Abstract
RSC is an essential, multisubunit chromatin remodeling complex. We show here that the Rsc4 subunit of RSC interacted via its C terminus with Rpb5, a conserved subunit shared by all three nuclear RNA polymerases (Pol). Furthermore, the RSC complex coimmunoprecipitated with all three RNA polymerases. Mutations in the C terminus of Rsc4 conferred a thermosensitive phenotype and the loss of interaction with Rpb5. Certain thermosensitive rpb5 mutations were lethal in combination with an rsc4 mutation, supporting the physiological significance of the interaction. Pol II transcription of ca. 12% of the yeast genome was increased or decreased twofold or more in a rsc4 C-terminal mutant. The transcription of the Pol III-transcribed genes SNR6 and RPR1 was also reduced, in agreement with the observed localization of RSC near many class III genes. Rsc4 C-terminal mutations did not alter the stability or assembly of the RSC complex, suggesting an impact on Rsc4 function. Strikingly, a C-terminal mutation of Rsc4 did not impair RSC recruitment to the RSC-responsive genes DUT1 and SMX3 but rather changed the chromatin accessibility of DNases to their promoter regions, suggesting that the altered transcription of DUT1 and SMX3 was the consequence of altered chromatin remodeling.
Transcription occurs in the crowded context of the nucleus in which genes are wrapped in chromatin. The first step in gene expression involves the modification and/or the remodeling of repressive chromatin by specialized complexes. For polymerase II (Pol II)-transcribed genes, these steps are followed by the recruitment of Mediator, the general transcription factors (GTFs) and the Pol II itself, although in an order that can vary from one promoter to another (9, 34). The pathway leading from silent chromatin to transcription by Pol I and Pol III has not been studied as thoroughly but is globally similar, with an additional contribution of cognate GTFs. In yeast and human cells, the Pol III-specific transcription factor TFIIIC has been found to be required for the proper nucleosomal organization of Pol III genes (4, 23, 32). In the case of Pol I transcription, the mammalian termination factor TTF-I is able to activate transcription by promoting chromatin remodeling in synergy with ATP-dependent cofactors in vitro (24). Transcription initiation is not the only step at which chromatin might interfere with transcription. Nucleosomes residing in the transcribed region can inhibit the movement of RNA polymerases during elongation. To contend with this, the FACT complex helps human Pol II transcribe through nucleosome-induced blocks (28, 38). These observations suggest that factors that relieve the repressive effect of nucleosomes might act in conjunction with the transcription machinery at the successive stages of the transcription cycle.
The repressive effect of nucleosomes is overcome by two cooperative mechanisms. The first involves the covalent modification of the histones, including the acetylation of specific histone tail lysines by acetyl transferases (reviewed by Strahl and Allis[48]). The second mechanism is affected by multiprotein complexes termed remodelers that reposition the nucleosomes and/or modify their association with the DNA (reviewed by Vignali et al. [51] and Narlikar et al. [35]). The RSC complex belongs to a group of remodelers, the prototype of which is the yeast SWI/SNF complex (7). RSC comprises 15 subunits, 5 of which are highly similar to SWI/SNF subunits and 2 of which are shared with SWI/SNF complex (1, 6-8, 11, 26, 43, 50). In contrast to SWI/SNF, RSC is essential for yeast cell survival and is 10 times more abundant (7).
DNA microarray studies have been performed to analyze the consequences of mutations in Rsc3, Rsc30, and Rsc4 subunits of the RSC complex on transcription by Pol II (1, 22). Together, these three subunits control, either positively or negatively, the transcription of ∼10% of the yeast genome. Strikingly, these subunits controlled the transcription of distinct sets of genes, suggesting that they might direct the recruitment of RSC to particular locations. The number and distribution of RSC gene targets in the genome has been investigated by microarray hybridization of DNA enriched by chromatin immunoprecipitation of RSC subunits (11, 36). RSC was found to bind 700 to 1,400 targets in the yeast genome, usually in intergenic regions. One of the striking features was that about one-quarter of the RSC targets were adjacent to genes transcribed by Pol III, suggesting a role for RSC in the transcription of some class III genes (36). Recently, it has been shown that Rsc4 tandem bromodomains interact with histone H3 acetylated at Lys 14 (22). Mutations in Rsc4 bromodomains affected viability, modified histone H3 recognition and impaired Pol II transcription of numerous genes. However, it is not known whether Rsc4 bromodomains are important for targeting RSC to specific locations through interactions with modified histones or if they are important for regulating nucleosome remodeling, or both.
Although the in vitro activity of RSC has been well documented (30, 31, 41, 44), few studies have addressed the role of RSC in chromatin remodeling in vivo. Hsu et al. reported that RSC mutations altered chromatin organization at the centromeres and chromosome segregation (20). In the case of the CHA1 gene, it was found that the depletion of the RSC catalytic subunit, Sth1, led to open chromatin structure and active transcription, indicating that RSC was required for the maintenance of a repressive nucleosomal state (33).
The large size and the multisubunit structure of RSC suggest that RSC might interact with a variety of partners distinct from histones to be targeted to specific locations on the genome or to regulate its activity. In the present study, we found that Rsc4 subunit of RSC interacts with Rpb5, a subunit shared by the three forms of RNA polymerases, via its C terminus. We studied the functional consequences of Rsc4 mutations affecting its C terminus on viability, gene transcription, RSC recruitment, and chromatin remodeling.
MATERIALS AND METHODS
Media, plasmids, and strains.
Yeast strains are described in Table 1. Three hemagglutinin (HA)- or thirteen Myc-tagged strains were constructed as described by Longtine et al. (29). The Escherichia coli strains used were DH5α [endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR (φ80d lacZΔ M15)] and BL21 [F− ompT hsdS(rB− mB−)]. Bacteria were grown in LB medium supplemented with kanamycin (50 μg/ml) or ampicillin (100 μg/ml) for, respectively, pENTR or pDEST20 and pDEST17 plasmids.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| MW3461 | MATaade2-101 lys2-801 leu2-Δ1 his3-Δ200 ura3-52 trp1-Δ63 rpc160-Δ1::HIS3 pC160-240[TRP1 CEN4 RPC160-240] rsc4::13MYC::Kan-MX6 | This study |
| MW3522 | MATaura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 RPA190::3HA::HIS3-MX6 | This study |
| MW3720 | MATaade2-101 lys2-801 leu2-Δ1 his3-Δ200 ura3-52 trp1-Δ63 rpc160-Δ1::HIS3 pC160-6 [URA3 CEN4 RPC160] rsc4::13MYC::Kan-MX6 RPA190::3HA::HIS3-MX6 | This study |
| MW3730 | MATaura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4::13MYC::Kan-MX6 | This study |
| MW3922 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4-Δ3::HIS3 | This study |
| MW3993 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4-Δ4::HIS3 | This study |
| MW4019 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4-Δ4::HIS3 STH1::13MYC::Kan-MX6 | This study |
| MW4023 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 STH1::13MYC::Kan-MX6 | This study |
| YPH499 | MATaura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 | 47 |
| YPH500 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 | 47 |
| Y190 | MATagal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL→lacZ LYS2::GAL(UAS)→HIS3 cyhR | 18 |
| YCB1 | MATaura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rpb5::ura3::LEU2 rsc4-Δ4::HIS3 pFL44-RPB5 (RPB5 URA3 2μ) | This work |
| YFN2 | MATaura3-52 his3-Δ200 ade2-1 trp1-Δ63 lys2-801 leu2-Δ1 rpb5::ura3::LEU2 pFL44-RPB5 (RPB5 URA3 2μ) | Zaros et al., unpublished |
| D439-2a | MATα ura3-52 trp1-Δ63 lys2-801 leu2-Δ1 rpb1-1 | 3 |
| MW4407 | MATaura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 RPA190::3HA::KanMX6 | This study |
| MW4409 | MATaura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4-Δ4::HIS3 RPA190::3HA::KanMX6 | This study |
| MW4415 | MATaura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 RPC160::3HA::KanMX6 | This study |
| MW4417 | MATaura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4-Δ4::HIS3 RPC160::3HA::KanMX6 | This study |
| MW4424 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 STH1::13MYC::Kan-MX6 RPA190::3HA::TRP1 | This study |
| MW4428 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 STH1::13MYC::Kan-MX6 RPC160::3HA::TRP1 | This study |
| MW4430 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4-Δ4::HIS3 STH1::13MYC::Kan-MX6 RPA190::3HA::TRP1 | This study |
| MW4434 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4-Δ4::HIS3 STH1::13MYC::Kan-MX6 RPC160::3HA::TRP1 | This study |
Yeast genetic techniques and media have been described by Sherman (46). Standard molecular biology techniques were as described by Sambrook et al. (42). Two-hybrid interactions were tested in strain Y190 (18) as described previously (14, 53).
