Abstract
Influenza virus infection activates cytolytic T lymphocytes (CTL) that contribute to viral clearance by releasing perforin and granzymes from cytoplasmic granules. Virus-specific, perforin-dependent CD8+ CTL were detected in freshly isolated cells from the mouse lung parenchyma but not from the mediastinal lymph nodes (MLN), where they are primed, or from the spleen during primary influenza virus infection. To determine whether this difference was due to the low frequency or incomplete maturation of effector CTL in MLN, we measured expression of perforin, granzymes A, B, and C, and IFN-γ mRNAs in CD8+ populations and single cells immediately after isolation from virus-infected mice. Quantitative PCR revealed significant expression of perforin, granzyme A, granzyme B, and IFN-γ in activated CD8+ cells from MLN, spleen, and lung parenchyma. Granzyme C expression was not detected. Individual activated or nucleoprotein peptide/class I tetramer-binding CD8+ cells from the three tissues expressed diverse combinations of perforin, granzyme, and IFN-γ mRNAs. Although cells from lung expressed granzymes A and B at higher frequency, each of the tissues contained cells that coexpressed perforin with granzymes A and/or B. The main difference between MLN and lung was the elevated frequency of activated CD8+ T cells in the lung, rather than their perforin/granzyme expression profile. The data suggest that some CTL mature into perforin/granzyme-expressing effector cells in MLN but reach detectable frequencies only when they accumulate in the infected lung.
CD8+ cytolytic T lymphocytes (CTL) contribute to viral clearance and recovery from influenza A infection in mice and humans (1). The primary effector function of these cells is believed to be the antigen-specific lysis of infected respiratory epithelial cells, mediated mainly by exocytosis of granules containing perforin and granzymes (2, 3). CD8+ T cell-derived cytokines such as IFN-γ may influence leukocyte traffic and inflammation in the infected lung but are not essential for viral clearance (3–6). When a CTL recognizes viral peptide/MHC complexes on an infected target cell, it secretes cytokines and exocytoses its granule contents, which then are endocytosed by the target cell. Perforin is essential for the release of granzymes from the endosome into the target cell cytoplasm, from where they translocate to the nucleus and interact with death substrates to effect apoptotic death (7, 8). Although CTL granules contain at least eight distinct granzymes, granzymes A, B, and C are the most abundant (9). Granzyme B is essential for the rapid apoptotic cell death detected in 51Cr-release assays of CTL activity in some antiviral and antitumor responses (10, 11) whereas granzyme A triggers another slower pathway of target cell apoptosis and is important in the clearance of ectromelia virus (12, 13). Recent data suggest that the relative contributions of granzymes A and B depend on properties of the target cell (14). The function of granzyme C is unknown, although a role in cytotoxicity of a T cell clone has been reported (15).
We recently have reported that the genes for perforin and granzymes A, B, and C each are induced with distinct kinetics after polyclonal activation of naive CD8+ T cells in vitro, suggesting that CTL maturation is a progressive process (16). In influenza virus infection, CTL precursors are primed in the draining mediastinal lymph nodes (MLN) within a few days of infection (17, 18) but, because they can be detected at this site only after restimulation in vitro, it is not known whether they have effector activity in vivo. On the other hand, effector CTL can be detected directly in the lung parenchyma and bronchoalveolar lavage fluid at the peak of the primary T cell response to influenza virus without further expansion or maturation in vitro (3, 5, 19–21). Because of the insensitivity of the 51Cr-release assay and the lack of a single-cell assay for effector CTL, it is not clear whether these cells mature in the MLN or require further differentiation after reexposure to the infected respiratory epithelium and other signals in the lung.
In a new approach to these questions, we have used RT-PCR to detect perforin, granzymes A, B, and C, and IFN-γ mRNA expression in single T cells and populations from influenza virus-infected mice, without restimulation in vitro. We show that influenza infection causes significant induction of perforin and both granzymes A and B, but not granzyme C, in activated CD8+ T cells in vivo and that individual, virus-specific cells are highly diverse in their perforin/granzyme expression profiles. The data suggest that all three tissues contain effector CTL and that the selective detection of virus-specific CTL activity in the lung mainly reflects their elevated frequencies at this site, rather than their maturation status.
Materials and Methods
Virus Infection.
