Abstract
Cytochrome c oxidase (COX) is a 13-subunit protein complex that catalyzes the last step in mitochondrial electron transfer in mammals. Of the 10 subunits encoded by nuclear DNA (three are mtDNA products), some are expressed as tissue- and/or development-specific isoforms. For COX subunit VIII, previous work showed that expression of the contractile muscle-specific isoform gene, COX8H, is absent in humans and Old World monkeys, and the other isoform gene, COX8L, is expressed ubiquitously. Here, we show that COX8H is transcribed in most primate clades, but its expression is absent in catarrhines, that is, in Old World monkeys and hominids (apes, including humans), having become a pseudogene in the stem of the catarrhines. The ubiquitously expressed isoform, COX8L, underwent nonsynonymous rate acceleration and elevation in the ratio of nonsynonymous/synonymous changes in the stem of anthropoid primates (New World monkeys and catarrhines), possibly setting the stage for loss of the heart-type (H) isoform. The most rapidly evolving region of VIII-L is one that interacts with COX I, suggesting that the changes are functionally coadaptive. Because accelerated rates of nonsynonymous substitutions in anthropoids such as observed for COX8L are also shown by genes for at least 13 other electron transport chain components, these encoded amino acid replacements may be viewed as part of a series of coadaptive changes that optimized the anthropoid biochemical machinery for aerobic energy metabolism. We argue that these changes were linked to the evolution of an expanded neocortex in anthropoid primates.
Cytochrome c oxidase (COX) is a dimeric holoenzyme that functions as the terminal complex of the mitochondrial electron transport chain (ETC). COX both catalyzes the transfer of electrons from cytochrome c to oxygen and modulates the rate at which this crucial step in aerobic energy metabolism occurs (1). In mammals, each COX monomer contains 13 subunits (2–5). The three largest subunits are encoded by mtDNA, the rest by nuclear DNA. The known catalytic functions of COX are performed by mitochondrial-encoded subunits I and II, whereas the functions of the nuclear-encoded subunits have not yet been clearly established; however, they probably involve assembly of the holoenzyme and some role in regulation of COX activity and cellular energy production (6–15).
Among the nuclear-encoded COX subunits, VIa, VIIa, and VIII have H (heart-type) and L (liver-type) isoforms. For each subunit with these isoforms, H shows developmental stage and tissue specificity in expression pattern, being primarily expressed postnatally in mature contractile muscles (16–20). In contrast, L is ubiquitously expressed in all developmental stages and tissues, although at low levels in mature contractile muscles when H predominantly is being expressed there (21–23). Phylogenetic analyses of comparative sequence data for subunits VIa, VIIa, and VIII show that the H and L isoforms of each of these COX subunits originated from gene duplication events that preceded the radiation of placental mammals (24–26). Thus, it would seem that by the time of the placental mammalian radiation, the H isoforms were well adapted to the energy requirements of contractile muscle tissues. Nevertheless, in previous work (27, 28), the H isoform of COX subunit VIII could not be found in humans or in the one Old World monkey (a long-tailed macaque) that was also examined.
Following up on this earlier finding concerning the loss of COX VIII-H, we have gathered results that address the following questions. When, during humankind's evolutionary history, did the loss of COX VIII-H occur? What was the nature of the mutational change that resulted in inactivation of the gene (COX8H) encoding COX VIII-H? Was this mutational change selectively neutral or, instead, favored by positive Darwinian selection? Were there positively selected changes to COX8L? If so, they might help to explain why COX8L could become the sole gene encoding subunit VIII.
At the start of our study, we considered alternative possibilities as to when inactivation of COX8H occurred: in a common ancestor of all living primates (lemuriforms, loriforms, tarsiers, and anthropoids), in the stem lineage of haplorhines (tarsiers and anthropoids), in the common ancestral lineage of New World monkeys and catarrhines (i.e., the stem lineage of anthropoid primates), or, closer to the present, in the stem lineage of catarrhines (Old World monkeys and apes, including humans). We conjectured that if inactivation of COX8H occurred during anthropoid primate evolution (such as in either stem anthropoids or stem catarrhines), this inactivation could be considered part of a series of coadaptive changes that presumably optimized the biochemical machinery for aerobic energy metabolism. This series of coadaptive changes includes accelerated amino acid replacement rates for subunits of COX and other ETC components. These accelerations involved cytochrome b (29), cytochrome c (refs. 30 and 31; D.E.W., M. Uddin, M.G., and L.I.G., unpublished data), NADH-ubiquinone oxidoreductase subunit B14.5b, hinge protein, ATP synthase subunit F6 (32), and COX subunits I (33, 34), II (35), IV (36–38), Va, VIb, VIc, VIIa-H, VIIc (refs. 26, 32, and 39; J. W. Doan, A.G., D.E.W., T.R.S., M.H., M.G., M.L.W., and L.I.G., unpublished data), and VIII-L (ref. 32 and this report).