Immunoprecipitations.
Whole-cell extracts were prepared from 100 ml of cells growing exponentially in yeast extract-peptone-dextrose (YPD) medium. Cells were collected at an optical density at 600 nm of 0.6 to 0.8, washed twice with water and twice with lysis buffer (50 mM HEPES [pH 7.5], 100 mM NaCl, 20% glycerol, 1 mM dithiothreitol [DTT], 0.5 mM EDTA, 0.05% NP-40) supplemented with a protease inhibitor cocktail (Complete; Roche) and 1 mM phenylmethylsulfonyl fluoride, and resuspended in 0.5 ml of the same buffer. Lysis was performed in the presence of glass beads (0.2 ml, 425 to 600 μm) by vortexing for 30 min at 4°C. The cell debris were eliminated by centrifugation (15 min at 4°C at 18,000 × g, twice). The supernatant was collected and stored at −80°C.
During the immunoprecipitation procedure, the incubations and washes were performed at 4°C with agitation. A total of 2 × 107 anti-mouse immunoglobulin G (IgG)-magnetic beads (Dynal M450) were washed with 0.1% bovine serum albumin in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) and preincubated for 30 min in 0.1% bovine serum albumin in PBS. The 12CA5 anti-HA antibody (70 ng/μl), 9E10 anti-Myc antibody (30 ng/μl), or 8WG16 antibody (200 ng/μl) were added for 1 h, after which the beads were washed three times for 5 min and twice shortly with incubation buffer (the same as lysis buffer). The protein extracts (15 μg/μl, 100 μl) were incubated with the beads for 3 h, after which the beads were washed three times (5 min) with incubation buffer. The affinity-purified proteins were released from the beads by boiling them for 10 min. Eluted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with 12CA5 anti-HA, 9E10 anti-Myc, anti-Rsc4, anti-Rsc6 (B. Cairns, unpublished material), or 8WG16 anti-CTD of Rpb1 (Covance).
In vitro interaction studies.
To overexpress Rpb5, Rsc4, Rsc4Δ4, Rsc4-C93, and Rsc4-C93-Δ4 proteins, the RPB5 open reading frame (ORF), the RSC4 ORF, or the RSC4 C-terminal domain, as well as the RSC4 ORF or the RSC4 C-terminal domain lacking codons for the four last amino acids, were amplified by PCR of genomic DNA and cloned in the pENTR/D-TOPO vector (Invitrogen). The oligonucleotide sequences are available on request. The resulting pENTR plasmids were verified by DNA sequence analysis. The plasmids for the glutathione S-transferase (GST) fusion proteins Rsc4, Rsc4Δ4, Rsc4-C93, and Rsc4-C93-Δ4 and the hexahistidine (His6)-Rpb5 fusion protein expression were obtained by an LR Gateway reaction between the pENTR plasmids and the pDEST20 or pDEST17 plasmids (Invitrogen).
Rsc4, Rsc4Δ4, Rsc4-C93, and Rsc4-C93-Δ4 were expressed as GST fusion proteins in High Five insect cells by using Bac-to-Bac baculovirus expression system (Invitrogen). Cell extracts were prepared as described previously (12) except that sonication was performed in PBS (pH 7.4) containing 20% glycerol, 5 mM 2-mercaptoethanol, and protease inhibitor cocktail (Complete). Then, 1-ml lysates (6 to 8 mg of total protein) were diluted to final volume of 20 ml in loading buffer (PBS [pH 7.4], 1 mM phenylmethylsulfonyl fluoride, 10 mM DTT, 0.1% Triton X-100). and loaded onto 0.5 ml of glutathione-Sepharose 4 Fast Flow columns (Amersham Biosciences) equilibrated with the same buffer plus 1% Triton X-100. The columns were washed sequentially with 15 ml of (i) loading buffer; (ii) PBS, 1 M NaCl, and 10 mM DTT; and (iii) PBS plus 10 mM DTT. GST fusion proteins were eluted with 5 ml of elution buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 10 mM DTT, 20 mM reduced glutathione, 5% glycerol), and 0.5-ml fractions were collected. Peak fractions were pooled, dialyzed against dialysis buffer (10 mM HEPES [pH 7.5], 50 mM NaCl, 10 mM DTT, 10% glycerol), frozen in small aliquots in liquid nitrogen, and stored at −80°C. SDS-PAGE, followed by Coomassie brilliant blue staining, showed that GST fusion proteins were purified to near homogeneity. GST alone and GST-Anc1(Taf14) were purified from E. coli BL21 transformed with recombinant plasmids (M. Kabani, unpublished material).
A total of 0.3 nmol of GST-Rsc4, GST-Rsc4Δ4, GST-Rsc4-C93, or GST-Rsc4-C93-Δ4 or as controls GST or GST-Anc1(Taf14) were incubated with 50 μl of glutathione-Sepharose 4 Fast Flow (50% slurry) in GST-binding buffer (20 mM HEPES, [pH 6.8], 100 mM KCl, 5 mM MgCl2, 0.1% NP-40, 2% glycerol, 1 mM DTT, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) and rotated for 1 h at 4°C in a total volume of 900 μl. A 40-μl aliquot of extracts of E. coli BL21 expressing His6-Rpb5 (140 μg of total protein) was added, and reaction mixtures were rotated for 2 h at 4°C. By Western blotting with anti-Rpb5 antibodies (21) compared to serial dilutions of homogeneous yeast Pol III preparation, we estimated that this amount of the extract contained ca. 1.4 nmol of His6-Rpb5 protein. Reactions were centrifuged for 2 min at 14,000 rpm, the supernatant was removed, and the pellet was washed three times with 500 μl of GST-binding buffer. The pellet fractions were boiled for 5 min in 50 μl of SDS-PAGE sample buffer. The supernatant and pellet fractions (10 μl) were then resolved by SDS-PAGE (12% polyacrylamide) and analyzed by Western blotting with polyclonal anti-GST antibodies (Sigma) and anti-Rpb5 antibodies (21).
Microarray analysis.
Gene expression was monitored with DNA microarray manufactured by the Service de Génomique Fonctionnelle (CEA/Evry, France) as described previously (13) except that an indirect cDNA labeling protocol of the targets was used (adapted from P. Brown [http://cmgm.stanford.edu/pbrown/protocols/aadUTPCouplingProcedure.htm]; see Materials and Methods in the supplemental material).
DNase I analysis of chromatin structure.
Preparation of yeast nuclei and chromatin analysis with DNase I were performed as described previously (16). Yeast strains MW4023 and MW4019 were grown in YPD medium at 25°C and then incubated for 6 h at 30°C or 37°C. The rpb1-1 mutant (strain D439-2a, the RY260 isolate obtained by backcrosses with the YPH499 wild-type strain (3), and the wild-type strain YPH499 were grown in YPD medium at 25°C and then shifted to 37°C for 1 h. Next, 250-ml portions of the cell cultures at an optical density at 600 nm of 0.8 were collected and used for nucleus preparation. Nuclear pellets were further digested with different concentrations of DNase I (Invitrogen). Typically, samples corresponding to 0.5, 1, 2, and 4 U of DNase I per ml were purified and selected for secondary digestion with the appropriate restriction enzyme: SphI for the DUT1 gene, BanII for the SMX3 gene, and SpeI for the HTA3 gene. Further, DNA was fractionated on a 1.2% agarose gel, blotted onto nylon membrane, and hybridized to the corresponding radioactively labeled probe. To prepare the probes, PCR fragments obtained by amplification on yeast genomic DNA were labeled by using a Prime-It II random primer labeling kit (Stratagene). The DUT1 PCR fragment corresponds to the region from 222 to 526 bp downstream from the initiation codon, the SMX3 fragment covers the region from 390 to 643 bp downstream from the ATG, and the HTA3 fragment corresponds to the region from 331 to 798 bp downstream from the initiation codon.