Specific pathogen-free female BALB/c or C57BL/6 mice (6–8 weeks old) were obtained from the Animal Resources Centre, Perth, Western Australia. Mice were anesthetized with methoxyflurane (Penthrane, Abbott), and BALB/c mice were infected intranasally with 8 × 105 plaque-forming units/50 μl PBS of the reassortant virus Mem71, bearing the hemagglutinin of A/Memphis/1/71 (H3) and the neuraminidase of A/Bellamy/42 (N1) (22). C57BL/6 mice were infected similarly with 104.5 plaque-forming units of the H3N2 virus HKx31 (23).
Cell Preparation.
Lung parenchymal cells were obtained by mincing, collagenase/DNase I digestion, and centrifugation over a 20–55% discontinuous Percoll gradient as described (22). Cell suspensions from LN, spleen, and lung were stained for 20 min on ice with rat mAb to CD8α (53-6.72; Becton Dickinson), CD44 (IM7; PharMingen), CD11a (I21/7.7; hybridoma supernatant), and CD62L (Mel-14; hybridoma supernatant), followed by FITC-conjugated rabbit anti-rat Ig (Vector Laboratories) and Tricolor-conjugated streptavidin (Caltag, South San Francisco, CA). Alternatively, cells from C57BL/6 mice were stained with anti-CD8α mAb and a tetrameric complex of H-2Db molecules with the HKx31 influenza virus nucleoprotein epitope ASNENMETM (NP366–374) labeled with phycoerythrin (23). Propidium iodide was added at 1 μg/ml immediately before cell sorting. Cell suspensions were separated by using a fluorescence-activated cell sorter (FACSVantage; Becton Dickinson) with lysis ii or cellquest (Becton Dickinson) software after gating on viable lymphocytes based on forward and side scatter and exclusion of propidium iodide-stained cells. Subsets of CD44high CD62Llow and CD44low CD62Lhigh cells were collected while gating on live CD8+ cells. CD44high CD62Llow cells typically constituted 8%, 7%, and 34%, and CD44low CD62Lhigh cells were 16%, 7%, and 17% of live MLN, spleen, and lung CD8+ cells, respectively (means of three experiments). Cell purities were routinely >95% on reanalysis immediately after sorting.
51Cr-Release Cytotoxicity Assay.
Target cells of the H-2d mastocytoma P815 were incubated at 37°C in 5% CO2 for 1 h in DMEM with 2% FCS and 180 hemagglutinating units of Mem71 virus or with 10% FCS with or without 10 μg of the synthetic peptide TYQRTRALV, representing a major CTL epitope of the Mem71 nucleoprotein NP147–155 (provided by David Jackson, University of Melbourne), then with 100 μCi of Na51CrO4 (Amersham Pharmacia) for 90 min. Labeled cells were cultured at 5 × 103 per well with serially diluted effector cells in 200-μl volumes in round-bottomed, 96-well plates. In some experiments, effector cells were incubated for 2 h at 37°C with 10 or 100 ng/ml concanamycin A (Sigma) or similarly diluted diluent (DMSO) before addition of target cells. The assay was incubated for 4 h at 37°C, after which supernates were collected for measurement of γ-emission (24). Percent specific lysis was calculated as: 100 × [(mean sample release − mean spontaneous release)/(mean maximal release B mean spontaneous release)].
Quantitative Competitive RT-PCR (QC-PCR).
RNA was isolated from fluorescence-activated cell sorter-purified samples of 104 cells by using RNAzol B total RNA isolating reagent (Tel-Test, Friendswood, TX) according to the manufacturer's instructions, then reverse-transcribed by using AMV reverse transcriptase (Promega) (25). Mixtures of a fixed amount of test cDNA (1% of the sample, from the equivalent of 100 cells) were coamplified with 5-fold serial dilutions of competitor cDNA containing small deletions (25) by using primers and probes described elsewhere (16, 25, 26). After agarose electrophoresis to verify product size, amplified DNA was quantified by ELISA, using hybridization to internal FITC-labeled probes specific for either the exogenous competitor products or the endogenous DNA products. The number of mRNA molecules in the sample was determined from the equivalence point. Sensitivity of detection of endogenous and competitor cDNA was similar. QC-PCR detected 4–20 input competitor molecules by agarose gel electrophoresis and ≈4 molecules by ELISA for the products measured here. Both intra- and interassay variation was low in replicate assays of a given RNA sample, with SD generally <15% of the means.
Single-Cell RT-PCR.
