Abstract
Rabex-5 is an exchange factor for Rab5, a master regulator of endosomal trafficking. Rabex-5 binds monoubiquitin, undergoes covalent ubiquitination, and contains an intrinsic ubiquitin E3 ligase activity, all of which require an N-terminal A20 zinc finger and an immediately C-terminal helix. The structure of the N-terminal portion of Rabex-5 bound to ubiquitin at 2.5 Å resolution shows that Rabex-5:ubiquitin interactions occur at two sites. The first site is a new type of ubiquitin binding domain, an inverted ubiquitin interaction motif (IUIM), that binds with ~29 μM affinity to the canonical Ile44 hydrophobic patch on ubiquitin. The second is a diaromatic patch on the A20 zinc finger, which binds with ~22 μM affinity to a polar region centered on Asp58 of ubiquitin. The A20 zinc finger diaromatic patch mediates E3 ligase activity by directly recruiting a ubiquitin-loaded ubiquitin conjugating enzyme.
The Rab GTPases are central regulators of vesicular trafficking and organelle identity in all eukaryotes 1,2. The Rab family is the largest branch of the Ras superfamily, comprising more than 60 members in mammalian cells. Like other small GTPases, the localization and activity of the Rab proteins is regulated by GTPase activating proteins (GAPs), guanine nucleotide dissociation inhibitors (GDIs), and guanine nucleotide exchange factors (GEFs) 3,4. Rab GEFs promote the binding of GTP to Rab proteins, which in turn converts them to their active signaling conformation, and stabilizes their binding to cellular membranes. The founding member of the Rab5 GEF family is the yeast vacuolar sorting protein Vps9 5. Vps9 is the yeast ortholog of the human Rab5 GEF Rabex-5. All Rab5 GEFs have in common a catalytic unit comprising a helical bundle and a Vps9 homology domain 6. Most Rab5 GEFs do not function alone, but rather as components of larger multiprotein complexes, as exemplified by the Rabaptin-5-Rabex-5 complex 7–10.
Covalent monoubiquitination of proteins is a major regulatory signal in protein trafficking 11. In this process, the C-terminal carboxylate of a single molecule of the highly conserved 76-amino acid protein ubiquitin is covalently linked to a Lys residue in a substrate protein. This reaction is carried out by a series of enzymes known as E1, E2, and E3 12–14. Monoubiquitination of many transmembrane cargo proteins marks them for sorting into endosomal pathways 15–17. Monoubiquitin moieties on these proteins are recognized by specific ubiquitin binding domains in proteins of the trafficking machinery, including UIMs (ubiquitin interacting motifs), CUE (coupling of unfolded protein response to ER associated degradation), UEV (ubiquitin E2 variant) domains, and GAT (GGAs and TOM) domains 18. Furthermore, many trafficking proteins that contain ubiquitin binding domains are themselves monoubiquitinated in a manner that depends on both an E3 ubiquitin ligase and the presence of the binding domain 18. The monoubiquitination of these proteins is thought to regulate their activities.
The yeast counterpart of Rabex-5, Vps9, contains a C-terminal CUE domain and is a well-characterized example of the monoubiquitination of a monoubiquitin-binding protein 19–21. It had been anticipated that Rabex-5 might also contain a C-terminal ubiquitin binding domain 20, and a Rabex-5 ubiquitin interaction has been noted 21. However, the C-terminal segment of Rabex-5 does not bind to ubiquitin (Mattera, Tsai, Weissman, and Bonifacino, submitted). Rabex-5 interacts with ubiquitin, but does so through an N-terminal motif consisting of a zinc finger followed immediately by a 25-residue region predicted to form an α-helix (Mattera, Tsai, Weissman, and Bonifacino, submitted).
The Rabex-5 N-terminal zinc finger belongs to the A20 zinc finger (A20 ZnF) family and has E3 ubiquitin ligase activity (Mattera, Tsai, Weissman, and Bonifacino, submitted). The defining member of this family, A20, is a negative regulator of NF-κB signaling that has both deubiquitinating (DUB) enzyme and ubiquitin E3 ligase activities. A20 catalyzes the removal of a Lys63-linked polyubiquitin chain from the TNF receptor-1 binding protein RIP, followed by the ligation of a Lys48-linked polyubiquitin chain to RIP 22. The A20 protein contains seven A20 ZnF domains, and the fourth A20 ZnF is required for the ubiquitin ligase activity. E3 ubiquitin ligases identified previously fall into two classes. HECT domain ligases form covalent thiolesters with the ubiquitin C-terminal carboxylate, and directly transfer ubiquitin to the Lys residues of substrate proteins 12–14. RING ligases contain a zinc-binding RING finger that interacts with substrate and with an E2 ubiquitin conjugating enzyme, but does not form a covalent bond with the ubiquitin moiety to be transferred 12–14. The A20 ZnF domain represents a third class of ubiquitin E3 ligase 22. While the structural mechanisms for ubiquitin transfer through HECT 23 and RING 24 domain E3 ligases are established, no structural information has been available for A20 domain ligases.
To better understand the mechanisms of Rabex-5 ubiquitin recognition and its A20 ZnF domain-based E3 ligase activity, we determined the crystal structure of the complex between ubiquitin and the A20 ZnF and adjacent helix of Rabex-5. The structure, together with surface plasmon resonance (SPR) and isothermal titration calorimetric (ITC) analysis, shows that both the Rabex-5 A20 ZnF domain and the helix adjacent to it bind to ubiquitin with high affinity. This helix corresponds to an inverted UIM, hence we refer to it as the “IUIM” throughout the remainder of this report. We go on to map the determinants for the E3 ligase activity to a hydrophobic patch on the surface of the A20 ZnF, providing a structural template for understanding the A20 ZnF class of ubiquitin E3 ligases for the first time.