Restriction enzyme accessibility assay.
Restriction enzyme accessibility assays were performed essentially as described previously (40, 52). Cell nuclei prepared as described for DNase I analysis were digested with a saturating quantity of enzyme at 37°C for 1 h. A control without enzyme was included to monitor endonuclease activity. Purified DNA samples were quantified by SYBR Green real-time PCR using primers specific to the promoter region encompassing the corresponding restriction enzyme site. Additional control with primers to the region lacking restriction enzyme site was included. We expressed the accessibility as a percentage of the digestion on the promoter region normalized to the control region.
RESULTS
Interaction of the RSC complex with a shared subunit of RNA polymerases.
Previously, we screened the RNA polymerase subunits for interacting protein domains by using the two-hybrid system. The work centered on the interactions between RNA polymerase subunits and did not address the protein contacts between enzyme subunits and polypeptides belonging to other factors or complexes (14). We now investigate the significance of an unpublished interaction between Rpb5, one of the five subunits shared by the three nuclear RNA polymerases (45), and Rsc4, a subunit of the RSC remodeling complex (22). Using Rpb5 as a bait, two fragments were selected (one of which was found twice) that encoded the C terminus of Rsc4 subunit of the RSC chromatin remodeling complex. The interaction domains of the prey fusions spanned the C terminus of Rsc4 and began at either amino acid 533 or amino acid 558 and extended to the end of the protein at position 625, covering the last 93 or 68 amino acids of the subunit, respectively. The interaction of Rsc4 with a common polymerase subunit suggested that RSC could interact with all three RNA polymerases, a possibility we explored further.
To test this hypothesis, Sth1 was tagged at its C terminus by the insertion of a sequence encoding 13 Myc epitopes at the very end of the STH1 chromosomal ORF (29) in a wild-type strain (see Table 1 for a description of the strains used in the present study) or in strains in which the chromosomally expressed largest subunit of Pol I, A190, was tagged with three HA epitopes at its C terminus. The addition of the Myc or HA tags to these essential proteins did not affect the growth of the strain, indicating that the tag did not affect function. The Sth1-Myc expressing strain, in which the RNA polymerase subunits were not tagged, was used as a control for nonspecific adsorption. When A190-HA was immunoprecipitated with anti-HA antibodies (12CA5) in an RPA190-HA STH1-MYC strain, an anti-Myc reacting band was revealed (Fig. 1A, lane 2). No such band was observed when the immunoprecipitations (IP) were performed in strains in which Sth1 or A190 were not tagged (Fig. 1A, lanes 1 and 9, respectively), indicating that no anti-Myc reacting material was immunoprecipitated nonspecifically by 12CA5 antibody and that Sth1-Myc was not adsorbed nonspecifically to 12CA5 IgG magnetic beads. Similarly, the Myc-tagged Rsc4 subunit of RSC complex could be detected when A190-HA was immunoprecipitated from an RPA190-HA rsc4-MYC strain (Fig. S1, lane 2, in the supplemental material) but not from strains in which Rsc4-Myc or A190-HA were absent (Fig. S1, lanes 1 and 3, respectively, in the supplemental material). These observations suggest that Pol I and RSC interact specifically.
FIG. 1.
Coimmunoprecipitation of RSC with the RNA polymerases. (A) RSC coimmunoprecipitates with Pol I and Pol III. Proteins were immunoprecipitated from crude extracts with anti-HA antibodies (top two panels). The presence of HA- or Myc-tagged alleles is indicated above each lane, as is the presence of the rsc4-Δ4 mutant allele. The immunoprecipitated proteins were revealed with 12CA5 antibodies (A190-HA or C160-HA) or 9E10 monoclonal antibodies (Sth1-Myc). The position of the proteins is indicated on the side of the Western blots. The amount of proteins present in the crude extracts (30 μg) was also analyzed by Western blotting with the same antibodies (bottom two panels). (B) RSC coimmunoprecipitates with Pol II. Extracts were incubated with IgG magnetic beads to which 8WG16 antibodies directed against Rpb1 CTD were bound (Rpb1) or not (IgG), and the immunoprecipitated proteins were revealed by Western blotting with 8WG16 or 9E10 antibodies. All strains expressed Sth1-Myc. The presence of rsc4-Δ4 allele is indicated. The position of Rpb1 or Sth1-Myc in the immunoprecipitate or crude extracts is indicated on the left side of the figure.
We next tested the interaction between Pol III and RSC. Using the anti-HA antibodies, Sth1-Myc coimmunoprecipitated with C160-HA in a RPC160-HA STH1-MYC strain (Fig. 1A, lane 6) but not when Sth1 or C160 were not tagged (Fig. 1A, lanes 5 and 9). We also tested the coimmunoprecipitation of chromosomally expressed Rsc4-Myc or Rsc1-Myc fusions with C160 tagged with three HA epitopes at its N terminus (HA-C160), expressed from a centromeric plasmid under the control of its own promoter. In this strain, the wild-type chromosomal copy of RPC160 was deleted. HA-C160, but not untagged C160, was immunoprecipitated by the anti-HA antibody (Fig. S1A, lanes 4 to 7, in the supplemental material). Strikingly, both Rsc1-Myc and Rsc4-Myc coimmunoprecipitated with HA-C160 but not with untagged Pol III. Furthermore, Rsc4-Myc and HA-C160 still coprecipitated when DNase was added during the incubation, indicating that their association was not dependent on the presence of DNA (data not shown). The observation that Sth1, Rsc1, and Rsc4 coimmunoprecipitated with C160 strongly suggests that RSC complex can be associated with Pol III.
To test for an interaction between Pol II and RSC, we immunoprecipitated Pol II with 8WG16 antibodies directed against the Rpb1 CTD in the STH1-MYC strain. Sth1-Myc coimmunoprecipitated with Rpb1 when 8WG16 antibodies were bound to the IgG magnetic beads (Fig. 1B, lane 1). However, Sth1-Myc or Rpb1 was not retained by the IgG beads alone (Fig. 1B, lane 2), suggesting that RSC specifically associates with Pol II. However, we note that only ∼0.1% of total cellular RSC was immunoprecipitated under these conditions. Altogether, these coimmunoprecipitation experiments indicate that the three RNA polymerases interact with the RSC chromatin remodeling complex in extracts.
rsc4 mutants.
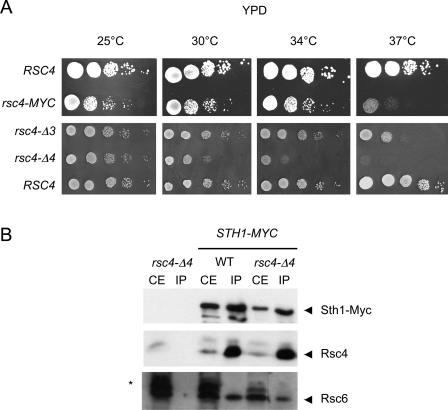
A strain bearing the rsc4-MYC allele exhibited a slight growth defect on YPD-rich medium at 30°C (Fig. 2A) and was unable to grow at 37 and 16°C (Fig. 2A and data not shown), suggesting that the presence of the Myc epitopes interfered with an essential function of the Rsc4 C terminus. In contrast, the addition of Myc epitopes to Sth1 or Rsc1 had no deleterious effect. To directly test for an essential function for the C terminus, a diploid strain was constructed in which one of the RSC4 alleles lacked the last 68 amino acids (rsc4-Δ68::Kanr). Tetrad dissection and complementation analysis indicated that Rsc4 C terminus is essential, in keeping with the sequence conservation of the last 40 amino acids of Rsc4 in various yeast species (data not shown).
FIG. 2.
