TABLE 2.
Maximum-likelihood results
| Criterion | Model chosen |  |
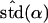 |
Shared-parameter MLEs | |
|---|---|---|---|---|---|
| yakuba–simulans | AIC/BIC | 10ii | 0.405 | 0 |
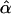 = 0.405 = 0.405 |
| AICc | 5iv | 0.428 | 0.351 |
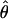 = 0.03, = 0.03, 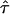 = 9.06 = 9.06 |
|
 = 0.00, = 0.00,  = 0.78 = 0.78 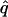 = 0.45 = 0.45 | |||||
| melanogaster–simulans | AIC | 10iv | 0.405 | 0.101 |
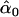 = 0.39, = 0.39,  = 0.98, = 0.98, 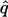 = 0.97 = 0.97 |
| BIC | 10ii | 0.436 | 0 |
 = 0.436 = 0.436 |
|
| AICc | 5iv | 0.404 | 0.375 |
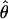 = 0.02, = 0.02, 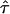 = 2.46 = 2.46 |
|
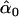 = 0.04, = 0.04,  = 0.79, = 0.79, 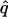 = 0.51 = 0.51 | |||||
| D. simulans lineage | AIC | 10iv | 0.436 | 0.097 |
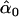 = 0.42, = 0.42,  = 0.98, = 0.98,  = 0.97 = 0.97 |
| BIC/AICc | 5iv | 0.430 | 0.324 |
 = 0.03, = 0.03,  = 0.63 = 0.63 |
|
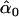 = 0.19, = 0.19,  = 0.87 = 0.87 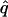 = 0.65 = 0.65 |
Likelihood models supported by the different model selection criteria are shown. Three sets of results are presented, with the divergence to D. simulans measured from D. yakuba, D. melanogaster, and the common ancestor of melanogaster and simulans. For the chosen models, the maximum-likelihood estimates of the across-locus mean and standard deviation of α are shown. These quantities, denoted 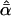 and
and 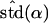 , were determined from the MLE parameters of pdf(α). Also shown are the estimated values of parameters common to all loci.
, were determined from the MLE parameters of pdf(α). Also shown are the estimated values of parameters common to all loci.