RESULTS
Structure of the Rabex-5 A20 ZnF and IUIM domains
The structure of Rabex-5 residues 9–73, comprising the A20 ZnF and IUIM domains (Fig. 1a), was determined in complex with ubiquitin in two different space groups: at 2.8 Å in a P61 lattice containing one complex per asymmetric unit, and at 2.5 Å in a C2 lattice containing three complexes per asymmetric unit. The structure was determined initially in the P61 lattice by molecular replacement (MR), using ubiquitin as the search model. Density modification was used to determine the structure of the A20 ZnF-IUIM portion of the complex (Fig. 1b). The structure of the complex in the P61 lattice was re-determined de novo by single anomalous dispersion (SAD) using the native Zn ion, and the resulting Fourier synthesis was found to be consistent with the MR solution. The C2 structure was determined by molecular replacement with the partially refined P61 structure. Together the two crystal forms present images of four copies of the complex.
Fig. 1. Structure of the Rabex-5 A20 ZnF domain and IUIM.

(a) The domain structure of Rabex-5 and the construct used in this study. The ZnF domain and the IUIM are colored magneta and green, respectively. (b) Density-modified Fourier synthesis from prime and switch phasing of the complex structure in space group P61. Rabex-5 and ubiquitin models are colored green and yellow, respectively. (c) Rabex-5 (ribbon model) contacts ubiquitin (surface models in orange and marine) at two sites in the crystal lattice. (d) Superposition of the four crystallographically independent molecules of the A20 ZnF domain and IUIM. The P61 molecule is colored green, and the three independent molecules in the asymmetric unit of the C2 lattice are colored pink, purple, and red. (e) The zinc binding site in the A20 ZnF domain. The Zn ion is colored red. (f) Ribbon and ball-and-stick representation showing exposed hydrophobic side-chains. (g) Surface of the A20 ZnF domain and IUIM colored by residue type: green (hydrophobic), red (acidic), blue (basic), and white (uncharged polar).
The structure reveals that the A20 ZnF and IUIM domains are fused into a single relatively rigid unit (Fig. 1c, d). Based on sequence analysis, the A20 ZnF has nominal boundaries from residues 13 to 47, with the IUIM comprising the remainder. The structure shows that residues 36–47 of the A20 ZnF, together with the entire IUIM, form a single contiguous helix. The non-helical N-terminal part of the A20 ZnF contains three of the zinc ligands: Cys19, Cys23, and Cys35. The fourth zinc ligand, Cys38, is located in the first turn of the helix (Fig. 1e). The zinc thus bridges the N-terminal non-helical part of the A20 ZnF with the helix, rigidly connecting the two. Trp31 of the non-helical part of the A20 ZnF stacks against Trp39 of the helix, further stabilizing the orientation of the helix with respect to the A20 ZnF (Fig. 1f).
The A20 ZnF has a striking hydrophobic patch on its surface, centered on a pair of highly solvent-exposed Tyr residues, Tyr25 and Tyr26 (Fig. 1f). Leu17 and Leu18 are contiguous with one side of the Tyr pair. The stacked aromatic residues Trp31, Trp39, and Tyr43 are contiguous with the other side, forming a hydrophobic strip of roughly 25 Å along its longest dimension. The hydrophobic strip is broken by a polar patch on the helix from His44 to Gln50. The surface of the C-terminal portion of the IUIM helix presents a second hydrophobic strip, consisting of the exposed residues Ile51, Trp55, Leu57, Ala58, Leu61 and Phe69 (Fig. 1f and g). This IUIM hydrophobic strip is flanked by the acidic residues Glu53, Asp54, Glu56, Glu59, Glu64, Glu65, Glu66, and Glu67.
Structure of the Rabex-5:ubiquitin complex
Ubiquitin and the Rabex-5 (9–73) fragment are present at a 1:1 stoichiometry in both crystal forms. The lattice in both crystal forms is packed such that the two molecules interact at two distinct interfaces. In the first interface, ubiquitin binds to Rabex-5 through extensive hydrophobic and polar interactions with the Rabex-5 IUIM. Ubiquitin binds through its surface hydrophobic patch centered on Ile44, Leu8, and Val70 (Fig. 2a). The IUIM binding site for ubiquitin is centered on Ile57 and Ala58, and includes all of the hydrophobic strip residues on the IUIM (Fig. 2b). The hydrophobic interactions are supplemented by hydrogen bonds between the side-chain of Rabex-5 Arg47 and the main chain carbonyl of ubiquitin Lys63 (Fig. 2c); the side-chain of Rabex-5 Asp54 and the main chain amides of ubiquitin Ala46 and Gly47; and a salt bridge between Rabex-5 Glu65 and ubiquitin Arg42 (Fig. 2d). Two potential sites of ubiquitin chain extension, Lys48 and Lys63, are near the interface (7 to 9 Å from Lys Nζ to nearest Rabex-5 atom; Fig. 2a). Neither side-chain participates directly in the interface, and both are solvent accessible in the complex. The shape complementarity score 25 is 0.76, which is considered highly complementary. The complex buries 780 Å2 and 710 Å2 of solvent accessible surface area, respectively, for Rabex-5 and ubiquitin.