The growth defects of the rsc4 mutants are not due to an RSC stability or assembly defect. (A) Growth of wild-type and rsc4 mutants at different temperatures. Serial dilutions of a wild-type strain (RSC4; YPH500) or rsc4-MYC (MW3730), rsc4-Δ3 (MW3922), or rsc4-Δ4 (MW3993) mutants were spotted on YPD medium and incubated for 3 to 4 days at the indicated temperature. (B) Rsc4-Δ4 mutant protein is correctly incorporated in the RSC complex. Protein extracts were prepared from a rsc4-Δ4 (MW3993), STH1-MYC (MW4023), or STH1-MYC rsc4-Δ4 strain (MW4019) grown for 6 h at 37°C. The protein extracts (750 μg) were incubated with anti-Myc antibodies bound to magnetic beads, and the immunoprecipitated proteins were eluted by boiling. Proteins from yeast crude extracts (CE, 30 μg) or immunoprecipitated with anti-Myc antibodies (IP) were separated by SDS-PAGE and blotted onto a nylon membrane. Proteins were revealed with antibodies against Myc, Rsc4, or Rsc6. The identity of the proteins is indicated on the right side of the blot. A nonspecific band revealed by the anti-Rsc6 antibodies in the crude extracts is indicated by an asterisk.
Next, the C terminus of Rsc4 was progressively deleted by inserting stop codons in the chromosomal copy of RSC4 gene. We found that deleting the last amino acid had no effect and that removal of five amino acids or more was lethal. Interestingly, deletion of three or four amino acids (rsc4-Δ3; rsc4-Δ4) led to a growth defect at various temperatures and a strong thermosensitive phenotype at 37°C (Fig. 2A), providing alleles useful for further study.
To investigate the role of Rsc4 C terminus on the structural integrity of the RSC complex, RSC was immunoprecipitated via chromosomally expressed Sth1-Myc in a wild-type or rsc4-Δ4 strain. The proteins immunoprecipitated with anti-Myc antibodies were analyzed by Western blotting and revealed by using anti-Myc, anti-Rsc4, or anti-Rsc6 antibodies. Rsc4-Δ4 was coimmunoprecipitated with RSC irrespective of the temperature at which the mutant was grown (25°C or 6 h at 30 or 37°C; data not shown and Fig. 2B). The abundance of Rsc4 relative to that of Sth1-Myc or Rsc6 was not affected by the mutation. Neither Rsc4-Δ4 nor Rsc6 was immunoprecipitated if Sth1 was not tagged. Rsc4-Δ4 could also be immunoprecipitated in similar experiments performed with a Rsc8-Myc-tagged strain grown at 30 or 37°C (data not shown). In addition, the Rsc4-Myc mutant protein coimmunoprecipitated with RSC (data not shown). We concluded that the growth defect of the mutant strains did not result from an assembly or stability defect of the mutant RSC complex.
We next tested whether the rsc4-Δ4 mutation impaired the interaction between the RNA polymerases and RSC. For that purpose, we performed the immunoprecipitation experiments as described above but in a rsc4-Δ4 background, immunoprecipitating either A190-HA, C160-HA, or Rpb1 in strains expressing either tagged or untagged Sth1. We found that Sth1-Myc coimmunoprecipitated Pol I or Pol II in rsc4-Δ4 strains (Fig. 1A, compare lanes 2 and 4; Fig. 1B, compare lanes 1 and 3). However, Sth1-Myc did not coimmunoprecipitate the C160-HA subunit of Pol III in the rsc4-Δ4 background (Fig. 1A, compare lanes 6 and 8).
rsc4 mutations impair interaction between Rsc4 C terminus and Rpb5.
To investigate whether the interaction between Rsc4 and Rpb5 was direct and to analyze the effect of the Rsc4-Δ4 mutation on the interaction with Rpb5, we performed GST pull-down assays. Rsc4, Rsc4-Δ4, Rsc4-C93 (corresponding to the fragment selected in the two-hybrid screen), and Rsc4-C93-Δ4 were produced as GST fusions in insect cells. The purified fusion proteins (0.3 nmol) were bound to glutathione-Sepharose beads and then incubated with a whole-cell extract of E. coli expressing Rpb5 (1.4 nmol). Unbound Rpb5 was washed away, and Rpb5 bound to the affinity beads was released by boiling and revealed by Western blotting (Fig. 3A). The beads alone, GST, or GST fused to Anc1, a subunit common to the general transcription factors TFIID and TFIIF, were used as a negative control in the pull-down experiments and did not retain Rpb5 (Fig. 3A, lanes 1 to 5, 10, and 11). In contrast, GST-Rsc4-C93 and GST-Rsc4 bound Rpb5, indicating that the interaction between the two proteins was direct (Fig. 3A, lanes 6, 7, 12, and 13). Moreover, the deletion of the last four amino acids of Rsc4-C93 markedly diminished the interaction with Rpb5 (Fig. 3A, lanes 8 and 9). However, the mutation did not impair the interaction of the complete Rsc4 protein with Rpb5 (Fig. 3A, lanes 14 and 15), suggesting that at least another segment of Rsc4 besides its C terminus contributed to the interaction with the RNA polymerase subunit.
FIG. 3.
The integrity of Rsc4 C terminus is essential for interaction with Rpb5. (A) Rsc4 interacts with Rpb5 in vitro. A total of 0.3 nmol of purified GST or GST fused to the last 93 amino acids of Rsc4 (GST-Rsc4-C93), to the complete Rsc4 protein (GST-Rsc4), to the corresponding mutants lacking the last 4 amino acids (GST-Rsc4-C93-Δ4 and GST-Rsc4-Δ4), or to the Anc1(Taf14) subunit of TFIIF and TFIID were bound to glutathione-Sepharose beads and then incubated with 140 μg of an E. coli extract expressing Rpb5 RNA polymerase common subunit (1.4 nmol). The supernatants (lanes S) were used as a positive controls. The beads were washed, and the bound material (lanes B) was released by boiling. To check for nonspecific binding, the procedure was also performed in the absence of extract (Beads). In each experiment, one-ninetieth of the supernatant and one-fifth of the bound material were loaded on SDS-PAGE gels and blotted onto a nylon membrane. As a positive control, extract from the E. coli strain expressing Rpb5 (0.011 nmol) was loaded in the first lane. Rpb5 was revealed with a polyclonal antibody (21). (B) Deletion of three or four amino acids from the Rsc4 C terminus abolished its interaction with Rpb5 in vivo. Two-hybrid interactions were tested in the Y190 strain (18), and the activation of the lacZ reporter was tested as previously described using an X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) agar overlay plate assay (53). Rpb5 is fused to the Gal4 DNA-binding domain (GDB-Rpb5). The Rsc4 last 93-amino-acid sequence (from 533 to 625; GAD-Rsc4-C93) was used as a positive control. The deletion of the three (GAD-Rsc4-C93-Δ3) or four (GAD-Rsc4-C93-Δ4) last amino acids abolished the interaction. (C) Wild-type and mutant GAD-Rsc4-C93 fusions were produced at similar levels. Equal amounts of protein extracts from Y190 transformants expressing the fusions were separated and blotted onto a nylon membrane. The fusions were revealed by using anti-HA antibodies.
We next tested whether the Rsc4-Δ4 mutant protein interacted with Rpb5 in vivo. We fused the Gal4 activation domain to fragments of Rsc4 beginning at amino acid 533 and ending at the last amino acid of the protein (GAD-Rsc4-C93) or three or four amino acids before the C terminus (GAD-Rsc4-C93-Δ3 and GAD-Rsc4-C93-Δ4, respectively). The complete Rsc4 subunit did not interact with Rpb5 in the two-hybrid system and was thus not used. Such a situation, where a domain of a protein but not the complete polypeptide interact, is not uncommon since we found previously that Rpb5 interacts in the two-hybrid system with C-terminal fragments of A190, Rpb1, and C160 but not with the complete RNA polymerase large subunits (14), even though these interactions were confirmed by the yeast Pol II crystal structure (10). Strikingly, when tested in the two-hybrid system, the deletion of three or four C-terminal amino acids led to a complete loss of the interaction between Rsc4-C93 and Rpb5 (Fig. 3B) even though the fusion proteins were produced at the same level as GAD-Rsc4-C93 (Fig. 3C). The addition of 13 Myc epitopes also abolished the interaction between Rpb5 and Rsc4 C terminus in the two-hybrid system (data not shown), indicating a close correlation between the observed loss of interaction in vivo and the growth phenotypes of the mutants.