Single cells were placed in 96-well plates by using the automated cell deposition unit attached to a FACS Vantage, lysed with Nonidet P-40, reverse-transcribed, and amplified by two rounds of 40-cycle nested PCR as described (16, 26). Products were agarose-electrophoresed and visualized by ethidium bromide DNA staining. Each PCR run included a set of 10-fold serially diluted cloned cDNAs for each product and a minimum of six negative control samples. The sensitivity of each reaction was typically 2.5 molecules of DNA. Data are presented only for cells that yielded a CD3ɛ PCR product in experiments in which no contamination was detected in any negative control sample.
Results
Induction of Cytolytic Activity in the Respiratory Tract by Influenza Virus Infection.
We previously detected virus-specific cytolytic activity in freshly isolated leukocytes from lung parenchyma and bronchoalveolar lavage, but not MLN, of mice at the peak of the primary response to infection with influenza virus (5, 20). Fig. 1A shows that the cytolytic activity of lung parenchymal cells is mediated by CD8+ cells. Freshly isolated CD8+ and CD8− T cells from MLN, spleen, and lung parenchyma of 7-day Mem71 influenza virus-infected BALB/c mice were incubated with virus-infected or NP147–155 peptide-coated syngeneic target cells. Significant specific lysis was observed only in CD8+ cells from lung parenchyma. Lysis was mediated mainly by the Ca2+-dependent perforin pathway because it was markedly reduced when 2.5–10 mM EGTA was included in the 51Cr-release assay (data not shown) and when effector cells were preincubated with concanamycin A, a vacuolar-type H+-ATPase inhibitor that inhibits perforin-mediated cytotoxicity (27, 28) (Fig. 1B). Further separation of lung CD8+ cells showed that lytic activity was enriched in the CD44high CD62Llow (activated/memory) cell fraction, which could account for all of the activity detected in the unfractionated lung population (Fig. 1C).
Figure 1.
Phenotype and perforin dependence of cytolytic cells from influenza virus-infected mice. (A) CD8+ and CD8− cells from the indicated tissues were purified from 7-day Mem71-infected BALB/c mice and assayed in duplicate in 51Cr-release assays with Mem71-infected (○), NP147–155-coated (●), or untreated (▵) P815 target cells. (B) Lung parenchymal cells from 7-day Mem71-infected BALB/c mice were incubated with concanamycin A at 100 ng/ml (▵) or 10 ng/ml (▿) or with (○) or without (●) diluent at the higher dose for 2 h, then assayed in triplicate in 51Cr-release assays with Mem71-infected or NP147–155-coated P815 target cells as indicated. The symbols with a broken line (Left) show lytic activity of untreated lung cells against untreated target cells. (C) Unfractionated leukocytes and CD8+ cells (24% of the leukocyte population) of naive (CD44low CD62Lhigh; 28% of the CD8+ population) and activated (CD44high CD62Llow; 36%) phenotype from lung parenchyma of 7-day Mem71-infected BALB/c mice were assayed in duplicate in 51Cr-release assays with Mem71-infected (○), NP147–155-coated (●), or untreated (▴) P815 target cells.
Expression of Perforin, Granzyme, and IFN-γ mRNAs by CD8+ T Cells from Infected Mice.
QC-PCR was used to measure levels of perforin, granzymes A, B, and C, and IFN-γ mRNA in freshly isolated CD8+ cells from MLN, spleen, and lung parenchyma. Kinetic analyses showed that perforin, granzymes A and B, and IFN-γ were expressed in all tissues, peaking at days 5–10 after influenza infection (data not shown). Granzyme C mRNA was not detected at any time in CD8+ cells from any of the tissues, although control granzyme C cDNA was detected with similar sensitivity to the other products measured here. Maximum levels of perforin and IFN-γ expression were comparable in all tissues whereas expression of granzymes A and B reached 5- to 60-fold-higher levels in lung than in MLN or spleen.
The relationship between cytolytic mediator expression and activation marker phenotype is shown in Fig. 2 Left. CD8+ cells of naive (CD44low CD62Lhigh) and activated/memory (CD44high CD62Llow, termed “activated” for brevity) phenotype were purified from MLN, spleen, and lung of 7-day virus-infected mice or from peripheral LN, spleen, and lung of uninfected control mice and analyzed by QC-PCR. Peripheral LN from control mice were used because MLN were difficult to detect in the absence of infection. Cells of naive phenotype from control mice expressed low or undetectable levels of each mRNA species except CD3ɛ, consistent with evidence that naive CD8+ T cells do not express these genes (16, 29, 30). Again, granzyme C was not detected in any population despite detection of control cDNA in the same assay. Expression of each of the other products was generally higher in activated LN and spleen CD8+ cells from infected mice than from control mice, but was comparable in activated lung parenchymal cells. Their expression was similar in the activated fraction of all three tissues from infected mice.