Fig. 2. Ubiquitin recognition by Rabex-5.

(a) Ribbon and ball-and-stick representation of ubiquitin (orange) shown bound to a surface model of Rabex-5 IUIM. (b) Ribbon and ball-and-stick representation of Rabex-5 IUIM bound to a surface model of ubiquitin. (c) Hydrogen bonds near the N-terminus of the IUIM. (d) Hydrogen bonds near the C-terminus of the IUIM. (e) Ribbon and ball-and-stick representation of ubiquitin (marine) shown bound to a surface model of Rabex-5 A20 ZnF domain (magenta). (f) Ribbon and ball-and-stick representation of Rabex-5 A20 ZnF domain (magenta) bound to a surface model of ubiquitin (marine). (g) The ubiquitin Asp58 hydrogen bonds with Rabex-5 A20 ZnF Ser36. (h) Ubiquitin binds to Rabex-5 through two different non-overlapping surfaces on ubiquitin. Surfaces contacting IUIM are colored orange, and those contacting the A20 ZnF are colored marine.
The A20 ZnF of Rabex-5 forms a second interface with a molecule of ubiquitin that is distinct from the one bound to the IUIM (Fig. 2e, f, h). At the closest point of approach, these two ubiquitin molecules are 20 Å apart. On the A20 ZnF, the two hyperexposed Tyr residues 25 and 26 at the center of the hydrophobic patch are the main locus of the interaction. Several other residues of the A20 domain also interact: Asn28, Trp31, Ser36, and Lys37 (Fig. 2f). The interaction site on ubiquitin consists of the polar residues Arg54, Thr55, Ser57, Asp58, Tyr59, and Asn60 (Fig. 2e). Aliphatic and aromatic carbon atoms on these side-chains interact with the di-Tyr motif on the A20 ZnF domain. Rabex-5 Ser36 forms two hydrogen bonds with ubiquitin Asp58 (Fig. 2g), one of which is a short-range and presumably strong interaction between the two side-chains. The shape complementarity score is 0.74, nearly identical to the high score obtained for the ubiquitin/IUIM interaction. Roughly 420 Å2 of solvent accessible surface area is buried on each molecule.
Rabex-5 contains an inverted UIM
The IUIM of Rabex-5 binds to the Ile44 patch on ubiquitin in a manner very akin to the binding of the Vps27 UIM-1 26. UIMs are ~25 residue, single helix motifs that were first discovered through bioinformatics analysis of the sequence of the polyubiquitin binding site in the proteasome subunit S5a 27. UIMs typically bind monoubiquitin with 200 – 2000 μM affinity 28. UIMs within the endocytic proteins epsin, eps15, eps15R, and Vps27 29–32 both bind to monoubiquitin and promote the monoubiquitination of the proteins that contain them. This monoubiquitination requires an E3 ligase, as UIMs are not themselves E3 ligases.
The Rabex-5 and Vps27 UIM-1 (pdb entry 1Q0W) complexes were compared by superimposing the ubiquitin molecules, which differ by 1.0 Å r.m.s.d. over the first 73 Cα positions (Fig. 3a). Vps27 residues 257–275 are helical and in contact with the same surface on ubiquitin as Rabex-5 residues 49–67. The conserved central Ala266 of the Vps27 UIM-1 corresponds to the central Ala58 of the Rabex-5 IUIM (Fig. 3b-d). There is a one-to-one correspondence between ubiquitin-interacting residues in the C-terminal two-thirds of the Rabex-5 IUIM and the N-terminal two-thirds of the Vps27 UIM-1. The close relationship breaks down only for the N-terminal five residues of Rabex-5. The C-terminal acidic cluster of Rabex-5 matches the N-terminal acidic cluster of Vps27 UIM-1, for example. Asp-54 of Rabex-5 makes interactions with the main chain of ubiquitin residues 46 and 47 much like those reported for its counterpart in Vps27, the conserved Ser270 26. For this reason, we designated the Rabex-5 ubiquitin binding helix as an “inverted ubiquitin interacting motif” (IUIM).
Fig. 3. Conservation of IUIMs and relationship to UIMs.
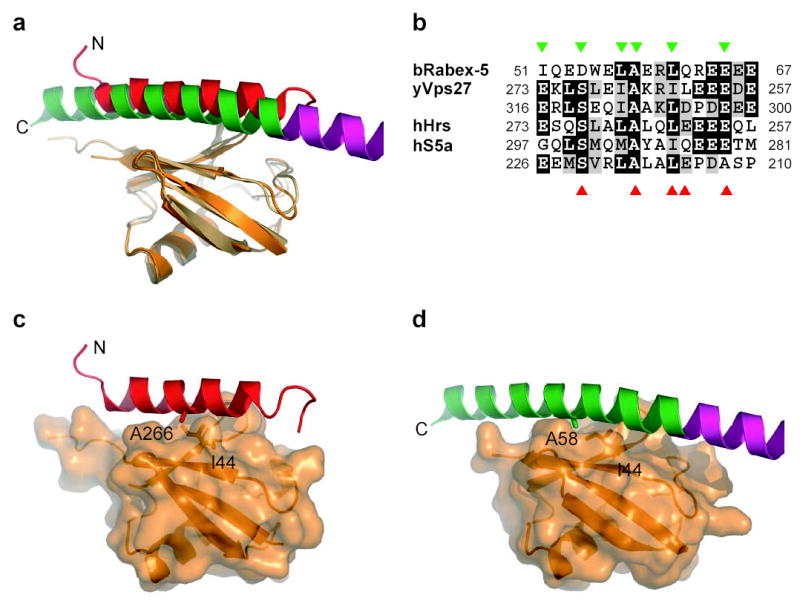
(a) Superposition of Rabex-5 IUIM and Vps27 UIM (PDB ID 1Q0W) based on overlay of ubiquitin molecules. (b) Sequence of Rabex-5 IUIM aligned with secondary structure elements and annotated by function. Sequences of UIMs of yeast Vps27, human Hrs (Vps27 ortholog), and human S5a are shown below ordered from C terminal to N-terminal. Residues contacting ubquitin are marked with orange (IUIM) and marine (Vps27 UIM-1) triangles, respectively. (c) Vps27 UIM (ribbon) bound to ubiquitin (translucent surface with underlying ribbon). (d) Rabex-5 IUIM (ribbon) in the same orientation as panel (b) for comparison.
