pc)1 alone) or the unpaired arrangement (N = 162, P = 0.48 vs. esc6 alone, P = 0.30 vs. E(Pc)1 alone). Repeating this analysis using the matched control method gave a similar result for both the paired and the unpaired configurations. Here the distributions of E(Pc)1/CyO vs. E(Pc)1/esc6 virtually overlapped in both the paired and the unpaired chromosomal arrangements (Figure 3C). The likelihood that the joint distribution of the single and double mutants was the same was 0.35 and 0.36 in the paired and unpaired analyses [N = 131 (paired) and 106 (unpaired) for E(Pc)1/CyO, N = 390 (paired) and 342 (unpaired) for E(Pc)1/esc6]. We concluded that the E(Pc)1 allele did not enhance the esc phenotype in this assay. Together with the earlier observation that mutations in Psc have no effect in the DSB repair assay, this suggests that esc and E(Pc) affect the balance between homologous recombination and nonhomologous end-joining by a mechanism distinct from that used by these genes to modulate gene expression.
Rpd3 masks esc6 and E(Pc)1 radiation sensitivity:
A second possibility was that the esc and E(Pc) proteins interacted with a third protein that affects the balance between the two major DSB repair pathways. Recent results from yeast show that the levels of histone H3 and H4 acetylation change dynamically over the course of DSB repair (Tamburini and Tyler 2005). Mutant forms of Esa1 or Yng2, key proteins in the Tip60/NuA4 histone acetylase complex, which contains the E(Pc) protein, cause defects in nonhomologous end-joining in yeast (Choy et al. 2001; Bird et al. 2002). Mutations in Rpd3 also cause defects in the same pathway (Jazayeri et al. 2004; Tamburini and Tyler 2005). Furthermore, studies in yeast show that modulation of H4 acetylation status differentially affects nonhomologous end-joining and homologous recombination; nonhomologous end-joining occurs only if all four lysines on the H4 tail are acetylated, while homologous recombination requires acetylation of only a subset of these lysine residues (Bird et al. 2002). Therefore, the balance between these two pathways could be changed if histone H4 became hypoacetylated in the region of the DSB. We focused on the Rpd3 gene because it has a DSB repair defect in yeast and the Rpd3 protein associates with the esc/E(z) protein complex (Tie et al. 2003; Jazayeri et al. 2004). We first tested a deficiency that includes the Rpd3 gene, Df(3L)GN24, for sensitivity to ionizing radiation to see if there was a DNA repair defect in Drosophila similar to that seen in yeast. The results shown in Table 4 demonstrate a moderate sensitivity to ionizing radiation following irradiation with 30 Gy; only 47% of Df(3L)GN24/+ larvae survived to adulthood compared to ∼66% survival of the wild-type control larvae. Note that virtually all of the larvae that are heterozygous for either of two alleles of the fly RAD51 ortholog [spnA1 or Df(3R)X3F] fail to survive to adulthood. We next tested the Rpd3 deficiency in combination with the esc6 or E(Pc)1 mutations. There was a striking difference between the esc6 and E(Pc)1 heterozygotes, both of which show at least 85% survival under the same conditions, and these mutations in combination with Rpd3. Larvae heterozygous for both esc6 and Df(3L)GN24 or E(Pc)1 and Df(3L)GN24 had nearly the same frequency of eclosion to adulthood as did the flies carrying only the Rpd3 deficiency, indicating that the deficiency for Rpd3 masked the ability of both the esc6 and E(Pc)1 mutations to enhance survival following exposure to ionizing radiation. We suggest that Rpd3 is acting before the esc and E(Pc) genes in a pathway that directly or indirectly affects histone acetylation status at the DSB.
TABLE 4.