Figure 2.
Perforin, granzyme, and IFN-γ expression by resting and activated CD8+ cells from uninfected and influenza virus-infected mice. CD8+ cells of CD44low CD62Lhigh (naive) and CD44high CD62Llow (activated) phenotype were purified from pooled inguinal, brachial, and axillary LN, spleen, and lung parenchyma of control uninfected BALB/c mice (A) and MLN, spleen, and lung parenchyma of 7-day Mem71-infected BALB/c mice (B) (two to three mice per group). Cells were lysed immediately for determination of CD3ɛ, perforin (Per), granzymes (Gr) A–C, and IFN-γ mRNA levels by QC-PCR. (Left) Numbers of mRNA molecules in 100-cell equivalents; broken lines indicate the threshold of detection. (Right) Average numbers of total, naive, and activated CD8+ cells per mouse in each tissue.
Additional differences between control and infected mice were apparent when the impact of infection on cell composition was considered (Fig. 2 Right). The absolute number and frequency of CD8+ cells with a naive phenotype were lower in all tissues of infected mice than control mice, whereas the number and frequency of lung cells with an activated phenotype were 4.4- and 2.7-fold higher, respectively. The majority of other CD8+ cells in all populations were cells with intermediate CD44 and high CD62L levels (data not shown). Activated cells were about 6-fold more frequent in the lung than in MLN or spleen of infected mice. Multiplication of average mRNA levels per cell (Fig. 2 Left) by cell numbers (Fig. 2 Right) showed that total expression was higher in the activated CD8+ population in lung compared with MLN of infected mice (perforin, 5.2-fold; granzyme A, 33-fold; granzyme B, 41-fold; IFN-γ, 41-fold). The data suggest that the difference in lytic activity between MLN, spleen, and lung shown in Fig. 1 can be attributed, at least in part, to the higher frequency of activated cells in lung.
Single-Cell Analysis of Perforin, Granzyme, and IFN-γ mRNA Expression.
We previously showed that two-round nested RT-PCR can detect cytokine, perforin, and granzyme transcripts at high frequency in single T cells after activation in vitro (16, 31, 32). We therefore used nested RT-PCR to determine whether activated CD8+ T cells from lymphoid tissues and lung differed in their patterns of expression of perforin, granzymes A–C, and IFN-γ at the single-cell level. CD44high CD62Llow CD8+ cells were isolated from MLN and lung of 7-day virus-infected mice and assayed without restimulation in vitro (Fig. 3). Individual cells from both tissues expressed various combinations of mRNAs, as we have observed previously in CD8+ T cells activated in vitro (16). Each product was found both with and without the other product, and many cells expressed granzymes A and/or B without perforin. Frequencies of perforin, granzyme A, granzyme C, and IFN-γ were similar between the two tissues, and only granzyme B was expressed at a significantly higher frequency in lung than in MLN (2.6-fold, P < 0.009 by z test). As in Fig. 2, however, activated cells comprised a smaller fraction of the CD8+ population in MLN (8%) than in lung (36%). Although the PCR method used in this analysis did not quantify mRNA levels per cell, the granzyme C PCR products obtained from 6–8% of cells in Fig. 3 (and some cells in Fig. 4) were of low abundance, suggesting that mRNA levels per positive cell were substantially lower for this granzyme than for the other mediators.
Figure 3.
Single-cell analysis of perforin, granzyme, and IFN-γ mRNA expression by activated CD8+ cells from influenza virus-infected mice. Single CD44high CD62Llow CD8+ cells purified from MLN and lung parenchyma of 7-day Mem71-infected BALB/c mice (n = 4) were lysed immediately for analysis of CD3ɛ, perforin, granzymes A–C, and IFN-γ expression by two-round nested RT-PCR. Detection of a perforin, granzyme, or IFN-γ PCR product is indicated by shading of the boxes. The number of cells with each expression pattern is shown at the right; the frequency expressing each product is shown along the bottom.
Figure 4.
Single-cell analysis of perforin, granzyme, and IFN-γ mRNA expression by NP/class I tetramer-binding CD8+ cells from influenza virus-infected mice. Single NP366–374/H-2Db tetramer-binding and nonbinding CD8+ cells purified from MLN, spleen, and lung parenchyma of 8-day HKx31-infected C57BL/6 mice (n = 3) were lysed immediately for analysis of CD3ɛ, perforin, granzymes A–C, and IFN-γ expression by two-round nested RT-PCR. Data are displayed as described in the legend to Fig. 3.