A template for the A20 ZnF family
The A20 ZnF is a compact unit built around two pairs of Cys residues and a single zinc ion. The coordination of the zinc by four Cys (Fig. 1e), and the helical conformation of the last Cys, fit the pattern of the classic CCCC finger 33. This class of zinc finger is found in the GATA, LIM, and PHD domain-containing DNA-binding proteins, C1 and FYVE domain-containing peripheral membrane proteins, and represents the first zinc site in the RING domain E3 ligases 33. The A20 ZnF is unique in that the first two Cys are separated by three residues, as compared to two residues in other CCCC fingers. The hyperexposed Tyr-Tyr pair on the Rabex-5 A20 ZnF is its most prominent surface feature. These two aromatic residues are often Phe or Tyr in the sequences of other A20 ZnF domains (Fig. 4a,b). The Tyr-Tyr pair is surrounded by other conserved residues (Fig. 4b). The A20 ZnF can be overlaid on the Cbl RING structure based on the similarity between the A20 zinc site and the first zinc site in the RING domain. The E2 binding site on the Cbl RING domain is known from the crystal structure of its complex with UbcH7 24. This site overlays a non-conserved polar surface on A20 that is distal to the Tyr-Tyr patch. This is consistent with the classification of the A20 ZnF E3 ligases as a separate group from the RING E3 ligases. Another zinc finger domain, the NZF domain, also binds ubiquitin. The NZF domain contains no α-helix and binds ubiquitin through its Ile44 patch34, and thus has little in common with the A20 zinc finger.
Fig. 4. Conservation of A20 ZnF domains.

(a) Structure-based sequence alignment of A20 zinc fingers, labeled b, bovine; m, murine; h, human. Conserved Cys residues are indicated as black triangles at bottom, and ubiquitin-binding residues are indicated with blue triangles at bottom. Sequence numbering is for bovine Rabex-5. (b) The surface of the Rabex-5 A20 ZnF colored by conservation. Ubiquitin-binding residues are labeled marine.
Mutational analysis of ubiquitin binding sites
In order to evaluate the energetic contribution of the A20 ZnF and IUIM ubiqiutin interaction sites observed in the structure to ubiquitin binding, these regions were mutagenized alone and in combination. The effects on binding were measured in two ways. Isothermal titration calorimetry (ITC) was used to determine the effect on binding to ubiquitin in free solution (Fig. 5a). Surface plasmon resonance (SPR) was used to determine the effect on binding to ubiquitin immobilized on a planar surface (Fig. 5b). Wild-type Rabex-5 (9–73) binds to ubiquitin in solution with an apparent affinity of KD = 12 μM when the ITC data are fit assuming there is only one type of binding site. These data fits had relatively high residuals, suggesting that the single site model was inappropriate. It was not possible to obtain consistent and statistically significant improvement by fitting the ITC data to a two-site model, so we pursued a mutational approach to estimating the ubiquitin affinities of the two sites. The mutations L57D and A58D in the IUIM reduced affinity to KD = 22 and 29 μM, respectively. The mutation Y26A reduced affinity to KD = 28 μM. The double mutants Y25A/A58D completely abolished binding at achievable concentrations. From these results we infer that the most deleterious point mutants in each site are capable of completely abrogating binding to that site. We infer that the A20 site has an affinity of ~ 29 μM and the IUIM site has an affinity of ~ 28 μM in free solution.
Fig. 5. The ubiquitin:Rabex-5 interaction in vitro.

(a) ITC analysis of the binding of ubiquitin to the Rabex-5 (9–73) constructs in vitro. The inset shows the raw heat change elicited by successive injections of ubiquitin into a solution of Rabex-5 (9–73) wild type. The main Fig. depicts the normalized integration data (kcal/mol of ubiquitin) as a function of the molar ratio of ubiquitin to the various Rabex-5 (9–73) constructs. The results shown are representative of four independent experiments carried out with the wild-type construct and two independent experiments carried out with each individual mutant. (b) Surface plasmon resonance sensorgrams for selected Rabex-5(9–73) mutants with GST-ubiquitin wild-type immobilized. (c-e) Histograms showing relative affinities of Rabex-5(9–73) mutants with GST-ubiquitin wild-type (c), I44D (d) and D58A (e). KA values were obtained by taking inverse value of KD values from Table 2 and normalized so that KA for Rabex-5(9–73) wild-type was set to 100. A two-site model was used to fit the data for GST-ubiquitin wild-type (c) and one-site model for GST-ubiquitin I44D (d) and D58A (e). Values greater than 100 in panels c-e probably reflect the limitations of the simple one and two site models used to fit the data, rather than real increases in affinity.