Ionizing radiation analysis of the Rpd3 deficiency at 30 Gy
| Genotype | Survivors | Total | Proportion | Range (Na) |
|---|---|---|---|---|
| E(Pc)1/+ | 519 | 586 | 0.89 | 0.83–1 (6) |
| esc6/+ | 605 | 707 | 0.86 | 0.75–0.93 (5) |
| Df(3L)GN24/+ | 119 | 252 | 0.47 | 0.47 (1) |
| Df(3L)GN24/E(Pc)1 | 77 | 133 | 0.58 | 0.58 (1) |
| Df(3L)GN24/esc6 | 65 | 150 | 0.43 | 0.43 (1) |
| red1 e1 | 518 | 766 | 0.68 | 0.62–0.75 (3) |
| y1 w67c23 | 498 | 750 | 0.66 | 0.51–0.77 (3) |
| Df(3R)X3F/+ | 11 | 681 | 0.016 | 0.0025–0.036 (2) |
| spnA1/+ | 20 | 1076 | 0.019 | 0.0090–0.023 (4) |
Number of replicates.
DISCUSSION
Eukaryotes use both homologous recombination and nonhomologous end-joining to repair DSBs; survival is compromised severely when both are inactivated (Couedel et al. 2004). In our homologous recombination assay, every cell in the adult head had a DSB generated at the white locus by excision of the P element. This was true for flies heterozygous for either the esc6 or the E(Pc)1 mutation and for wild-type flies (data not shown). These breaks were repaired either by homologous recombination using the allelic white gene or by nonhomologous end-joining (Gloor et al. 2000). We found that heterozygous mutants of any of three genes, E(Pc), Pcl, or esc, increased the likelihood that homologous recombination was used to repair a DSB made by P-element excision in cells of the developing eye-antennal imaginal disc. Thus, although we did not measure nonhomologous end-joining directly, we were able to conclude that the increase in pigmentation indicated an increase in the repair of DSBs by homologous recombination at the expense of nonhomologous end-joining and that heterozygosity for null alleles of either gene changed the balance between these two pathways. Further studies will be needed to determine if there is a defect in nonhomologous end-joining that is compensated by an increase in homologous recombination, or if nonhomologous end-joining is unaffected and homologous recombination is enhanced.
The repair bias toward homologous recombination was observed when either paired or unpaired allelic templates were used. This suggested that the effects of the esc and E(Pc) mutants were not related to any chromosome-pairing-dependent activities of these genes.
Alterations in DSB repair capacity or DSB pathway choice were not restricted to DSBs made by P-element excision at the white locus. Heterozygous mutants of either esc or E(Pc) showed a significant increase in genome integrity and a significant decrease in apoptosis following exposure to ionizing radiation. Not surprisingly, these animals were somewhat resistant to high doses of ionizing radiation.
The increase in homologous recombination and the increase in genome integrity seen in esc6 heterozygous animals was suppressed by a P{esc+} transgene. This demonstrated that the effects were specific to the esc mutations and were not caused by genetic background effects. While we did not suppress the effect of the E(Pc)1 mutant with a transgene, we did observe that a large deficiency that included the E(Pc) gene had a similar effect to that of E(Pc)1 in the homologous recombination assay.
Notably, the increase in homologous recombination was not seen in animals heterozygous for either single mutant of Psc. Furthermore, Psc, Pc, or Scm heterozygous animals are not resistant to ionizing radiation (data not shown). The esc, Pcl, E(Pc), and Psc genes produce proteins that are localized to three different complexes. The proteins produced from the esc and Pcl genes are members of the esc/E(z) histone H3 methyltransferase complex (Tie et al. 2003), the E(Pc) protein is found in the Tip60/NuA4 histone acetyltransferase complex (Doyon et al. 2004), and the Psc, Pc, and Scm proteins are components of the PRC1 complex (Francis et al. 2001). Interestingly, the esc and E(Pc) mutants alter the choice of repair pathway by a common mechanism since the esc6/E(Pc)1 double mutant lacked an additive effect in the homologous recombination assay over either single mutant. This suggests that the effects that we observe on the choice of repair pathway are not linked to the methylation activity of the esc/E(z) complex or to the methyl-H3-binding activity of PCR1, but rather are linked to some other function that connects the esc/E(z) and Tip60/NuA4 complexes.