The specificity of the cells analyzed in Fig. 3 was not known. We therefore used NP366–374/H-2Db tetramers to detect specific T cells in a similar model of infection with HKx31 influenza virus in C57BL/6 mice (23). These tetramers correspond to one of two major CD8+ T cell specificities defined in this response (23, 33–35). At days 7 and 8 of infection, as in the Mem71 system used above, freshly isolated lung parenchymal cells displayed strong lytic activity against syngeneic target cells infected with HKx31 virus or loaded with NP366–374, whereas those from MLN and spleen did not (data not shown). Tetramer-binding CD8+ cells were purified from the MLN (0.2–1.4% of CD8+ cells in three experiments), spleen (0.5–2%), and lung parenchyma (3–5%) of C57BL/6 mice 8 days after infection and assayed for perforin, granzyme, and IFN-γ expression (Fig. 4). The proportion of cells positive for one or more products was higher among tetramer-binding than tetramer-nonbinding cells in MLN (84% vs. 41%), spleen (84% vs. 11%), and lung (95% vs. 74%). Frequencies of tetramer-binding cells expressing perforin, granzyme B, or IFN-γ were similar in all tissues whereas granzyme A was more frequently expressed in the lung and spleen than in MLN (1.8-fold, P < 0.009 by z test, and 1.5-fold, P < 0.05, respectively). Granzyme C was rarely expressed. By contrast, there were marked differences between the tetramer-nonbinding cells from the three tissues, with 30–50% of lung cells but very few MLN or spleen cells expressing perforin, granzyme B, or IFN-γ. These data suggest that the tetramer-nonbinding cells from MLN and spleen were mainly naive or resting cells whereas most of those from lung were activated. Tetramer-nonbinding lung cells are expected to include cells specific for other influenza virus epitopes (34) as well as bystander cells.
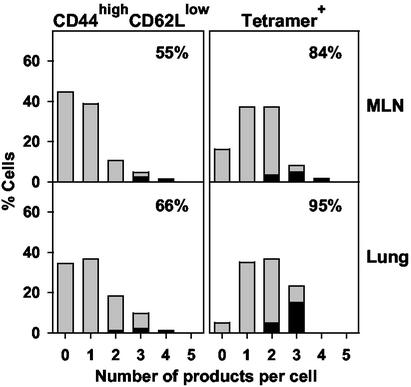
Collectively, these single-cell analyses showed that a high proportion of freshly isolated, activated, or epitope-specific CD8+ T cells from all tissues expressed perforin, granzyme A, granzyme B, and/or IFN-γ in various combinations. In addition to increased frequencies of granzyme A or B expression, the main differences noted in the lung compared with MLN were modest increases in the frequencies of positive cells (P = 0.05 for tetramer-binding cells), particularly those expressing multiple products such as the predicted cytolytic phenotype of perforin with granzymes A and/or B (Fig. 5).
Figure 5.
Distribution of the number of mRNA species detected per cell isolated from the MLN and lungs of influenza virus-infected mice. The data in Figs. 3 and 4 were used to determine the percentage of activated (Left) or tetramer-binding (Right) CD8+ cells expressing 0–5 of the PCR products for perforin, granzymes A–C, and IFN-γ (gray bars). Percentages of cells expressing perforin with granzymes A and/or B are superimposed in the black bars. In the upper right of each graph is the percentage of cells positive for one or more products.
Discussion
CTL can be detected readily by 51Cr-release assays of cells from the lung parenchyma or airways, but not MLN or spleen, of influenza virus-infected mice without further clonal expansion and/or maturation in vitro (5, 17–21). This probably reflects low antigen loads in the lymphoid tissues because viral replication is largely limited to the respiratory epithelium (3). It therefore has been unclear whether effector CTL complete their differentiation in the draining LN or after migration into the infected lung, where they are reexposed to viral antigen and other signals. The latter possibility is supported by our earlier finding that some activated CD8+ cells from lungs of influenza virus-infected mice have the potential to alter their cytokine profiles in response to a new signal (36).