Wild-type Rabex-5 (9–73) binds to immobilized ubiquitin with apparent affinities of KD = 1.3 μM and 37 μM when the SPR data are fit with a two site model (Fig. 5c, Table 2). GST-ubiquitin was immobilized to the sensor chip. Since GST is a dimer, two ubiquitin molecules are expected to be presented to the Rabex-5 molecule on average. The higher apparent affinity of wild-type Rabex-5 (9–73) for dimeric immobilized ubiquitin as opposed to ubiquitin in solution suggests that avidity plays a role in the first case. The most deleterious mutant tested in the A20 ZnF domain, Y25A, had KD = 29 μM, while the most deleterious mutant in the IUIM, A58D, had KD = 22 μM. The double mutant Y25A/A58D had essentially no detectable binding. The SPR data thus suggest that the A20 ZnF and IUIM sites have KD values of ~22 and 29 μM, respectively. The agreement between the ITC and SPR results for the two sites is remarkably good in view of the differences in the presentation of the molecules in the two experiments.
Table 2.
Binding affinities of Rabex-5(9–73) for ubiquitin
| SPRa |
ITCb | ||||
|---|---|---|---|---|---|
| GST-Ubiquitin WT | GST-Ubiquitin I44D | GST-Ubiquitin D58A | Ubiquitin WT | ||
| Rabex-5(9–73) | Kd(1) (μM) | Kd(2) (μM) | Kd(app)c (μM) | Kd(app)c (μM) | Kd(app)c (μM) |
| WT | 1.3 ± 0.2 | 37 ± 6.7 | 23 ± 0.8 | 28 ± 3.3 | 12 ± 1 |
| L18D | 1.4 ± 0.1 | 52 ± 8.1 | 49 ± 1.3 | 17 ± 3.3 | |
| Y25A | 29 ± 4.8 | N.A.d | N.D.e | 26 ± 2.8 | |
| Y25P | 26 ± 3.1 | N.A.d | N.D.e | 24 ± 4.7 | |
| Y26A | 30 ± 1.5 | N.A.d | N.D.e | 20 ± 11 | 29 ± 1 |
| Y26P | 24 ± 1.4 | N.A.d | N.D.e | 21 ± 1.9 | |
| P29D | 1.8 ± 0.1 | 66 ± 9.3 | 42 ± 2.8 | 30 ± 2.8 | |
| Y43A | 1.6 ± 0.1 | 56 ± 10 | 8.8 ± 0.3 | 27 ± 2.8 | 14 |
| R47A | 1.8 ± 0.05 | 42 ± 2.5 | 7 ± 0.4 | 21 ± 8.9 | |
| I51D | 7.1 ± 1.5 | 79 ± 12 | 35 ± 1.2 | N.D.e | |
| D54A | 4.3 ± 0.6 | 25 ± 5.8 | 11 ± 6.4 | 83 ± 8.9 | 18 ± 1 |
| L57D | 15 ± 0.3 | N.A.d | 14 ± 0.4 | N.D.d | 22 ± 3 |
| A58D | 22 ± 0.4 | N.A.d | 20 ± 0.4 | N.D.d | 21 ± 1 |
| L61D | 4.4 ± 0.5 | 43 ± 9.1 | 8.1 ± 0.3 | N.D.d | 15 |
| D54A | 19 ± 1 | ||||
| E65A | 2.1 ± 0.1 | 50 ± 12 | 7.7 ± 0.2 | 53 ± 1.3 | 29 ± 1 |
| Y25A/Y26A | 36 ± 3 | N.A.d | N.D.e | 42 ± 5.3 | |
| Y25A/L57D | N.D.e | N.D.e | N.D.e | N.D.e | |
| Y25A/A58D | N.D.e | N.D.e | N.D.e | N.D.e | N.D.e |
| Y26A/L57D | N.D.e | N.D.e | N.D.e | N.D.e | |
For SPR experiments, human ubiquitin wild-type (WT), I44D and D58A as glutathione S-transferase (GST) fusion were immobilized on a CM5 chip via covalent linkage to the N-terminus of GST. Rabex-5(9–73) wild-type (WT) and mutants were in the mobile phase. Standard deviations are calculated from three independent experiments.
For ITC experiments, bovine ubiquitin (Sigma) was titrated to the Rabex-5(9–73) wild-type (WT) and mutants. Results shown represent the means and standard deviations of four independent experiments for wild-type and two independent experiments for mutants, except for Y43A and L61D, which were measured once.
Apparent dissociation constant calculated from fitting the data to one-site model.
N.A, binding appears to be most appropriately fit by a single site model, so no Kd(2) was determined.
N.D., either binding was not detectable or the Kd not determined because binding is too weak to obtain reliable quantitation.