The results gained in our study show that COX8H is a functional gene in most primate clades but became a pseudogene, and thus nonexpressed, in the stem of the catarrhines. Also, on enlarging the COX8L dataset of Osada (32) by adding to it New World monkey, tarsier, loriform (galago, slow loris), and lemur coding sequences, we show that before silencing of COX8H expression the rate of nonsynonymous substitutions in COX8L accelerated markedly in the stem of the anthropoid primates. We argue that this accelerated COX8L nonsynonymous rate, as part of a series of coadaptive changes in the anthropoid ETC genes, was positively selected. By optimizing the molecular machinery for aerobic energy metabolism, it became advantageous for the descendant catarrhines to have COX8L as the sole gene encoding COX subunit VIII in all developmental stages and tissues, including heart and other contractile muscles. Thus, we further argue that the mutational change that converted COX8H into a pseudogene in the stem catarrhines was also positively selected.
Materials and Methods
DNA and RNA Sequences and Samples.
Genomic DNA and/or cDNA sequences from Pan paniscus (bonobo), Gorilla gorilla (gorilla), Pongo pygmaeus (orangutan), Hylobates agilis (agile gibbon), Theropithecus gelada (gelada baboon), Papio anubis (olive baboon, also known as anubis baboon), Macaca silenus (lion-tailed macaque), Trachypithecus cristatus (silvered leaf monkey), Saimiri sciureus (squirrel monkey), Ateles belzebuth (white-bellied spider monkey), Tarsius syrichta (Philippine tarsier), Perodicticus potto (potto), Otolemur crassicaudatus (fat-tailed galago), Nycticebus coucang (slow loris), and Eulemur fulvus (brown lemur) have been placed in GenBank. Published sequences from GenBank for COX8H used in this analysis are Mus musculus (mouse) (NM_007751), Rattus norvegicus (rat) (NM_012786), Bos taurus (cow) (U15540), and Homo sapiens (human) (AF015416); for COX8L, published sequences include human (NM_004074, AC023920), Pan troglodytes (common chimpanzee) (AB072322, AB072323), Macaca fascicularis (crab-eating macaque) (AB072015), mouse (NM_007750), rat (AI409704), cow (J05201), and Sus scrofa (pig) (BF702874).
DNA and RNA were prepared from a variety of tissues including lung, liver, kidney, and heart. RNA was isolated either by using a standard phenol-chloroform method (40) or with the Qiagen (Valencia, CA) Midi kit. In some species, total RNA was obtained from the Center for Reproduction of Endangered Species, San Diego. DNA was isolated by using the Promega Wizard genomic DNA purification kit or was from laboratory stock solutions.
COX8H PCR Amplifications.
PCR amplification of COX8H genomic DNA.
A region spanning ≈1 kb from exon 2 of COX8H to the terminal exon of PSMD13 (the gene encoding the p40.5 subunit of the 26S proteasome) was amplified for potto, galago, brown lemur, tarsier, gelada baboon, and cow (Fig. 1). Primers in exon 1 and in or downstream of exon 2 were used to complete sequencing of exon 2 for potto, galago, tarsier, and gelada baboon. Gelada baboon exon 1 and 5′-flanking region were amplified by using a one-way PCR protocol (M.H. and L.I.G., unpublished data). Anubis baboon exons 1 and 2 were amplified separately by using primers upstream and downstream of each exon. For human, bonobo, gorilla, and spider monkey, ≈3.4 kb upstream of exon 2 was amplified and sequenced; for orangutan and gibbon, a portion of this region was amplified and sequenced (Fig. 1). Primers and PCR amplification conditions for both COX8 genes are available on request.
Figure 1.
Organization of the COX8H locus. (A) Cow COX8H. The sizes of the intergenic regions between exon 2 of COX8H and exon 13 of PSMD13 were determined from sequences included in this study. (B) Species retaining the organization of the active cow gene. Other species with this gene structure and the regions analyzed are shown. (C) Species that maintain the cow gene structure but in which COX8H is a pseudogene. (D) Organization of COX8H in human. The exon 1 region has been invaded by gene-disrupting repeat sequences (see text). (E) Other hominid species and regions analyzed. All contain the human-type gene structure. Empty bars, pseudogene; filled bars, expressed gene.
COX8H 5′ and 3′ RACE.
RACE PCR was performed (41) to generate full-length cDNA sequences for spider monkey, tarsier, and brown lemur COX8H. After obtaining 3′ cDNA sequences, gene-specific reverse primers were designed for 5′ RACE PCR, which was carried out largely as described (42). To examine whether transcription of COX8H occurs in catarrhine primates, we used COX8H primers specific for human exon 2 and anubis baboon exons 1 and 2 and performed 3′ RACE PCR with cDNAs prepared from heart and other human tissues, as well as from anubis baboon heart.