We have shown here that NP-specific CTL were enriched in the activated (CD44high CD62Llow) CD8+ population that accumulated in the lung parenchyma during the first week after infection. Cells with this phenotype comprised 30–40% of lung CD8+ cells but were only a minor fraction (<10%) in MLN and spleen of the same animals and could not be purified in sufficient numbers for 51Cr-release assays. To bypass this limitation, we have exploited the sensitivity of RT-PCR to measure the expression of perforin and granzyme mRNAs by CD8+ cells from MLN, spleen, and lung parenchyma. Several new findings were obtained.
First, by quantitative and single-cell PCR, we found that the activated CD8+ fractions from MLN, spleen, and lung parenchyma of infected mice all contained mRNAs for the mediators required for lytic activity, namely, perforin, granzyme A, and/or granzyme B. Some NP epitope-specific CD8+ T cells from all three tissues in another similar model of influenza virus infection also expressed these combinations of mediators. The failure to detect lytic activity in CD8+ populations from MLN and spleen without in vitro restimulation therefore most likely was due to the low frequency, rather than the absence, of mature CTL.
Second, single-cell analysis revealed marked diversity in the perforin, granzyme, and IFN-γ expression profiles of individual activated and NP-specific CD8+ T cells from infected mice. Our recent study of naive CD8+ T cells activated in vitro with polyclonal stimuli (antibodies to CD3, CD8, and CD11a) and IL-2 examined coexpression of perforin and granzymes at the single-cell level and also found that expression profiles were highly diverse (16). Many cells in both studies expressed perforin without granzyme A or B mRNA and vice versa. Because perforin and granzymes are stored as inactive proteins in granules until target cell-induced exocytosis, these stores may be present in the absence of the corresponding mRNA. Further regulation is achieved by cleavage of the proform of perforin and granzymes to yield the active proteins (37, 38). Reagents are not yet available to measure each of these proteins in their inactive or activated forms in single cells, but the frequencies obtained here by single-cell RT-PCR provide minimal estimates of the number of cells able to coexpress these products.
Third, we noted significant differences between the patterns of perforin and granzyme expression induced in response to influenza infection in this study and to polyclonal activation in vitro (16). In vitro, the proportion of cells expressing multiple products increased markedly between days 3 and 7, suggesting that they progressively acquired the ability to express additional granzymes as they underwent clonal expansion (16). By comparison, the minor shift in numbers of mediators expressed by lung CD8+ cells compared with MLN in the influenza response (Fig. 5) is consistent with the idea that the maturational state of CTL in the MLN and lung was not markedly different at the time points studied.
CD8+ T cells from the two responses also differed in their relative expression of granzymes A, B, and C. The present study analyzes perforin and granzyme expression patterns in influenza virus infection and shows that infection was associated with elevation of perforin and granzymes A and B, but not granzyme C, mRNAs. We did not detect granzyme C mRNA by quantitative PCR in any of the CD8+ populations assayed from MLN, spleen, or lung of infected or control mice. By single-cell PCR, some CD8+ cells from MLN and lung of infected mice expressed this granzyme but at low frequency and low abundance. By comparison, only a minority of CD8+ T cells activated polyclonally for 7 days in vitro expressed granzyme A whereas most expressed granzymes B and C (16). Others also have reported that granzyme C expression is low in T cells primed in vivo but is strongly induced in polyclonal and cloned T cells in vitro (9, 39). It is not known whether specific inducers or inhibitors are responsible for these differential expression patterns. The physiological functions of granzyme C also are not yet known, although antisense oligonucleotides were reported to block cytolytic activity of a T cell clone (15). The poor expression detected in the present study argues against participation of this granzyme in CTL activity in influenza virus-infected mice.
IFN-γ synthesis is widely used as a surrogate marker for CTL function in CD8+ T cells, and it was established many years ago that CTL can secrete IFN-γ when they interact with their specific target cell (40). Although IFN-γ expression generally paralleled expression of perforin and granzymes A and B at the population level, the single-cell analyses suggested that its expression was regulated independently of any of the cytolytic mediators. It is significant, however, that at least 20% of activated or tetramer-binding CD8+ cells from all tissues expressed this cytokine ex vivo, without the usual restimulation used in IFN-γ ELISpot or intracellular staining assays. Because expression of this cytokine in vitro normally occurs as a transient response to TCR triggering over ≈6–24 h (41, 42), the data suggest that at least 20% of the cells had been triggered in the hours before isolation.