In order to determine which interfaces of ubiquitin were involved in functional interactions, the ubiquitin mutants I44D and D58A were generated. The binding of these mutants to wild-type and mutant Rabex-5 constructs was assessed to determine which interfaces were functional, an approach known as “double mutant cycle” analysis. The I44D mutant was made to disrupt the canonical hydrophobic patch on the surface of ubiquitin that interacts structurally with the IUIM. The ubiquitin D58A mutant was designed to disrupt the two hydrogen bonds between ubiquitin Asp58 and the Rabex-5 Ser36 side-chain and main-chain. Ubiquitin I44D binds to wild-type Rabex-5 and its IUIM mutant A58D with very similar affinity (KD = 20–23 μM; Fig. 5d; Table 2). This is consistent with the expectation that either mutant should completely abrogate the Rabex-5 IUIM/ubiquitin Ile44 patch interaction, yet have no effect on the A20 ZnF/ubiquitin Asp58 patch interaction. Ubiquitin I44D binds with sharply reduced affinity to Rabex-5 A20 ZnF domain mutant Y25A, and not at all to Y25A/Y26A. This confirms that the residual interaction of I44D with Rabex-5 requires an intact A20 ZnF domain binding site. Ubiquitin D58A binds to wild-type Rabex-5 and its A20 ZnF domain mutant Y25A with very similar affinity (KD = 26–28 μM; Fig. 5e; Table 2), consistent with the structural finding that these two regions directly interact. In contrast, mutating the Rabex-5 IUIM (A58D) almost completely blocks binding. Thus the residual Rabex-5 binding of the D58A mutant occurs entirely through the IUIM. The double mutant cycle results show that both of the ubiquitin:Rabex-5 interfaces seen in the crystal structure are functional.
Mechanism for E3 ligase activity
In order to probe the mechanism for ubiquitin E3 ligase activity by Rabex-5, we tested the ability of Rabex-5 (9–73) and its A20 ZnF domain mutant Y25A/Y26A to recruit ubiquitin conjugating enzymes (Ubcs; Fig. 6a). Both ubiquitin loaded and unloaded UbcH5 and Ubc7 were generated and tested. To assess specificity, Rabex-5 was compared to a known Ubc7-binding protein, gp78 35 and a known UbcH5-binding E3, RNF25 36. Ubiquitin-loaded UbcH5 binds to wild-type Rabex-5 (9–73), but not to the A20 ZnF domain Y25A/Y26A mutant nor to gp78 (Fig. 6a). Unloaded UbcH5 does not bind to Rabex-5 (9–73). Ubc7 binds to gp78 with roughly equal affinity in both its ubiquitin-loaded and unloaded states (Fig. 6a). Thus Rabex-5 specifically recruits at least one Ubc, UbcH5, in a manner that depends on the interaction between ubiquitin and the A20 ZnF domain. The presence of ubiquitin alone is insufficient for recruitment, at least at concentrations tested, since ubiquitin-loaded Ubc7 is not recruited. The E3 ligase activity with respect to GST-Rabex-5 (9–73) was tested for wild-type and mutant constructs. The A20 ZnF domain Tyr mutants tested blocked catalytic activity, consistent with the requirement for E2 binding (Fig. 6b). The IUIM mutant A58D did not block activity, but rather enhanced activity several-fold.
Fig. 6. E2 recruitment and E3 ligase activity of Rabex-5 and its mutants.

(a) Detection of [35S]-labeled E2 enzymes shows that ubiquitin-loaded UbcH5 selectively binds to Rabex-5 with an intact A20 ZnF domain, but not to the Y25A/Y26A Rabex-5 mutant. The E2 enzyme Ubc7 binds to the Ubc-binding domain of gp78 independent of its ubiquitin loading, and does not bind to Rabex-5. (b) Wild-type Rabex-5 has ubiquitin E3 ligase activity as indicated by detection of Flag-ubiquitin. Tyr mutations in the A20 ZnF domain block this activity, while the A58D IUIM mutant enhances activity. The arrow marks the position corresponding to the molecular weight of unmodified GST-Rabex-5 (9–73).
DISCUSSION
The structure of the Rabex-5 IUIM is a striking illustration of convergent evolution. Since there is no known genetic mechanism for inverting the orientation of a protein sequence, the Rabex-5 IUIM must have evolved independently of conventional UIMs. The GATA-like CCCC zinc fingers, of which the A20 ZnF is a subtype, all contain a C-terminal α-helix. The extension of this helix for Rabex-5 seems a likely mechanism for the origin of the IUIM. It is remarkable that the use of particular amino acid side-chains to interact with particular ubiquitin residues is so well preserved. Key Rabex-5 IUIM Ala and Leu residues have Ala and Leu/Ile counterparts in conventional UIMs that make nearly identical interactions. The cluster of acidic residues at the C-terminus of the IUIM is matched by a similar cluster at the N-terminus of the UIM. The conserved Ser of the conventional UIM is replaced by an Asp in the IUIM, in one of the few differences. However, the Ser and the Asp both interact with the same main-chain NH groups on ubiquitin. It is equally striking that the orientation of the helix, apart from its opposite N to C direction, with respect to the ubiquitin surface is so similar. This suggests that there may be very few ways for a single α-helix to bind to ubiquitin with significant affinity.
Most monoubiquitin binding domains bind free ubiquitin with KD values ranging from 100–500 μM 18. By the standards of monoubiquitin binding domains, the Rabex-5IUIM is an unusually strong binder with its apparent KD of ~ 29 μM. The Rabex-5 IUIM joins the Vps9 CUE domain 19,37 as a “champion” monoubiquitin binder. It is probably no coincidence that the two of the most potent monoubiquitin binding proteins described are each other’s orthologs. What is more surprising is that Rabex-5 and Vps9, despite their similar catalytic domains and biological functions, have evolved completely different high affinity ubiquitin binding domains located in completely different parts of their sequences. How does the Rabex-5 IUIM achieve such high affinity binding as compared to conventional UIMs and most other ubiquitin binding domains? The surface area buried in the ubiquitin:IUIM interface, over 700 Å2 for each protein, is comparable to that buried in the high affinity Vps9 CUE:ubiquitin interface, and larger than that of most other structurally characterized ubiquitin:domain interfaces. As compared to the conventional UIM, the IUIM interacts via one additional turn of helix that forms hydrogen bonds with the Lys63 region of ubiquitin. These interactions could plausibly contribute 2 kcal/mol and thereby account for the ~10-fold gain in affinity as compared to the Vps27 UIM-1.