COX8L PCR Amplifications.
PCR amplification of COX8L genomic DNA.
Exons 1 and 2 of COX8L were amplified separately for bonobo, gorilla, orangutan, gibbon, and gelada baboon by using primers based on the human COX8L genomic sequence (GenBank accession no. AC023920).
COX8L 5′ and 3′ RACE.
Full-length cDNA sequences for tarsier, brown lemur, and fat-tailed galago COX8L were generated by using RACE PCR. cDNA sequences for silvered leaf monkey, lion-tailed macaque, anubis baboon, spider monkey, squirrel monkey, and slow loris were generated by RT-PCR.
PCR Product Purification, Cloning, Sequencing, and Alignment.
PCR products were isolated by agarose gel electrophoresis (1%) and QIAquick DNA purification columns (Qiagen). Purified products were either sequenced directly with amplification primers or cloned into the pGEM-T easy vector (Promega) and sequenced with SP6 and T7 primers. Nucleotide and inferred amino acid sequences were aligned with CLUSTALW and refined by eye. Repeat elements such as SINE and LINE, and other repetitive elements were detected with REPEATMASKER (http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker). Our examination of COX8H for repeat elements encompassed between 1 and 3.4 kb of nucleotide sequence upstream of exon 2 for the various species.
Phylogenetic Analysis and Rates of Nucleotide Substitution.
Branch and bound maximum parsimony gene trees were inferred from the COX8L coding sequences in PAUP* (Version 4.0b10; ref. 43). Measures of topological support were obtained from 250 replicates of branch and bound bootstrapping with replacement. The tree most consistent with a commonly accepted primate species tree (44) was modified to be completely consistent with that phylogeny, and the number of nucleotide substitutions necessary for such modification was determined.
Ancestral coding region sequences were reconstructed in PAUP* by using the deltran algorithm, and the Pamilo–Bianchi–Li method (45, 46) was used to calculate Ka/Ks between nodes by using the program FENS (47). Ka stands for nonsynonymous substitutions/nonsynonymous site; similarly, Ks stands for synonymous substitutions/synonymous site. Rates of Ka and Ks were calculated by using divergence dates from the combined fossil and molecular data (44). Because the mitochondria-targeting presequences are highly conserved (39) and not incorporated into the COX holoenzyme, the codons that code for these presequences were removed from calculations of Ka. However, to more adequately sample the synonymous (presumably selectively neutral) sites, these presequence-encoding codons were retained along with the mature polypeptide-encoding codons for the calculations of Ks. By this procedure, the difference between the small number of synonymous sites and larger number of nonsynonymous sites is not as large as the case when only the mature polypeptide-encoding codons are used to calculate both Ka and Ks.
Results
Organization of COX8H.
Primate taxa that express COX8H have the same gene organization seen in such nonprimate mammals as murine rodents and cow (Fig. 1 A and B). COX8H in mammals consists of two exons, the second located <1 kb from the 3′ end of the terminal exon of a gene encoding the 26S proteasome p40.5 subunit (PSMD13). In hominids (apes including humans), exon 2 is largely intact, but a 2.4-kb region upstream of exon 2, including exon 1, has been replaced by elements of repeat sequence families (Fig. 1 D and E). These elements consist of simple (TCCA)n repeats, truncated L2 LINEs, Ricksha_a DNA elements, and a SINE belonging to the AluSp subfamily. Unlike the hominids, gelada baboon and anubis baboon retain an intact intron–exon structure, although there are point mutations in the exons. The regions analyzed in each species are indicated (Fig. 1).
Transcription of COX8H.
COX8H is transcribed in the major primate clades except catarrhines. In three noncatarrhine primate species (spider monkey, tarsier, and lemur), transcription was confirmed by generating full-length cDNA sequences with 5′ and 3′ RACE PCR. The 5′ RACE PCR products for brown lemur are typical (Fig. 2) and show a single major band after amplification with nested primers. The nucleotide sequences of the RACE products can be reliably aligned with that of cow without any alignment gaps in the coding region (Fig. 3).
Figure 2.

RACE PCR performed with brown lemur RNA and COX8H primers. RNA was extracted from liver tissue (see Materials and Methods). Lane 1, 5′ RACE PCR product using nested primers displayed on a 1% agarose gel; lane 2, 100-bp ladder.
Figure 3.
Alignment of COX8H sequences. cDNA sequences for brown lemur, tarsier, and spider monkey and genomic sequences for potto, galago (exon 2), and gelada and anubis baboons are aligned with cow and human. Start and stop codons (bold) and the putative polyadenylation sites (underlined) are noted. For potto and galago, such polyadenylation sites are located ≈40 bp downstream of those found in other species. Bonobo and gorilla (data not shown) share exon 2 coding region deletions with other catarrhines (see text). Coding sequence is shaded; the interruption in the shading between residues H and I indicates the end of the mitochondria-targeting presequence. The vertical line indicates the start of exon 2; dots indicate identity and dashes indicate insertions or deletions. The amino acid sequence for cow is shown.