Collectively, the data support a scenario in which at least some activated CD8+ cells acquire full CTL effector function in the MLN but immediately emigrate rather than accumulate at that site. The elevated numbers of CTL in the lung therefore could reflect preferential migration, retention, and/or cell division in situ. Other cells may leave the MLN before acquiring full effector activity and complete their differentiation when reexposed to viral antigen or other signals in the infected respiratory tract. Completion of the maturation process in the infected lung then could allow perforin and granzyme expression, like the cytokine response (36), to be shaped by local signals. The diversity of perforin/granzyme expression patterns observed here in single activated CD8+ T cells from both tissue sites may reflect these different maturation states.
Acknowledgments
We thank Dr. David Jackson for the generous provision of peptides, Grace Chojnowski and Paula Hall for assistance with flow cytometry, and Dr. Michael Rudd for assistance in establishing the infection model. This work was supported by the Cooperative Research Centre for Vaccine Technology, the Australian National Health and Medical Research Council, and the Queensland Cancer Fund.
Abbreviations
- CTL
cytolytic T lymphocyte(s)
- MLN
mediastinal lymph node
- NP
nucleoprotein
- QC-PCR
quantitative competitive RT-PCR
References
- 1.Ada G L, Jones P D. Curr Top Microbiol Immunol. 1986;128:1–54. doi: 10.1007/978-3-642-71272-2_1. [DOI] [PubMed] [Google Scholar]
- 2.Topham D J, Tripp R A, Doherty P C. J Immunol. 1997;159:5197–5200. [PubMed] [Google Scholar]
- 3.Doherty P C, Topham D J, Tripp R A, Cardin R D, Brooks J W, Stevenson P G. Immunol Rev. 1997;159:105–117. doi: 10.1111/j.1600-065x.1997.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 4.Graham M B, Dalton D K, Giltinan D, Braciale V L, Stewart T A, Braciale T J. J Exp Med. 1993;178:1725–1732. doi: 10.1084/jem.178.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgarth N, Kelso A. J Virol. 1996;70:4411–4418. doi: 10.1128/jvi.70.7.4411-4418.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price G E, Gaszewska-Mastarlarz A, Moskophidis D. J Virol. 2000;74:3996–4003. doi: 10.1128/jvi.74.9.3996-4003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smyth M J, Kelly J M, Sutton V R, Davis J E, Browne K A, Sayers T J, Trapani J A. J Leukocyte Biol. 2001;70:18–29. [PubMed] [Google Scholar]
- 8.Metkar S S, Wang B, Aguilar-Santelises M, Raja S M, Uhlin-Hansen L, Podack E, Trapani J A, Froelich C J. Immunity. 2002;16:417–428. doi: 10.1016/s1074-7613(02)00286-8. [DOI] [PubMed] [Google Scholar]
- 9.Ebnet K, Chluba-de Tapia J, Hurtenbach U, Kramer M D, Simon M M. Int Immunol. 1991;3:9–19. doi: 10.1093/intimm/3.1.9. [DOI] [PubMed] [Google Scholar]
- 10.Heusel J W, Wesselschmidt R L, Shresta S, Russell J H, Ley T J. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 11.Simon M M, Hausmann M, Tran T, Ebnet K, Tschopp J, ThaHla R, Müllbacher A. J Exp Med. 1997;186:1781–1786. doi: 10.1084/jem.186.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shresta S, Graubert T A, Thomas D A, Raptis S Z, Ley T J. Immunity. 1999;10:595–605. doi: 10.1016/s1074-7613(00)80059-x. [DOI] [PubMed] [Google Scholar]
- 13.Müllbacher A, Waring P, ThaHla R, Tran T, Chin S, Stehle T, Museteanu C, Simon M M. Proc Natl Acad Sci USA. 1999;96:13950–13955. doi: 10.1073/pnas.96.24.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardo J, Balkow S, Anel A, Simon M M. Eur J Immunol. 2002;32:1980–1985. doi: 10.1002/1521-4141(200207)32:7<1980::AID-IMMU1980>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 15.Bailey N C, Kelly C J. Eur J Immunol. 1997;27:2302–2309. doi: 10.1002/eji.1830270926. [DOI] [PubMed] [Google Scholar]