Monoubiquitin-binding domains in many proteins, including eps15, epsin, Vps27, Vps9, and Rabex-5, are required for the covalent monoubiquitination of these same proteins. These domains are not themselves ubiquitin ligases. The mechanism by which monoubiquitin binding domains promote ubiquitination is unknown, although several models have been proposed 18. In one popular model 29, the UIM directly recruits a covalent HECT domain E3 ligase-ubiquitin thiolester adduct by binding to the ubiquitin moiety. In other models, the ubiquitin binding domain has a less direct role. In these indirect models, the ubiquitin binding domain interacts with other ubiquitinated factors important to the reaction and recruits and/or allosterically activates these factors. In a variant of the indirect model, the ubiquitin binding domain is involved in allosterically regulating the structure of the protein that contains it such that substrate Lys residues are exposed.
The Rabex-5 A20 ZnF and IUIM system sheds new light on this question as the first example of a crystal structure in which a monoubiquitin binding domain, an E3 ligase, and ubiquitin are present. The A20 ZnF domain E3 ligase presumably functions like the RING E3 ligase as an adaptor and activator of the E2 ubiquitin conjugating enzyme. Thus we can ask if it is plausible for the IUIM and the A20 ZnF to cooperate in recruiting a ubiquitin-thiolester adduct of an E2. In this scenario, the IUIM would bind to the ubiquitin moiety while the A20 ZnF domain would bind to the E2. The structure shows that the C-terminus (with the last three residues 74–76 modeled) of the ubiquitin bound to the IUIM is 50 Å away from the E2 binding site on the A20 ZnF domain. In the structure of the Cbl/UbcH7 complex 24, there is only a 15 Å distance between the active site Cys86 of UbcH7 and the closest point on the RING domain, and the farthest point of the UbcH7 surface from Cys86 is only 27 Å. The solution of the Rabex-5 A20 ZnF –IUIM domains in four crystallographically independent states represents the modest range of flexibility between the two domains, and the range of movement is insufficient to bring the IUIM ubiquitin binding site into proximity with the putative E2 binding site. This is consistent with the observation that the A58D IUIM mutant does not impair E3 ligase activity (Mattera, Tsai, Weissman, and Bonifacino, submitted, and Fig. 6b). The observation that Rabex-5 A58D enhances ligase activity suggests a possible role for the IUIM in limiting ubiquitin chain extension, a possibility that will be important to resolve in further analysis. Therefore simple distance constraints appear to rule out a direct E2 monoubiquitin thiolester recruitment model for the IUIM of Rabex-5.
This study has shed considerable light on the little characterized mechanism of the A20 ZnF domain E3 ligase. We have found that the A20 ZnF domain binds strongly to a novel region, the Asp58 patch, on the surface of ubiquitin. It has long been anticipated that regions of the ubiquitin surface other than Ile44 would have important roles in ubiquitin function. This is the first demonstration of a ubiquitin binding domain that does not interact with the Ile44 region. The A20 ZnF domain uses the interaction between its diaromatic patch and the Asp58 patch on ubiquitin to recruit a ubiquitin-loaded Ubc. The diaromatic patch is conserved in most A20 ZnF domains, including ZnF-4 of A20 itself, which has been implicated in the ligase activity of that protein. There is a strong preference for some Ubcs over others, so the recruitment is not a function of ubiquitin binding alone. The ubiquitin:A20 ZnF complex exposes considerable hydrophobic surface area to solution, including one side each of the side-chains of the diaromatic patch. A model of a possible A20 ZnF:ubiquitin:E2 complex was generated by overlaying the SUMO molecule in the Ubc9-SUMO-RanGAP1-Nup358 complex38 onto the A20-ZnF domain-bound ubiquitin molecule (Supplementary Fig. 1). The Ubc in this model is 20 Å away from the closest point of contact with the A20 ZnF domain. However, the exposed faces of Tyr25 and Tyr26 are the closest point to the docked Ubc, and there is no apparent obstacle to a rotation of the Ubc to directly contact the A20 ZnF domain.
Supplementary Figure 1.
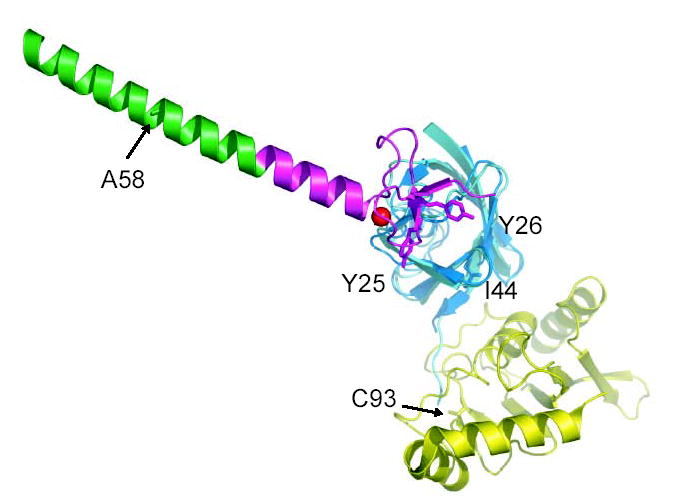
A speculative model for ubiquitin-loaded E2 binding to Rabex-5(9–73).