To further document the physiological importance of the interaction between Rpb5 and Rsc4, we tested for synthetic phenotypes between rsc4-Δ4 and three rpb5 thermosensitive alleles (Fig. 4). This was done by transforming the rsc4-Δ4 rpb5 YCB1 strain, in which the rpb5 deletion was complemented by the wild-type RPB5 allele borne on a URA3 plasmid, or with TRP1 centromeric plasmids bearing various RPB5 alleles. Two of these, rpb5-H147R and rpb5-R200E, changed conserved residues (Zaros et al., unpublished data). The other conditional mutation was a chimera in which the hinge region, linking the N and C termini of Rpb5 (amino acids 120 to 146) was substituted by a mutant version of the human protein that changed H146 to K (rpb5-Chi7K). All rpb5 mutants were able to grow at 25°C, as was rsc4-Δ4. Strikingly, while rpb5-H147R and rpb5-Chi7K were unable to grow at 25°C in the presence of rsc4-Δ4, no synthetic phenotype was apparent when rsc4-Δ4 was combined with rpb5-R200E. Whereas the GST pull-down and two-hybrid experiments indicated that the interaction between Rpb5 and Rsc4 C terminus was direct and required its last four amino acids, the synthetic lethality experiments with specific RPB5 alleles supported the idea of a specific functional interaction.
FIG. 4.
Allele specific lethality between rsc4-Δ4 and rpb5 mutations. The various rpb5 mutant strains with (top plates) or without (bottom plates) rsc4-Δ4 mutations were serially diluted after growth on Casamino Acids medium supplemented with adenine and uracil (CAU) to allow the loss of the RPB5 URA3 plasmid, spotted on CAU or CAU plus 5-fluoorotic acid (CAU+5FOA) medium, and incubated for 2 to 3 days at 25°C. The identity of the RPB5 allele borne on the TRP1 pGEN vector with which the YCB1 strain (top plates) or YFN2 strain (bottom plates) were transformed is indicated on the left side. The “-” denotes the transformation by the pGEN vector alone as a negative control.
Effect of rsc4 mutations on Pol II and Pol III transcription.
The effect of rsc4 C-terminal mutations on Pol II transcription was investigated by using DNA microarray hybridization. RNAs were extracted from a wild-type or an rsc4-Δ4 mutant strain growing exponentially on YPD-rich medium at 25°C and then shifted to 30 or 37°C for 6 h. Two independent RNA preparations were analyzed by hybridization to microarrays. The mean expression ratios in the mutant versus the wild-type strain were computed. At the permissive temperature of 30°C, of the 4,396 genes that were analyzed (see Materials and Methods), 204 were induced and 124 were repressed twofold or more in the mutant (Table 2; see list in the supplemental material). At 37°C, of 4,330 analyzed genes, 338 genes were induced and 168 genes were repressed twofold or more. Thus, Rsc4 appears to control the transcription of ca. 11.7% of all yeast genes.
TABLE 2.
Summary of rsc4-Δ4 transcriptome analysis
| Gene category | No. (%) of genes expressed ata:
|
||||
|---|---|---|---|---|---|
| 30°C | 37°C | 30 and 37°C | 30°C specific | 37°C specific | |
| Expressed genes | 4,396 (79.7)* | 4,330 (78.5)* | |||
| Genes induced ≥2-fold | 204 (4.6)† | 338 (7.8)† | 112 | 92 | 226 |
| Genes repressed ≥2-fold | 124 (2.8)† | 168 (3.9)† | 57 | 67 | 111 |
*, the percentage expressed relative to the total number of spotted genes (n= 5,516); †, the percentage expressed relative to the number of expressed genes.
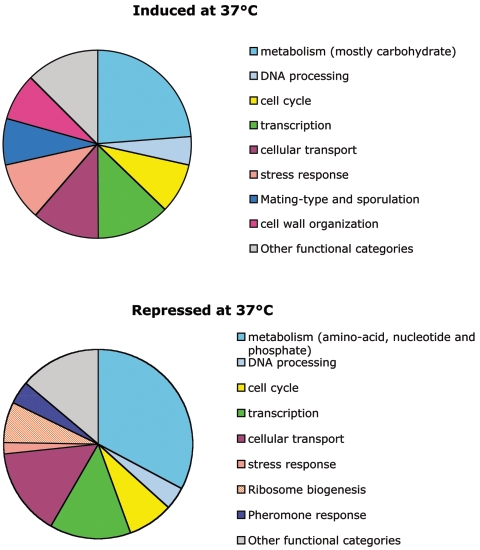
Genes misregulated in the rsc4 mutant partitioned into various Munich Information Center for Protein Sequences (MIPS) functional categories including metabolism, cell wall organization and biogenesis, cell cycle, stress response, transport or amino acid biosynthesis, etc. (Fig. 5; see also the supplemental material). Repressed genes included ribosomal proteins, proteins involved in rRNA processing, and genes involved in pheromone response, and genes from these functional categories were induced. Previous transcriptome analysis of RSC3 and RSC30 mutants indicated a bias toward similar functional categories even though the genes affected were different (1; see Discussion).
FIG. 5.
Effect of rsc4-Δ4 mutation on Pol II transcription. Genes induced and repressed twofold or more in the rsc4-Δ4 mutant grown at 37°C were classified according to MIPS categories. Pie charts representing their percentages are shown. Genes of unknown function were not taken into account to compute the percentages. A complete list of induced and repressed genes can be found in the supplemental material section, together with a breakup of the transcription and metabolism categories.
Since RSC and Pol III coimmunoprecipitated, we also investigated the effect of an rsc4 thermosensitive mutation on transcription by Pol III in vivo. Total RNAs from a wild-type or rsc4-MYC strain were extracted, separated by gel electrophoresis, and blotted onto a membrane. The amounts of U6 snRNA, RPR1 RNA, RNase P RNA, and two different tRNAs (tRNA3Leu and tRNA2Lys) and their short-lived precursors were quantified by Northern blot analysis and phosphorimaging (Fig. 6 and data not shown). Since the U6 RNA transcript is stable, we analyzed the accumulation of the unstable Maxi-U6 derivative in which 59 nucleotides were inserted (5, 32). Although transferring a wild-type strain to 37°C had no effect on transcription of Maxi-U6, the presence of the rsc4-MYC mutation decreased its transcription threefold (Fig. 6A).
FIG. 6.
Analysis of U6 and RPR1 transcription by Pol III in the rsc4-MYC mutant. (A) Northern analysis of Maxi-U6 transcripts. Total RNAs were extracted from a wild-type (RSC4) or mutant (rsc4-MYC) strain grown exponentially at 30°C or transferred for 7 h at 37°C. Then, 10 μg of the total RNAs was separated on a polyacrylamide denaturing gel and transferred to a nylon membrane. Even lane loading and transfer of the RNAs was verified by staining the nylon membrane with methylene blue (not shown). The effect of the rsc4-MYC mutation on Maxi-U6 (5, 32) transcription was investigated by Northern hybridization with a U6 probes. (B) Northern analysis of RPR1 precursor and mature transcripts. The position of the precursor and mature RPR1 RNA, represented by an open box, is indicated. The same blot was probed with tRNA3Leu DNA. Since tRNA3Leu levels did not change in rsc4-Δ4 mutant, it was used as a loading control.
At the permissive temperature, the RPR1 precursor was already 1.8-fold less abundant in the mutant strain compared to the wild-type (Fig. 6B). After 7 h of incubation at 37°C, transcription of RPR1 RNA was further diminished threefold in the rsc4-MYC mutant strain but only twofold in the wild-type strain, showing that its transcription was also impaired by the mutation. A diminution of RPR1 transcription was also seen in the rsc4-Δ4 mutant (data not shown). Accordingly, it was found previously that RPR1 transcription is particularly sensitive to Pol III machinery defects (27). In contrast, the transcription of two short-lived tRNA precursors (tRNA3Leu and tRNA2Lys ) was not affected (data not shown). The amount of mature tRNA3Leu did not change and was used as a load control (Fig. 6B). This latter observation indicates that altered RPR1 and SNR6 does not result from a general Pol III transcription defect due to the temperature shift. Altogether, these results indicated that the synthesis of at least some Pol III transcripts is dependent on the integrity of Rsc4.