Using cDNAs prepared from heart tissue, we were unable to generate COX8H cDNA sequences for representative catarrhine primates (human and anubis baboon), a finding that supports previous results for humans (27, 28) and an Old World monkey (28). We also examined genomic DNA sequences for the COX8H locus in human, bonobo, gorilla, gelada baboon, and anubis baboon. Exon 2 in all of these catarrhine primates has been disrupted by insertions and deletions and by elimination of the normal stop codon. In baboon exon 1, the initiating ATG has been replaced by ATT (Fig. 3). These mutations, together with the insertion of DNA repeats in all hominids examined, preclude expression of functional COX8H in catarrhines.
Phylogenetic Analysis of COX8L.
COX8L, like COX8H, consists of two exons separated by an intron. Each of the aligned COX8L coding sequences is 207 nt in length, and no alignment gaps are necessary. Maximum parsimony analysis generated 630 lowest nucleotide-substitution-length bifurcating trees (56 trees when zero-length branches are collapsed), each with a tree length of 161 when all substitutions are given equal weight. The bootstrap consensus tree supported murine (bootstrap value 100%), loriform (98%), anthropoid (99%), New World monkey (100%), catarrhine (96%), Old World monkey (97%), macaque/baboon (82%), gelada/anubis baboon (96%), ape (85%), human/chimpanzee (65%), and bonobo/common chimpanzee (71%) clades.
The accepted primate species tree (Fig. 4) has a length of 162. The tree that is both at the lowest nucleotide substitution length (161) and is most congruent with the accepted species tree differed from this species tree by placing the brown lemur at the base of the Anthropoidea rather than at the base of the Loriformes. To fit this possible gene phylogeny into the primate species phylogeny by the method of Goodman et al. (48) requires two gene duplication and six gene expression events. Thus, as the accepted primate species tree at nucleotide substitution length 162 does not require any gene duplication or expression events, this tree is actually the most parsimonious and most probably represents the correct gene phylogenetic tree.
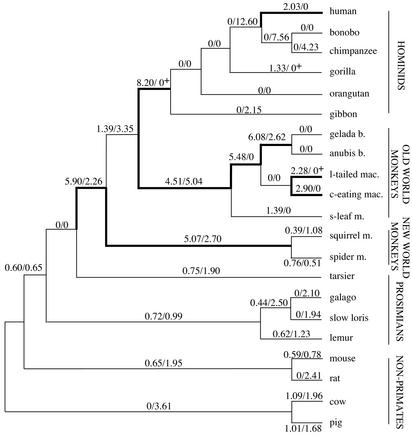
Figure 4.
Evolution of COX8L in primates. Rates of Ka (nonsynonymous substitutions/nonsynonymous site/year × 10−9) and Ks (synonymous substitutions/synonymous site/year × 10−9) are shown as numerator and denominator, respectively, on branches of the tree consistent with the overall evidence on primate phylogeny (44). Bold lines designate branches with elevated Ka rates. +, method of Nei and Gojobori as implemented in FENS (47) was used instead of the Pamilo–Bianchi–Li method to determine Ka and Ks values (see Materials and Methods). gelada b., gelada baboon; anubis b., anubis baboon; l-tailed mac., lion-tailed macaque; c-eating mac., crab-eating macaque; s-leaf m., silvered leaf monkey; squirrel m., squirrel monkey; spider m., spider monkey.
Accelerated Rates of Nonsynonymous Substitution in COX8L.
Rates of nonsynonymous and synonymous substitutions are shown on the primate species tree (Fig. 4). Our examination of COX8L shows that it underwent accelerated rates of nonsynonymous substitutions before, as well as after, the silencing of COX8H. The Ka rate shows marked elevations on the anthropoid and hominid stem lineages and subsequent decreases from the last common ape ancestor to the present, except on the terminal human lineage, where the Ka rate is elevated. During Old World monkey evolution, the Ka rate shows elevations on the Old World monkey, macaque/baboon, and baboon stem lineages and also on each of the two terminal macaque lineages. Also the stem lineage of New World monkeys shows an elevated Ka rate followed by marked decreases in Ka rate in both squirrel monkey and spider monkey. In contrast to the elevated rates in anthropoid primates, the Ka rate is uniformly low in nonanthropoid primates and nonprimate mammals. Much of the difference between anthropoids and nonanthropoids is concentrated in the 12 N-terminal amino acid residues of the mature polypeptide. There, we find an elevated amino acid replacement rate almost five times higher in anthropoids than in nonanthropoids (Figs. 5 and 6). These N-terminal amino acid residues encompass the sites that interact with the mitochondria-encoded COX I subunit of the holoenzyme (4). During evolution of the anthropoids, this N-terminal region evolved at a faster rate than did the remaining 32 aa residues of the mature polypeptide (Figs. 5 and 6).