Since the esc/E(z) complex often associates with a histone chaperone and the Rpd3 histone deacetylase (Furuyama et al. 2003; Tie et al. 2003) and the E(Pc) protein is a component of a histone acetyl transferase known to be required for DSB repair in yeast (Bird et al. 2002; Choy and Kron 2002; Allard et al. 2004; Doyon and Cote 2004), we tested the hypothesis that histone acetylation forms a link between these two complexes and found that a heterozygous mutation for Rpd3 blocked the ability of heterozygous esc6 or E(Pc)1 alleles to confer resistance to ionizing radiation. This suggests that the Rpd3 gene product acts upstream of the esc and E(Pc) proteins in a pathway that influences the choice of which mechanism is used to repair DSBs.
Histone acetylation and DSB repair:
In yeast, the acetylation status of histone H4 plays a crucial role in determining whether a DSB is repaired by nonhomologous end-joining or by homologous recombination (Bilsland and Downs 2005), and this role is distinct from its role in regulation of gene expression (Boudreault et al. 2003). Recent work in yeast and Drosophila shows that the NuA4/Tip60 histone acetyltransferase complex [which includes the yeast E(Pc) ortholog] is recruited to DSB sites and acetylates the tail lysines of histone H2A and H4 (Bird et al. 2002; Kusch et al. 2004; Bilsland and Downs 2005). Histone acetylation is thought to neutralize the positively charged histone tail, thereby reducing the affinity between DNA and histones and loosening the compaction of the chromatin (Doyon and Cote 2004). The homologous recombination or nonhomologous end-joining repair machinery can then gain access to the damage and facilitate repair.
Furthermore, Bird et al. (2002) have found that nonhomologous end-joining requires the acetylation of all four lysine residues on the H4 histone tail, whereas homologous recombination requires only partial acetylation of these lysines. Likewise, mutations in the nonessential NuA4 subunit, Yng2, result in global hypoacetylation of histone H4 (Choy et al. 2001) and are synthetic lethal with the YKU70 gene, further supporting the argument that nonhomologous end-joining requires complete acetylation of histone H4 (Choy and Kron 2002). Finally, yeast with hypoacetylated histone H4 not only are proficient in homologous recombination but also show enhanced recombination in a sister-chromatid exchange assay (Choy and Kron 2002). These data suggest a compensatory relationship between nonhomologous end-joining and homologous recombination in Saccharomyces cerevisiae.
A model for the genetic interaction among Rpd3, esc, and E(Pc):
The yeast data, together with our results, suggest a model for the gene dose-dependent interaction among Rpd3, esc, and E(Pc) in Drosophila. We propose that, similar to S. cerevisiae, nonhomologous end-joining in Drosophila somatic cells is dependent upon full acetylation of histone H4 tails, but that homologous recombination is not. In this model, a heterozygous E(Pc) mutation would cause a decrease in E(Pc) protein levels, resulting in a decrease in activity of the Tip60 histone acetyltransferase at the DSB and a shift toward homologous recombination. Likewise, a heterozygous esc mutation would result in less of the esc/E(z) complex. Since the Rpd3 protein is found in several different complexes (Jazayeri et al. 2004; Robert et al. 2004; Tamburini and Tyler 2005) aside from the esc/E(z) complex (Furuyama et al. 2003; Tie et al. 2003), a decrease in the amount of esc/E(z) complex would result in more free Rpd3 protein in the cell. The excess free Rpd3 protein could deacetylate more of the histone H4 at the DSB and thus shift DSB repair toward homologous recombination. In either instance, a decrease in the levels of Rpd3 protein in the presence of mutations in either esc or E(Pc) might be expected to result in more complete histone H4 acetylation and to restore the normal balance between homologous recombination and nonhomologous end-joining.
It is becoming increasingly clear that PcG proteins have functions beyond the regulation of homeotic genes. Many recent studies have identified deregulation of different PcG proteins (Ezh2, Pcl, Bmi-1) in tumorigenesis (Bracken et al. 2003; Pasini et al. 2004a; Sanchez-Beato et al. 2004; Wang et al. 2004), suggesting increased proliferation as a possible mechanism. If esc or E(Pc) proteins were highly expressed in mammalian cancer, one might expect more frequent nonhomologous end-joining and, consequently, genome instability. Conversely, loss of one copy of either gene may provide a survival advantage under challenge with ionizing radiation or radiomimetic drugs. It is thus possible that the increased proliferation of tumor cells with mutations in these genes is, in part, not a direct result of increased expression of these genes, but rather a secondary effect of genome instability caused by decreases in gene conversion and increases in error-prone nonhomologous end-joining.