Effect of rsc4-Δ4 mutation on RSC recruitment and chromatin structure.
RSC recruitment to promoters of several Pol II promoters and to several Pol III genes was investigated by using chromatin immunoprecipitation analysis. Myc tags were fused to the Sth1 subunit of RSC in RSC4 or rsc4-Δ4 yeast strains (Table 1). Nonspecific DNA-binding of the tagged protein was evaluated by control PCR experiments with primers specific for POL1 coding region which was previously shown to be devoid of RSC (36). We analyzed RSC occupancy of the promoter regions of Pol II genes repressed at 30 and 37°C (BAR1 and DUT1) or only at 37°C (SMX3 and STE2) or induced (DDR2, OYE3, GPH1, and CDC26) in the rsc4-Δ4 strain. RSC occupancy of the 35S rRNA gene promoter (Pol I-transcribed gene) and Pol III-transcribed genes [SNR6, RPR1, and tF(GAA)P2] was also analyzed. HTA3 promoter region was used as positive control since RSC binding to this region was previously observed by chromatin immunoprecipitation analysis (36) and since HTA3 transcription was not affected in the rsc4-Δ4 mutant. The recruitment of RSC complex was readily detected at most of the promoter regions tested (Fig. S2 in the supplemental material). However, no significant difference was observed in RSC occupancy between the wild-type and rsc4 mutant strains at either permissive or restrictive temperatures. These results indicated that the observed effect of rsc4-Δ4 mutation on transcription was not the consequence of decreased RSC recruitment to target promoter regions.
Even though RSC recruitment was normal in the rsc4-Δ4 mutant, chromatin remodeling could still be impaired if the mutation affected RSC function. To examine this possibility, we analyzed the chromatin structure of the DUT1 gene, the transcription of which is repressed 3.6-fold at 30°C in rsc4-Δ4 mutant and further reduced 6.1-fold at 37°C relative to the wild type. Yeast nuclei were isolated from RSC4 and rsc4-Δ4 strains grown at 30°C or shifted to 37°C for 6 h, treated with various concentrations of DNase I, and the DUT1 nucleosomal organization was assessed by indirect end labeling. Well-positioned nucleosomes could be identified on the DUT1 coding region and upstream from the ATG initiation codon in the wild-type strain (Fig. 7A), and a strong hypersensitive region was located close to the potential TATA box in the 5′ noncoding region (15). Strikingly, the hypersensitivity of this region was markedly decreased in the mutant at 30 and 37°C, in line with the repressive effect of the mutation on DUT1 transcription at both temperatures.
FIG. 7.
Chromatin organization at DUT1 and SMX3 loci is altered in rsc4-Δ4 mutant. (A) Effect of the rsc4-Δ4 mutation on DUT1 nucleosomal organization. Nuclei from RSC4 wild-type (MW4023) and rsc4-Δ4 mutant strains (MW4019) incubated for 6 h at 30 or 37°C were digested with the indicated amounts of DNase I (0.5, 1, 2, and 4 U/ml) for 20 min at 37°C. DNA was isolated, digested with SphI, analyzed on a 1.2% agarose gel, blotted, and hybridized with probe corresponding to the region from 222 to 526 nucleotides downstream from the ATG initiation codon. The positions of marker fragments are indicated. The schematic on the left shows the derived positions of nucleosomes, together with the position of the DUT1 gene from the ATG to the TAA codon, the potential TATA box (15), and the SphI restriction site. (B) MseI accessibility of the DUT1 promoter region in RSC4 and rsc4-Δ4 yeast strains. Yeast nuclei from RSC4 and rsc4-Δ4 strains grown at 30°C or shifted to 37°C for 6 h were digested with MseI enzyme at a saturating concentration. Purified DNA was subjected to real-time PCR using the primer pair specific to the region from −205 to −25 of the DUT1 promoter that encompassed the MseI site (−70 to −67). A region from positions +126 to +206 of the DUT1 lacking an MseI site was used as a negative control. The MseI accessibility was expressed as the percentage of digestion of the promoter region normalized to the control region. Observed experimental errors are indicated. (C) Effect of the rsc4-Δ4 mutation on SMX3 nucleosomal organization. DNase I footprinting was performed as in panel A except that the DNA was digested with BanII and that the DNA probe hybridized to positions +390 to +643 relative to the SMX3 ATG.
To confirm the decreased accessibility of DUT1 TATA box region, we analyzed the chromatin structure by the restriction enzyme accessibility assay and real-time PCR analysis (40). The MseI site (positions −70 to −67) located on the potential DUT1 TATA box was chosen for this purpose. Nuclei from RSC4 and rsc4-Δ4 strains grown at 30°C or shifted to 37°C for 6 h were digested with excess MseI enzyme, and purified DNA was subjected to real-time PCR with the primer pair specific to the region from positions −205 to −25 of the DUT1 promoter that included the MseI site. The amount of PCR product generated was inversely proportional to the level of digestion occurring across the region amplified by the primers. As a control of DNA recovery, the region from positions +126 to +206 of DUT1 lacking an MseI site was also amplified. The MseI accessibility was expressed as the percentage of digestion on the promoter region normalized to the control region. We observed that the DUT1 promoter region was significantly less accessible to MseI in the rsc4-Δ4 mutant at 30 and 37°C (Fig. 7B), a finding in agreement with our DNase I accessibility results. Thus, two independent approaches revealed a decreased accessibility of the DUT1 promoter region in the rsc4-Δ4 background.
We also analyzed the nucleosomal organization of SMX3 gene, the transcription of which was repressed less than twofold at 30°C and fourfold at 37°C in the rsc4-Δ4 background. DNase I mapping of the SMX3 region indicated the presence of several positioned nucleosomes with strong hypersensitive sites between nucleosomes −2 and −1 and nucleosomes −1 and +1 in the promoter region and between nucleosomes +3 and +4 in the terminator region (Fig. 7C). At the permissive temperature, the chromatin organization in the rsc4-Δ4 mutant was very similar to that of the wild type. In contrast, at the restrictive temperature, the DNase I accessibility of the hypersensitive sites between promoter nucleosomes was strongly reduced.
As a control, we analyzed the chromatin structure of HTA3 gene since it is one of the genes that is the most strongly enriched by the RSC chromatin immunoprecipitation assay (36) and since its transcription was not impaired by the rsc4-Δ4 mutation. As expected, the chromatin structure of HTA3 is not altered in the rsc4-Δ4 background either at 30 or at 37°C (Fig. S3 in the supplemental material). We also analyzed the chromatin structure of GPH1 and CDC26, two genes that were induced (threefold at 30°C and sevenfold at 37°C for GPH1; fourfold at 30 and 37°C for CDC26) in the rsc4-Δ4 mutant. Their chromatin structure was unchanged in the mutant irrespective of the temperature (data not shown). These data suggested a role for Rsc4 in chromatin remodeling that correlated with the efficiency of transcription of the RSC-responsive DUT1 and SMX3 genes.
Transcription inhibition does not alter DUT1 or SMX3 chromatin organization.
Since both transcription and chromatin organization were altered in the rsc4-Δ4 mutant, we could not distinguish whether altered chromatin organization was the cause or the consequence of diminished transcription at DUT1 and SMX3. We thus analyzed chromatin organization at these two genes in the rpb1-1 mutant that rapidly stops transcription after a temperature shift at 37°C (37). We measured the amount of DUT1 and SMX3 RNA in the wild-type and rpb1-1 strains by reverse transcription-PCR and found that the transcription of the two genes was not significantly altered at 25°C, whereas it was repressed 12.1-fold for DUT1 and 4.2-fold for SMX3 at 37°C. Strikingly, arresting transcription did not change the nuclease sensitivity of the two genes (Fig. 8), indicating that changes in chromatin organization at DUT1 and SMX3 are the consequence of altered remodeling by RSC in the rsc4-Δ4 mutant and not the indirect consequence of weaker transcription.
FIG. 8.
Inhibiting transcription did not alter chromatin organization at DUT1 or SMX3. (A) Chromatin organization at DUT1 locus was investigated in a wild-type or rpb1-1 mutant strain as in Fig. 7A. Chromatin organization was analyzed during exponential growth at 25°C, the permissive temperature for the rpb1-1 mutant, or after a 1-h shift at 37°C. (B) Chromatin organization at the SMX3 locus was analyzed in a wild-type or rpb1-1 mutant strain as in Fig. 7C.