Figure 5.
Amino acid alignment for COX VIII-L. Residues 1–25, comprising the mitochondria-targeting presequence, are not incorporated into the COX holoenzyme; residues 26–69 (bold) comprise the mature functional subunit VIII-L protein. The 12 residues interacting with COX subunit I are shaded (see text). See Fig. 4 legend for abbreviations.
Figure 6.
Rates of amino acid replacements during mammalian evolution for COX subunit VIII-L. Rates of replacement (with standard errors) are shown as number of amino acid replacements per residue per year ×10−9. Total years of anthropoid evolution is 228 million years; total years of nonanthropoid evolution is 468 million years.
Discussion
Mechanism and Timing of COX8H Inactivation.
A significant finding of this study is that COX8H is transcribed in most primate clades but not in the catarrhines. Our sequence data allow us to determine both the mechanism and timing of the silencing of COX8H expression. The shared presence of exon 2 coding region deletions, one of which removed the stop codon in humans, other hominids, and anubis and gelada baboons, provides compelling evidence that COX8H became a pseudogene in a common ancestor of Old World monkeys and hominids, i.e., on the catarrhine stem lineage. The shared presence of gene-disrupting DNA repeats in hominids, but not in Old World monkeys, indicates that insertion of these repeats into the COX8H locus postdated the hominid–Old World monkey split and therefore played no role in the silencing of COX8H expression.
Adaptive Evolution in COX8L.
Our results illustrate that in descent of anthropoids, on a range of stem lineages and on several terminal lineages, COX8L evolved under the force of positive Darwinian selection as judged by marked elevations of Ka rates and Ka/Ks values (Fig. 4). In contrast, in descent of nonanthropoids, COX8L evolved under the constraints of purifying selection as judged by uniformly low Ka rates and Ka/Ks values in the nonanthropoid lineages. Furthermore, a key functional region of COX VIII-L, the 12 N-terminal amino acid residues that are in close contact with the mtDNA-encoded COX I subunit, is a highly conserved region in nonanthropoids but the most rapidly evolving region in anthropoids (Figs. 5 and 6). Apparently, in anthropoids, selection acted to bring about amino acid replacements at functionally important positions of the mature COX VIII-L protein.
In phylogenetic analyses conducted up to now, accelerated rates of nonsynonymous substitutions in anthropoids, such as those observed for COX8L, are also shown by genes for at least 13 other ETC components (see Introduction). These encoded amino acid replacements may be viewed as part of a series of coadaptive changes that optimized the anthropoid biochemical machinery for aerobic energy metabolism.
Neocortical Expansion in Anthropoids Is a Selective Force in the Evolution of ETC Genes.
Combining the results of several types of studies indicates that the upsurges of amino acid replacements in anthropoid ETC proteins were not just coincident with enlargements of the brain but were causally linked to the brain's evolution. Paleontological studies of the primate fossil record show that cranial capacity is generally larger in later members of the suborder Anthropoidea than in earlier basal members and greater in anthropoids than in nonanthropoids (49). In addition, among living primates, the neocortical proportion of the brain is much larger in anthropoids than in nonanthropoids, and among anthropoids, it is largest in the catarrhines, especially in humans (50–52). Physiological studies show that the adult human brain, although contributing only 2% of the total body weight, fuels its energy expenditures by consuming ≈20% of the body's oxygen intake (ref. 53 and reviewed in refs. 54 and 55). Moreover, the human neonate devotes ≈65% of its metabolic energy to its rapidly growing brain (53). By inference, the human fetus devotes a high proportion of its metabolic energy to its rapidly enlarging brain. Comparative studies show that fetal, neonatal, and postnatal stages of life became more prolonged in anthropoids than in nonanthropoid primates and more prolonged in apes, especially so in humans, than in other anthropoids (56). The overriding importance of the brain to the life of anthropoids and the demands that anthropoid brains make for exceptionally high amounts of metabolic energy, indeed, implicate these enlarging anthropoid brains as a key selective agency in the evolution of anthropoid ETC genes.
In addition to the results of our ongoing study of the evolution of ETC genes, the results of a prior study of lactate dehydrogenase (LDH) isozymes (57, 58) point to a causal link between brain evolution and the evolution of the biochemical pathways for aerobic energy metabolism. In loriform and lemuriform brains, the skeletal muscle-type isozyme, M LDH, predominated over the heart-type isozyme, H LDH; just the opposite was the case in anthropoid brains. The shift from M to H LDH was most pronounced in cerebral cortical portions of the brain and also more pronounced in catarrhines than in New World monkeys (58). The view that M LDH suits anaerobic (low energy) metabolism whereas H LDH suits aerobic (high energy) metabolism (59, 60) led to the suggestion that brain tissues with a preponderance of H LDH can function at a higher energy level than brain tissues with a preponderance of M LDH (58).