- 16.Kelso A, Costelloe E O, Johnson B J, Groves P, Buttigieg K, Fitzpatrick D R. Int Immunol. 2002;14:605–613. doi: 10.1093/intimm/dxf028. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton-Easton A, Eichelberger M. J Virol. 1995;69:6359–6366. doi: 10.1128/jvi.69.10.6359-6366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tripp R A, Hou S, McMickle A, Houston J, Doherty P C. J Immunol. 1995;154:6013–6021. [PubMed] [Google Scholar]
- 19.Eichelberger M, Allan W, Zijlstra M, Jaenisch R, Doherty P C. J Exp Med. 1991;174:875–880. doi: 10.1084/jem.174.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumgarth N, Kelso A. Eur J Immunol. 1996;26:2189–2197. doi: 10.1002/eji.1830260934. [DOI] [PubMed] [Google Scholar]
- 21.Wijburg O L C, DiNatale S, Vadolas J, van Rooijen N, Strugnell R A. J Virol. 1997;71:9450–9457. doi: 10.1128/jvi.71.12.9450-9457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumgarth N, Brown L, Jackson D, Kelso A. J Virol. 1994;68:7575–7581. doi: 10.1128/jvi.68.11.7575-7581.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haanen J B A G, Toebes M, Cordaro T A, Wolkers M C, Kruisbeek A M, Schumacher T N M. Eur J Immunol. 1999;29:1168–1174. doi: 10.1002/(SICI)1521-4141(199904)29:04<1168::AID-IMMU1168>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 24.Kienzle N, Buttigieg K, Groves P, Kawula T, Kelso A. J Immunol. 2002;168:1672–1681. doi: 10.4049/jimmunol.168.4.1672. [DOI] [PubMed] [Google Scholar]
- 25.Fitzpatrick D R, Shirley K M, McDonald L E, Bielefeldt-Ohmann H, Kay G F, Kelso A. J Exp Med. 1998;188:103–117. doi: 10.1084/jem.188.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelso A, Groves P. Proc Natl Acad Sci USA. 1997;94:8070–8075. doi: 10.1073/pnas.94.15.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kataoka T, Shinohara N, Takayama H, Takaku K, Kondo S, Yonehara S, Nagai K. J Immunol. 1996;156:3678–3686. [PubMed] [Google Scholar]
- 28.Kataoka T, Togashi K, Takayama H, Takaku K, Nagai K. Immunology. 1997;91:493–500. doi: 10.1046/j.1365-2567.1997.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Sanz J A, MacDonald H R, Jenne D E, Tschopp J, Nabholz M. J Immunol. 1990;145:3111–3118. [PubMed] [Google Scholar]
- 30.Griffiths G M, Mueller C. Immunol Today. 1991;12:415–419. doi: 10.1016/0167-5699(91)90145-J. [DOI] [PubMed] [Google Scholar]
- 31.Troutt A B, Kelso A. Proc Natl Acad Sci USA. 1992;89:5276–5280. doi: 10.1073/pnas.89.12.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelso A, Groves P, Ramm L, Doyle A G. Int Immunol. 1999;11:617–621. doi: 10.1093/intimm/11.4.617. [DOI] [PubMed] [Google Scholar]
- 33.Flynn K J, Belz G T, Altman J D, Ahmed R, Woodland D L, Doherty P C. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 34.Belz G T, Xie W, Altman J D, Doherty P C. J Virol. 2000;74:3486–3493. doi: 10.1128/jvi.74.8.3486-3493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belz G T, Xie W, Doherty P C. J Immunol. 2001;166:4627–4633. doi: 10.4049/jimmunol.166.7.4627. [DOI] [PubMed] [Google Scholar]
- 36.Doyle A G, Buttigieg K, Groves P, Johnson B J, Kelso A. J Exp Med. 1999;190:1081–1091. doi: 10.1084/jem.190.8.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uellner R, Zvelebil M J, Hopkins J, Jones J, MacDougall L K, Morgan B P, Podack E, Waterfield M D, Griffiths G M. EMBO J. 1997;16:7287–7296. doi: 10.1093/emboj/16.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham C T, Ley T J. Proc Natl Acad Sci USA. 1999;96:8627–8632. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prendergast J A, Helgason C D, Bleackley R C. J Biol Chem. 1992;267:5090–5095. [PubMed] [Google Scholar]
- 40.Morris A G, Lin Y-L, Askonas B A. Nature. 1982;295:150–152. doi: 10.1038/295150a0. [DOI] [PubMed] [Google Scholar]
- 41.Kelso A, Metcalf D. Adv Immunol. 1990;48:69–105. doi: 10.1016/s0065-2776(08)60752-x. [DOI] [PubMed] [Google Scholar]
- 42.Slifka M K, Rodriguez F, Whitton J L. Nature. 1999;401:76–79. doi: 10.1038/43454. [DOI] [PubMed] [Google Scholar]