DISCUSSION
RSC is an essential, multisubunit complex that was shown to bind to 12 to 25% of yeast genes (36) and to remodel chromatin in vitro. However, a link between chromatin remodeling and transcription in vivo has only been shown for the CHA1 gene where the RSC catalytic subunit was found to be required to maintain a repressive chromatin state (33). In the present study, we found that the Rsc4 C terminus interacted with Rpb5, a shared subunit of RNA polymerases, and, as expected, RSC was found to interact with the three forms of RNA polymerases. Strikingly, the rsc4 C-terminal mutations that reduced the Rsc4-Rpb5 interaction led to strong growth defects and were correlated with altered Pol II and Pol III transcription. The transcription defects were markedly different from those due to mutations in two other RSC subunits (1). The loss of interaction between Rsc4 C terminus and Rpb5 did not affect RSC recruitment at Pol II and Pol III genes but rather altered the chromatin structure in the promoter region of two RSC-regulated genes, leading to impaired transcription. These observations led us to propose that the Rsc4 subunit has multiple transcriptional roles, including regulating interactions with RNA polymerases and histone H3 (22), as well as regulating chromatin remodeling and gene selectivity.
In the rsc4-Δ4 mutant grown at the restrictive temperature, 338 genes were induced while 168 genes were repressed twofold or more. rsc4 mutants of the protein bromodomains, required for the binding of acetylated Lys14 of histone H3, have been isolated previously (22). These mutants behaved somewhat differently than the rsc4-Δ4 mutant with respect to Pol II transcription. Of the 50 class II genes most down- or upregulated in the rsc4-2 mutant after a temperature shift at 37°C, we found that 32 and 40%, respectively, had their expression level altered in our rsc4-Δ4 mutant. This observation indicated that the bromodomains and the C terminus of Rsc4 did not have completely overlapping roles with respect to Pol II gene regulation. One could speculate that at those genes that depend on the Rsc4 C terminus, but not on its bromodomains, the alternative bromodomains residing in Sth1, Rsc1, or Rsc2 subunits might mediate the interaction of RSC with acetylated histone tails.
The number of genes that are affected in rsc4 mutants is comparable to the number of genes affected in rsc3 or rsc30 mutants (1, 22). Both induced and repressed genes were found, suggesting that RSC is required for the movement of nucleosomes to regulate transcription negatively or positively. Remarkably, very few genes affected in the rsc4 mutants were also affected in the mutants of Rsc3 or Rsc30 subunits. This observation strongly suggested a specific role for Rsc4 in gene regulation by RSC complex. Altogether, the three RSC subunits for which microarray transcription data are available control the expression of around 660 genes, directly or indirectly. The number of genes affected in rsc4-Δ4 mutant is roughly twice as large as that of the genes affected in a swi2 mutant, which lacks the catalytic subunit of the SWI/SNF remodeling complex (19, 49). The genome-wide location of the RSC complex using chromatin immunoprecipitation suggested that 700 to 1,400 genes are bound by the RSC complex (11, 36). These figures might underestimate the number of bound genes since the cross-linking efficiency of RSC to DNA was low and residency was only tested under nutrient replete conditions.
Although the effect of RSC mutations on transcription of certain genes could be indirect, the simplest explanation for the decreased transcription of DUT1 and SMX3 is impaired remodeling of their promoters. Conversely, the lack of chromatin alteration on genes that have elevated transcription in rsc4-Δ4 mutant could result from an analogous indirect effect. Nevertheless, since the sets of genes controlled by the various RSC subunits were largely distinct, the scope of RSC function might include a very significant proportion of the yeast genome. This observation leads to the hypothesis that the noncatalytic RSC subunits might be required for targeting the complex to specific genome locations via their interaction with distinct partners.
The analysis of the genome-wide localization of the RSC complex indicated that about one-fifth of its targets were adjacent to Pol III genes, suggesting that RSC might be involved in Pol III transcription (11, 36). In particular, RPR1 or SNR6 are highly occupied by RSC. Our finding that the shared subunit Rpb5 interacted specifically with Rsc4 in vivo and in vitro strongly argued for an interaction between the Pol III transcription machinery and RSC. The existence of a functional link is buttressed by our observation that Pol III transcription of RPR1 and SNR6 is affected in an rsc4 mutant that abolished the two-hybrid interaction between Rsc4 C terminus and Rpb5. The transcription of SNR6 and RPR1 was not altered in a strain in which the two Rsc4 bromodomains were mutated, further indicating that the role of the bromodomains and of the Rsc4 C terminus are not completely overlapping (22).
The effect of rsc4 mutations on Pol I transcription could not be investigated. The total amount of rRNAs diminished in the mutant at the permissive temperature, but Northern blot analyses indicated that the processing of the rRNA precursors was slowed down and that the 35S rRNA precursor accumulated (data not shown). This accumulation was possibly the result of a maturation defect, as has been previously observed for other rRNA processing mutants (17), in agreement with our observation that some of the genes involved in this maturation process are affected in the mutant (Fig. 5 and see also the supplemental material). Since the maturation defect may mask a possible effect on 35S synthesis, we could not conclude whether or not RSC stimulates Pol I transcription.
Here, we define the Rsc4 C terminus as one determinant important for interaction with Rpb5. Here, a four-amino-acid deletion abolished the interaction between Rsc4 C terminus and Rpb5 both in vivo and in vitro but did not affect the interaction between the complete Rsc4 subunit and Rpb5 in vitro (it could not be tested in the yeast two-hybrid system). This latter observation suggests that one or more additional domains of Rsc4 interact with Rpb5. Interestingly, whereas the Rsc4 C terminus mutations did not drastically diminish the rather weak interaction of RSC with Pol I or Pol II in extracts, the Rsc4 derivative lacking the C-terminal 4 amino acids abolished the interaction with Pol III, while the addition of Myc epitopes did not (Fig. 1; see Fig. S1 in the supplemental material). Additional two-hybrid data suggest that other subunits of RSC interact with RNA polymerases (unpublished data). For example, we selected the Rsc1 subunit of RSC in a two-hybrid screen with Rpb10, another subunit common to the three RNA polymerases. The Rsc1-Rpb10 protein-protein contact might stabilize, together with Rsc4, the association between RSC and RNA polymerases. Nevertheless, the contact between the Rsc4 C terminus and Rpb5 is not essential for RSC recruitment on promoters but is required for proper chromatin remodeling at selected genes.
Although the in vitro remodeling activity of RSC is supported by numerous experiments (30, 31, 41, 44), the effect of mutations or depletion of its subunits on the chromatin organization has been studied in vivo in only two cases. Hsu et al. (20) found that in an sfh1 mutant strain the nucleosomal structure of centromeric DNA is altered and chromosome segregation is inaccurate. As mentioned above, genome-wide transcription studies have indicated that RSC is required for wild-type levels of transcription of a significant portion of the genome. Nevertheless, only in the case of the CHA1 gene has a simultaneous effect of a RSC subunit depletion on transcription and chromatin organization been observed (33). In this case, the RSC defect resulted in the formation of an open chromatin at the CHA1 promoter and induced transcription. Conversely, we showed here that on DUT1 and SMX3 genes RSC activity was required both to support wild-type transcription levels and to induce DNase I-hypersensitive sites in the promoter region, indicating that RSC regulates positively the transcription of these genes by enhancing chromatin accessibility. We excluded the possibility that altered chromatin remodeling at these two genes in the rsc4-Δ4 mutant was the indirect result of poor transcription since its inhibition in a Pol II mutant did not change DNase I sensitivity. Together, these observations show that transcription levels can be controlled by RSC and that at least in some instances altered transcription can be the direct consequence of altered chromatin remodeling.