The emergence of a distinct fetal hemoglobin in anthropoid primates offers further support for the view of a causal link between evolution of the enlarged anthropoid brain and evolution of the molecular machinery for aerobic energy metabolism. Phylogenetic reconstructions show that nucleotide substitutions in cis-regulatory promoter elements of the γ-globin gene and nonsynonymous substitutions in this gene's exons shaped a fetal hemoglobin that differed from adult hemoglobin in both expression and structure (reviewed in ref. 61). With regard to the nonsynonymous substitutions, especially important were those in codons specifying the amino acid residues of the anthropoid γ-globin 2,3-bisphosphoglycerate binding site. The resultant amino acid replacements drastically reduced the 2,3-bisphosphoglycerate binding capacity of fetal hemoglobin, thus increasing the oxygen-binding capacity of fetal blood over that of maternal blood. This favored the transport of oxygen from mother to fetus and thereby, presumably, helped make possible the prolonged intrauterine fetal life and prenatal brain development of anthropoid primates.
Why Would Catarrhines Lose COX8H Expression?
Accepting that an important aspect of evolution in humankind's anthropoid ancestry selected for coadaptive changes in proteins involved in aerobic energy metabolism, why would loss of the COX VIII-H isoform be advantageous after it had already become specialized for functioning postnatally in heart and other contractile muscles? We assume that in the process of becoming so specialized the H isoform offered some small advantage and thus was favored by selection but was not essential for life, provided that the L isoform was available to be fully expressed postnatally as well as prenatally. Under these conditions, the advantages offered by COX VIII-L, after it underwent a burst of amino acid replacements in the early anthropoids, would override any disadvantages that would ensue if the H isoform were lost. Presumably, because the L isoform had become the better adapted for COX VIII's role in aerobic metabolism, selection favored the loss of the H isoform in heart (a highly aerobic tissue) and tolerated its loss in less aerobic contractile muscle tissues, provided the regulatory mutational change needed to fully express COX VIII-L in all adult tissues had already occurred.
Acknowledgments
We thank Drs. J. Rogers and K. Rice of the Southwest Regional Primate Research Center (San Antonio, TX) and Dr. W. Hylander and D. Haring of the Duke University Primate Center (Durham, NC) for primate tissues; Dr. O. Ryder and L. Chemnick of the San Diego Zoo (Center for Reproduction of Endangered Species) for nucleic acids; and Dr. M. Batzer of Louisiana State University (Baton Rouge, LA) for helpful discussion regarding DNA repeat elements. This work was supported by National Science Foundation Grants MCB-9816923 and BCS-9910679 and National Institutes of Health Grants GM 48517 and GM 65580.
Abbreviations
- COX
cytochrome c oxidase
- ETC
electron transport chain
- LDH
lactate dehydrogenase
Footnotes
References
- 1.Kadenbach B, Hüttemann M, Arnold S, Lee I, Bender E. Free Radical Biol Med. 2000;29:211–221. doi: 10.1016/s0891-5849(00)00305-1. [DOI] [PubMed] [Google Scholar]
- 2.Kadenbach B, Jarausch J, Hartmann R, Merle P. Anal Biochem. 1983;129:517–521. doi: 10.1016/0003-2697(83)90586-9. [DOI] [PubMed] [Google Scholar]
- 3.Kadenbach B, Kuhn-Nentwig L, Buge U. Curr Top Bioenerg. 1987;15:114–162. [Google Scholar]
- 4.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 5.Grossman L I, Lomax M I. Biochim Biophys Acta. 1997;1352:174–192. doi: 10.1016/s0167-4781(97)00025-0. [DOI] [PubMed] [Google Scholar]
- 6.Aggeler R, Capaldi R A. J Biol Chem. 1990;265:16389–16393. [PubMed] [Google Scholar]