Acknowledgments
We are extremely grateful to Faye Males for her excellent technical assistance and support. We thank Jacob Paul, Xeon Ramdewar, Sameer Mal, Esther Hsueh, Zamir Khan, Danesh Byayana, Vu Luong, Kadija Hersi, Michael Bertoia, Jennifer Baysarowich, and Kathryn Millar for technical support. Many thanks to Anthony Percival-Smith and David Litchfield for use of the dissecting fluorescence and fluorescence microscope. We are also thankful to the editor, M. Simmons, and the anonymous reviewers for their helpful suggestions. A.H. (née Coveny) was funded in part by an Ontario Graduate Scholarship. K.W. was funded in part by an Ontario Graduate Scholarship, a Natural Sciences and Engineering Research Council studentship, and an Ontario Graduate Scholarship in Science and Technology. This work was supported by an operating grant to G.G. from the Canadian Institutes of Health Research.
References
- Allard, S., J. Y. Masson and J. Cote, 2004. Chromatin remodeling and the maintenance of genome integrity. Biochim. Biophys. Acta 1677: 158–164. [DOI] [PubMed] [Google Scholar]
- Attwooll, C., S. Oddi, P. Cartwright, E. Prosperini, K. Agger et al., 2005. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J. Biol. Chem. 280: 1199–1208. [DOI] [PubMed] [Google Scholar]
- Bilsland, E., and J. A. Downs, 2005. Tails of histones in DNA double-strand break repair. Mutagenesis 20: 153–163. [DOI] [PubMed] [Google Scholar]
- Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon et al., 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415. [DOI] [PubMed] [Google Scholar]
- Boudreault, A. A., D. Cronier, W. Selleck, N. Lacoste, R. T. Utley et al., 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17: 1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken, A. P., D. Pasini, M. Capra, E. Prosperini, E. Colli et al., 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22: 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, M. H., J. J. Sekelsky, G. Tsang, R. S. Hawley and G. M. Rubin, 2000. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 14: 666–678. [PMC free article] [PubMed] [Google Scholar]
- Byrd, K., and V. G. Corces, 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casso, D., F. Ramirez-Weber and T. B. Kornberg, 2000. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 91: 451–454. [DOI] [PubMed] [Google Scholar]
- Choy, J. S., and S. J. Kron, 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 22: 8215–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy, J. S., B. T. Tobe, J. H. Huh and S. J. Kron, 2001. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 276: 43653–43662. [DOI] [PubMed] [Google Scholar]
- Ciapponi, L., G. Cenci, J. Ducau, C. Flores, D. Johnson-Schlitz et al., 2004. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr. Biol. 14: 1360–1366. [DOI] [PubMed] [Google Scholar]
- Couedel, C., K. D. Mills, M. Barchi, L. Shen, A. Olshen et al., 2004. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 18: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coveny, A. M., T. Dray and G. B. Gloor, 2002. The effect of heterologous insertions on gene conversion in mitotically dividing cells in Drosophila melanogaster. Genetics 161: 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon, Y., and J. Cote, 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14: 147–154. [DOI] [PubMed] [Google Scholar]
- Doyon, Y., W. Selleck, W. S. Lane, S. Tan and J. Cote, 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24: 1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukers, D. F., J. C. van Galen, C. Giroth, P. Jansen, R. G. Sewalt et al., 2004. Unique polycomb gene expression pattern in Hodgkin's lymphoma and Hodgkin's lymphoma-derived cell lines. Am. J. Pathol. 164: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W. R., D. M. Johnson-Schlitz, W. B. Eggleston and J. Sved, 1990. High-frequency P element loss in Drosophila is homolog dependent. Cell 62: 515–525. [DOI] [PubMed] [Google Scholar]
- Francis, N. J., A. J. Saurin, Z. Shao and R. E. Kingston, 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8: 545–556. [DOI] [PubMed] [Google Scholar]
- Furuyama, T., F. Tie and P. J. Harte, 2003. Polycomb group proteins esc and E(Z) are present in multiple distinct complexes that undergo dynamic changes during development. Genesis 35: 114–124. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., and V. G. Corces, 1998. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., K. Byrd and V. G. Corces, 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6: 1025–1035. [DOI] [PubMed] [Google Scholar]