The rsc4-Δ4 mutation alters chromatin remodeling but not RSC recruitment to promoter regions, suggesting a functional defect for the mutant complex. Indeed, preliminary experiments indicate that Pol II recruitment is affected at DUT1 and SMX3 promoters in the rsc4-Δ4 mutant. However, the sequence of events—whether recruitment of Pol II takes place before or after RSC-dependent chromatin remodeling—at DUT1 or SMX3 is not presently known, nor is the effect of the mutation on the recruitment of GTFs, Mediator, and other coactivators. Understanding these interrelationships will await further studies. Nevertheless, it seems reasonable to propose that RSC, in addition to its remodeling function, might act as a coactivator since deletion of the Rsc1 or the Rsc2 subunit of RSC leads to decreased Pol II recruitment at several genes under the control of Gcn4 transcription activator (39). The idea that complexes involved in chromatin organization may have a coactivator function independent of their catalytic activity is not unprecedented since coactivation by SAGA of GAL genes transcription does not require Gcn5 histone acetylase (2, 25).
Supplementary Material
Acknowledgments
We thank C. Boschiero for help with the experiments, O. Harismendy for help with the Northern analysis, M. Kabani for the GST and GST-Anc1 proteins, E. Favry for the homogeneous yeast Pol III preparation, and the DRIP for the monoclonal antibodies. We especially thank P. Thuriaux and A. Sentenac for reading and improving the manuscript.
V.B.-L.F. was supported by a grant from the Association pour la Recherche contre le Cancer. This study was supported by grants from HFSPO, the MENRT, and the ARC.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Angus-Hill, M., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and C. A. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik, S. R., and M. R. Green. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22:7365-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briand, J. F., F. Navarro, O. Gadal, and P. Thuriaux. 2001. Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnol, A.-F., F. Margottin, J. Huet, G. Almouzni, M.-N. Prioleau, M. Méchali, and A. Sentenac. 1993. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature 362:475-477. [DOI] [PubMed] [Google Scholar]
- 5.Burnol, A.-F., F. Margottin, P. Schultz, M.-C. Marsolier, P. Oudet, and A. Sentenac. 1993. Basal promoter and enhancer element of yeast U6 snRNA gene. J. Mol. Biol. 233:644-658. [DOI] [PubMed] [Google Scholar]
- 6.Cairns, B. R., H. Erdjument-Bromage, P. Tempst, F. Winston, and R. D. Kornberg. 1998. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell 2:639-651. [DOI] [PubMed] [Google Scholar]
- 7.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 8.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 9.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 10.Cramer, P., D. A. Bushnell, J. Fu, A. L. Gnatt, B. Maier-Davis, N. E. Thompson, R. R. Burgess, A. M. Edwards, P. R. David, and R. D. Kornberg. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640-649. [DOI] [PubMed] [Google Scholar]
- 11.Damelin, M., I. Simon, T. I. Moy, B. Wilson, S. Komili, P. Tempst, F. P. Roth, R. A. Young, B. R. Cairns, and P. A. Silver. 2002. The Genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell 9:563-573. [DOI] [PubMed] [Google Scholar]
- 12.Dumay-Odelot, H., J. Acker, R. Arrebola, A. Sentenac, and C. Marck. 2002. Multiple roles of the tau131 subunit of yeast transcription factor IIIC (TFIIIC) in TFIIIB assembly. Mol. Cell. Biol. 22:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fauchon, M., G. Lagniel, J. C. Aude, L. Lombardia, P. Soularue, C. Petat, G. Marguerie, A. Sentenac, M. Werner, and J. Labarre. 2002. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9:713-723. [DOI] [PubMed] [Google Scholar]
- 14.Flores, A., J.-F. Briand, O. Gadal, J.-C. Andrau, L. Rubbi, V. Van Mullem, C. Boschiero, M. Goussot, C. Marck, C. Carles, P. Thuriaux, A. Sentenac, and M. Werner. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. USA 96:7815-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gadsden, M. H., E. M. McIntosh, J. C. Game, P. J. Wilson, and R. H. Haynes. 1993. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 12:4425-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory, P. D., and W. Hörz. 1999. Mapping chromatin structure in yeast. Methods Enzymol. 304:365-376. [DOI] [PubMed] [Google Scholar]
- 17.Guglielmi, B., and M. Werner. 2002. The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J. Biol. Chem. 277:35712-35719. [DOI] [PubMed] [Google Scholar]
- 18.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 19.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, J. M., J. Huang, P. B. Meluh, and B. C. Laurent. 2003. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol. Cell. Biol. 23:3202-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huet, J., M. Riva, A. Sentenac, and P. Fromageot. 1985. Yeast RNA polymerase C and its subunits: specific antibodies as structural and functional probes. J. Biol. Chem. 260:15304-15310. [PubMed] [Google Scholar]
- 22.Kasten, M., H. Szerlong, H. Erdjument-Bromage, P. Tempst, M. Werner, and B. R. Cairns. 2004. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 23:1348-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kundu, T. K., Z. Wang, and R. G. Roeder. 1999. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 19:1605-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langst, G., P. B. Becker, and I. Grummt. 1998. TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J. 17:3135-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurent, B. C., X. Yang, and M. Carlson. 1992. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol. Cell. Biol. 12:1893-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefebvre, O., J. Rüth, and A. Sentenac. 1994. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5S RNA synthesis. J. Biol. Chem. 269:23374-23381. [PubMed] [Google Scholar]
- 28.LeRoy, G., G. Orphanides, W. S. Lane, and D. Reinberg. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282:1900-1904. [DOI] [PubMed] [Google Scholar]
- 29.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philipsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 30.Lorch, Y., M. Zhang, and R. D. Kornberg. 1999. Histone octamer transfer by a chromatin-remodeling complex. Cell 96:389-392. [DOI] [PubMed] [Google Scholar]
- 31.Lorch, Y., M. Zhang, and R. D. Kornberg. 2001. RSC unravels the nucleosome. Mol. Cell 7:89-95. [DOI] [PubMed] [Google Scholar]
- 32.Marsolier, M.-C., S. Tanaka, M. Livingstone-Zatchej, M. Grunstein, F. Thoma, and A. Sentenac. 1995. Reciprocal interferences between nucleosomal organization and transcriptional activation of the yeast SNR6 gene. Genes Dev. 9:410-422. [DOI] [PubMed] [Google Scholar]
- 33.Moreira, J. M., and S. Holmberg. 1999. Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J. 18:2836-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 35.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 36.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nonet, M., C. Scafe, J. Sexton, and R. Young. 1987. Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 7:1602-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orphanides, G., G. Leroy, C.-H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92:105-116. [DOI] [PubMed] [Google Scholar]
- 39.Qiu, H., C. Hu, S. Yoon, K. Natarajan, M. J. Swanson, and A. G. Hinnebusch. 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 24:4104-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao, S., E. Procko, and M. F. Shannon. 2001. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 167:4494-4503. [DOI] [PubMed] [Google Scholar]
- 41.Saha, A., J. Wittmeyer, and B. R. Cairns. 2002. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16:2120-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sengupta, S. M., M. VanKanegan, J. Persinger, C. Logie, B. R. Cairns, C. L. Peterson, and B. Bartholomew. 2001. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J. Biol. Chem. 276:12636-12644. [DOI] [PubMed] [Google Scholar]
- 45.Sentenac, A. 1985. Eukaryotic RNA polymerases. CRC Crit. Rev. 18:31-91. [DOI] [PubMed] [Google Scholar]
- 46.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 47.Sikorski, R. S., W. A. Michaud, S. Tugendreich, and P. Hieter. 1995. Allele shuffling: conjugational transfer, plasmid shuffling, and suppressor analysis in Saccharomyces cerevisiae. Gene 155:51-59. [DOI] [PubMed] [Google Scholar]
- 48.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 49.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsuchiya, E., M. Uno, A. Kiguchi, K. Masuoka, Y. Kanemori, S. Okabe, and T. Mikayawa. 1992. The Saccharomyces cerevisiae NPS1 gene, a novel CDC gene which encodes a 160 kDa nuclear protein involved in G2 phase control. EMBO J. 11:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinmann, A. S., S. E. Plevy, and S. T. Smale. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665-675. [DOI] [PubMed] [Google Scholar]
- 53.Werner, M., N. Chaussivert, I. M. Willis, and A. Sentenac. 1993. Interaction between a complex of RNA polymerase III subunits and the 70 kDa component of TFIIIB. J. Biol. Chem. 268:20721-20724. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.