- 7.Lightowlers R, Chrzanowska-Lightowlers Z, Marusich M, Capaldi R A. J Biol Chem. 1991;266:7688–7693. [PubMed] [Google Scholar]
- 8.Taanman J W, Capaldi R A. J Biol Chem. 1993;268:18754–18761. [PubMed] [Google Scholar]
- 9.Weishaupt A, Kadenbach B. Biochemistry. 1992;31:11477–11481. doi: 10.1021/bi00161a028. [DOI] [PubMed] [Google Scholar]
- 10.Anthony G, Reimann A, Kadenbach B. Proc Natl Acad Sci USA. 1993;90:1652–1656. doi: 10.1073/pnas.90.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taanman J W, Turina P, Capaldi R A. Biochemistry. 1994;33:11833–11841. doi: 10.1021/bi00205a020. [DOI] [PubMed] [Google Scholar]
- 12.Arnold S, Kadenbach B. Eur J Biochem. 1997;249:350–354. doi: 10.1111/j.1432-1033.1997.t01-1-00350.x. [DOI] [PubMed] [Google Scholar]
- 13.Arnold S, Goglia F, Kadenbach B. Eur J Biochem. 1998;252:325–330. doi: 10.1046/j.1432-1327.1998.2520325.x. [DOI] [PubMed] [Google Scholar]
- 14.Arnold S, Kadenbach B. FEBS Lett. 1999;443:105–108. doi: 10.1016/s0014-5793(98)01694-9. [DOI] [PubMed] [Google Scholar]
- 15.Ludwig B, Bender E, Arnold S, Hüttemann M, Lee I, Kadenbach B. Chembiochem. 2001;2:392–403. doi: 10.1002/1439-7633(20010601)2:6<392::AID-CBIC392>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 16.Ewart G D, Zhang Y Z, Capaldi R A. FEBS Lett. 1991;292:79–84. doi: 10.1016/0014-5793(91)80839-u. [DOI] [PubMed] [Google Scholar]
- 17.Bonne G, Seibel P, Possekel S, Marsac C, Kadenbach B. Eur J Biochem. 1993;217:1099–1107. doi: 10.1111/j.1432-1033.1993.tb18342.x. [DOI] [PubMed] [Google Scholar]
- 18.Schillace R, Preiss T, Lightowlers R N, Capaldi R A. Biochim Biophys Acta. 1994;1188:391–397. doi: 10.1016/0005-2728(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 19.Kim K, Lecordier A, Bowman L H. Biochem J. 1995;306:353–358. doi: 10.1042/bj3060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossman L I, Rosenthal N H, Akamatsu M, Erickson R P. Biochim Biophys Acta. 1995;1260:361–364. doi: 10.1016/0167-4781(94)00232-r. [DOI] [PubMed] [Google Scholar]
- 21.Van Beeumen J J, Van Kuilenburg A B P, Van Bun S, Van Den Bogert C, Tager J M, Muijsers A O. FEBS Lett. 1990;263:213–216. doi: 10.1016/0014-5793(90)81376-y. [DOI] [PubMed] [Google Scholar]
- 22.Kadenbach B, Stroh A, Becker A, Eckerskorn C, Lottspeich F. Biochim Biophys Acta. 1990;1015:368–372. doi: 10.1016/0005-2728(90)90042-3. [DOI] [PubMed] [Google Scholar]
- 23.Thames E L, Newton D A, Black S A, Bowman L H. Biochem J. 2000;351:133–142. doi: 10.1042/0264-6021:3510133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saccone C, Pesole G, Kadenbach B. Eur J Biochem. 1991;195:151–156. doi: 10.1111/j.1432-1033.1991.tb15688.x. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt T R, Jaradat S A, Goodman M, Lomax M I, Grossman L I. Mol Biol Evol. 1997;14:595–601. doi: 10.1093/oxfordjournals.molbev.a025798. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt T R, Goodman M, Grossman L I. Mol Biol Evol. 1999;16:619–626. doi: 10.1093/oxfordjournals.molbev.a026144. [DOI] [PubMed] [Google Scholar]
- 27.Van Kuilenburg A B P, Muijsers A O, Demol H, Dekker H L, Van Beeumen J J. FEBS Lett. 1988;240:127–132. doi: 10.1016/0014-5793(88)80353-3. [DOI] [PubMed] [Google Scholar]
- 28.Rizzuto R, Nakase H, Darras B, Francke U, Fabrizi G M, Mengel T, Walsh F, Kadenbach B, Dimauro S, Schon E A. J Biol Chem. 1989;264:10595–10600. [PubMed] [Google Scholar]
- 29.Andrews T D, Jermiin L S, Easteal S. J Mol Evol. 1998;47:249–257. doi: 10.1007/pl00006382. [DOI] [PubMed] [Google Scholar]
- 30.Baba M L, Darga L L, Goodman M, Czeluzniak J. J Mol Evol. 1981;17:197–213. doi: 10.1007/BF01732758. [DOI] [PubMed] [Google Scholar]
- 31.Grossman L I, Schmidt T R, Wildman D E, Goodman M. Mol Phylogenet Evol. 2001;18:26–36. doi: 10.1006/mpev.2000.0890. [DOI] [PubMed] [Google Scholar]