- Gil, J., D. Bernard and G. Peters, 2005. Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 24: 117–125. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B., N. A. Nassif, D. M. Johnson-Schlitz, C. R. Preston and W. R. Engels, 1991. Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253: 1110–1117. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B., J. Moretti, J. Mouyal and K. J. Keeler, 2000. Distinct P-element excision products in somatic and germline cells of Drosophila melanogaster. Genetics 155: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat, J., S. Huck, P. Bertrand, L. Pirzio, C. Desmaze et al., 2004. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14: 611–623. [DOI] [PubMed] [Google Scholar]
- Gutjahr, T., E. Frei, C. Spicer, S. Baumgartner, R. A. White et al., 1995. The Polycomb-group gene, extra sex combs, encodes a nuclear member of the WD-40 repeat family. EMBO J. 14: 4296–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J. E., 2000. Partners and pathways: repairing a double-strand break. Trends Genet. 16: 259–264. [DOI] [PubMed] [Google Scholar]
- Hendrickson, E. A., 1997. Cell-cycle regulation of mammalian DNA double-strand-break repair. Am. J. Hum. Genet. 61: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri, A., A. D. McAinsh and S. P. Jackson, 2004. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 101: 1644–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz, D. M., and W. R. Engels, 1993. P-element-induced interallelic gene conversion of insertions and deletions in Drosophila melanogaster. Mol. Cell. Biol. 13: 7006–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis, A., S. M. Bartley and P. J. Farnham, 2003. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol. Cancer Ther. 2: 113–121. [PubMed] [Google Scholar]
- Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser et al., 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306: 2084–2087. [DOI] [PubMed] [Google Scholar]
- Kuzmichev, A., R. Margueron, A. Vaquero, T. S. Preissner, M. Scher et al., 2005. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc. Natl. Acad. Sci. USA 102: 1859–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankenau, D. H., M. V. Peluso and S. Lankenau, 2000. The Su(Hw) chromatin insulator protein alters double-strand break repair frequencies in the Drosophila germ line. Chromosoma 109: 148–160. [DOI] [PubMed] [Google Scholar]
- Leung, C., M. Lingbeek, O. Shakhova, J. Liu, E. Tanger et al., 2004. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428: 337–341. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Manly, B., 1997. Randomization, Bootstrap and Monte Carlo Methods in Biology. Chapman & Hall/CRC, New York.
- Pasini, D., A. P. Bracken and K. Helin, 2004. a Polycomb group proteins in cell cycle progression and cancer. Cell Cycle 3: 396–400. [PubMed] [Google Scholar]
- Pasini, D., A. P. Bracken, M. R. Jensen, E. Lazzerini Denchi and K. Helin, 2004. b Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23: 4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst, F. M., C. J. Meijer, E. Fieret, T. Blokzijl, E. Mommers et al., 2003. Poorly differentiated breast carcinoma is associated with increased expression of the human polycomb group EZH2 gene. Neoplasia 5: 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy et al., 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Johnson-Schlitz, W. K. Benz et al., 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118: 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari, N., and T. Helleday, 2004. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 32: 3683–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Beato, M., E. Sanchez, J. F. Garcia, A. Perez-Rosado, M. C. Montoya et al., 2004. Abnormal PcG protein expression in Hodgkin's lymphoma. Relation with E2F6 and NFkappaB transcription factors. J. Pathol. 204: 528–537. [DOI] [PubMed] [Google Scholar]
- Tamburini, B. A., and J. K. Tyler, 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25: 4903–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie, F., J. Prasad-Sinha, A. Birve, A. Rasmuson-Lestander and P. J. Harte, 2003. A 1-megadalton esc/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol. Cell. Biol. 23: 3352–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., G. P. Robertson and J. Zhu, 2004. A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene 343: 69–78. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky, B., and G. Kroemer, 2004. Apoptosis and genomic instability. Nat. Rev. Mol. Cell Biol. 5: 752–762. [DOI] [PubMed] [Google Scholar]