- 32.Osada N, Kusuda J, Hirata M, Tanuma R, Hida M, Sugano S, Hirai M, Hashimoto K. Genomics. 2002;79:657–662. doi: 10.1006/geno.2002.6753. [DOI] [PubMed] [Google Scholar]
- 33.Wu W, Schmidt T R, Goodman M, Grossman L I. Mol Phylogenet Evol. 2000;17:294–304. doi: 10.1006/mpev.2000.0833. [DOI] [PubMed] [Google Scholar]
- 34.Andrews T D, Easteal S. J Mol Evol. 2000;50:562–568. doi: 10.1007/s002390010059. [DOI] [PubMed] [Google Scholar]
- 35.Adkins R M, Honeycutt R L. J Mol Evol. 1994;38:215–231. doi: 10.1007/BF00176084. [DOI] [PubMed] [Google Scholar]
- 36.Lomax M I, Hewett-Emmett D, Yang T L, Grossman L I. Proc Natl Acad Sci USA. 1992;89:5266–5270. doi: 10.1073/pnas.89.12.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu W, Goodman M, Lomax M I, Grossman L I. J Mol Evol. 1997;44:477–491. doi: 10.1007/pl00006172. [DOI] [PubMed] [Google Scholar]
- 38.Wildman D E, Wu W, Goodman M, Grossman L I. Mol Biol Evol. 2002;19:1812–1815. doi: 10.1093/oxfordjournals.molbev.a004005. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt T R, Goodman M, Grossman L I. Gene. 2002;286:13–19. doi: 10.1016/s0378-1119(01)00800-9. [DOI] [PubMed] [Google Scholar]
- 40.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 41.Frohmann M A. In: PCR Primer: A Laboratory Manual. Dieffenbach C W, Dveksler G S, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 381–409. [Google Scholar]
- 42.Hüttemann M. BioTechniques. 2002;32:730–736. doi: 10.2144/02324bm02. [DOI] [PubMed] [Google Scholar]
- 43.Swofford D L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2000. , Version 4.0b1. [Google Scholar]
- 44.Goodman M, Porter C A, Czelusniak J, Page S L, Schneider H, Shoshani J, Gunnell G, Groves C P. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 45.Li W H. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 46.Pamilo P, Bianchi N O. Mol Biol Evol. 1993;10:271–281. doi: 10.1093/oxfordjournals.molbev.a040003. [DOI] [PubMed] [Google Scholar]
- 47.De Koning A J P, Palumbo M, Messier W, Stewart C-B. fens: Facilitated Estimates of Nucleotide Substitutions. NY: Albany; 1998. , Version 1.0. [Google Scholar]
- 48.Goodman M, Czelusniak J, Moore G W, Romero-Herrera A E, Matsuda G. Syst Zool. 1979;28:132–163. [Google Scholar]
- 49.Martin R D. Primate Origins and Evolution: A Phylogenetic Reconstruction. London: Chapman & Hall; 1990. [Google Scholar]
- 50.Stephan H, Frahm H, Baron G. Folia Primatol (Basel) 1981;35:1–29. doi: 10.1159/000155963. [DOI] [PubMed] [Google Scholar]
- 51.Rilling J K, Insel T R. J Hum Evol. 1999;37:191–223. doi: 10.1006/jhev.1999.0313. [DOI] [PubMed] [Google Scholar]
- 52.Clark D A, Mitra P P, Wang S S. Nature. 2001;411:189–193. doi: 10.1038/35075564. [DOI] [PubMed] [Google Scholar]
- 53.Holliday M A. Pediatrics. 1971;47, Suppl. 2:169–179. [PubMed] [Google Scholar]
- 54.Foley R A, Lee P E. Philos Trans R Soc London B. 1991;334:223–232. doi: 10.1098/rstb.1991.0111. [DOI] [PubMed] [Google Scholar]
- 55.Aiello L C, Wheeler P. Curr Anthropol. 1995;36:199–221. [Google Scholar]
- 56.Harvey P H, Martin R D, Clutton-Brock T H. In: Primate Societies. Smuts B B, Cheney D L, Seyfarth R M, Wrangham R W, Struhsaker T T, editors. Chicago: Univ. Chicago Press; 1987. pp. 181–196. [Google Scholar]
- 57.Syner F N, Goodman M. Nature. 1966;209:426–428. doi: 10.1038/209426a0. [DOI] [PubMed] [Google Scholar]
- 58.Goodman M, Syner F N, Stimson C W, Rankin J J. Brain Res. 1969;14:447–459. doi: 10.1016/0006-8993(69)90121-8. [DOI] [PubMed] [Google Scholar]
- 59.Kaplan N O. In: Evolving Genes and Proteins. Bryson V, Vogel H J, editors. New York: Academic; 1965. pp. 271–272. [DOI] [PubMed] [Google Scholar]
- 60.Kaplan N O, Everse J, Admiraal J. Ann NY Acad Sci. 1968;151:400–412. doi: 10.1111/j.1749-6632.1968.tb11903.x. [DOI] [PubMed] [Google Scholar]
- 61.Goodman M. Am J Hum Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]