Ubiquitin bound to ZnF of Rabex-5(9–73) was superimposed to SUMO-1 of the SUMO-RanGAP1-Ubc9-Nup358 complex structure (PDB ID: 1Z5S). Green, IUIM; magenta, ZnF; red, zinc ion; marine, ubiquitin bound to ZnF; cyan, SUMO-1; yellow, Ubc9. Some key residues including Y25 and Y26 on ZnF, I44 on ubiquitin and the active site residue C93 on Ubc9 are labeled. Note that the C-terminus of SUMO-1 is in proximity to C93 of Ubc9.
In summary, this study has revealed for the first time the structures of two motifs involved in ubiquitination and monoubiquitin recognition: the A20 ZnF E3 ligase, and the IUIM. The juxtaposition of these two domains in space explains how they carry out their individual functions autonomously at the level of the isolated domains. The A20 ZnF and IUIM, while rigidly linked, are too far apart to directly cooperate in the transfer of a single monoubiquitin moiety. The mechanism by which monoubiquitin binding domains promote the monoubiquitination of proteins that contain them remains a major question in the field. We are one step closer to answering this question here by ruling out one of the major potential mechanisms, direct E2 ubiquitin thiolester recruitment, in the case of the Rabex-5 IUIM. Conversely, we have shown that the A20 ZnF domain immediately adjacent to the IUIM functions by just such a direct recruitment mechanism. Conservation of the A20 ZnF sequence in other proteins suggests that A20 and other related proteins will function by a similar mechanism.
METHODS
Cloning and sample preparation
DNA coding for the fragment of bovine Rabex-5 spanning residues 9–73 was cloned into parallel GST2 vector 39 using a PCR-based cloning strategy. Site-directed single and double mutants were generated using QuikChange Mutagenesis Kit (Stratagene). All the constructs were verified by DNA sequencing. Rabex-5(9–73) was overexpressed as an N-terminal glutathione S-transferase (GST) fusion protein in Escherichia coli strain BL21(DE3). ZnCl2 was added to terrific broth media at final concentration of 0.1 mM. Cells were grown at 37°C, induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside at optical density of 1.0 and further incubated at 20°C overnight. Harvested cells were resuspended in buffer A (50 mM TrisHCl, pH 7.4, 150 mM NaCl), lysed and centrifuged. Supernatant was applied to a glutathione-Sepharose column (Amersham) in buffer A and washed with buffer B (50 mM TrisHCl, pH 7.4, 500 mM NaCl). Tobacco etch virus protease was added to cleave GST and removed by passing through TALON resin (BD Bioscience) in the buffer A. The cleaved protein was concentrated and applied to a Superdex 200 column (Amersham) in the buffer C (50 mM TrisHCl, pH 7.4, 150 mM NaCl, 5 mM dithiothreitol). To form a complex with ubiquitin, bovine ubiquitin (Sigma) was added to the purified rabex-5(9–73) and the resulting complex was separated using a Superdex 200 column in the buffer C. The pooled complex solution was concentrated using a VivaSpin concentrator (Viva Science). All mutants were purified as described above except that no reducing agent was used in buffers. DNA coding for human ubiquitin was amplified by PCR and cloned into the pGST2 vector. Recombinant GST and GST-ubiquitin for SPR studies were produced from BL21 (DE3) cells and purified using glutathione-sepharose affinity column followed by a Superdex S200 (GE Health Sciences) gel filtration chromatography. GST and GST-ubiquitin samples were dialyzed against 10 mM HEPES (pH 7.4), 150 mM NaCl, and 0.5% surfactant P20 (HPS-P).
Crystallization, data collection and structure determination
Crystals of the complex were grown by hanging drop method at 22°C. The reservoir solution contained 0.1 M sodium citrate, pH 5.6, 20–25% polyethylene glycol 4000, and 0.2 M lithium sulfate. Diffraction-quality crystals were obtained by streak seeding. Crystals appeared in 1–2 days and were flash frozen under liquid nitrogen. Data were collected using a Riguaku rotating anode home source and an R-AxisIV detector at 100 K and processed with HKL2000 (HKL Research). An initial molecular-replacement (MR) solution was obtained using ubiquitin as a search model using MOLREP 40 in space group P61. Solvent flattening by the prime-and-switch algorithm in RESOLVE 41 yielded an electron density map sufficient to locate the missing Rabex-5(9–73). Single anomalous dispersion (SAD) at the Zn edge, λ = 1.28 Å, was used to re-determine the structure of the complex de novo using SOLVE/RESOLVE 42. The structure by SAD confirmed that the model by MR was correct. The structure in space group C2 was determined by molecular-replacement using the refined model in P61. Iterative manual model building and initial refinement were performed using O 43 and CNS 44, respectively. Final refinement was done using REFMAC5 45 with TLS parameters 46 incorporated. Statistics for data processing and refinement are listed in Table 1. All the structural figures were generated using PyMol (www.pymol.org).
Table 1.
Crystallographic data collection, phasing, and refinement statistics
| Crystal | A | B | C |
|---|---|---|---|
| Data collection | |||
| X-ray source | CuK_ | NSLS X12C | NSLS X12C |
| Wavelength (Å) | 1.5418 | 1.2820 | 1.2820 |
| Space group | P61 | P61 | C2 |
| Unit cell | a = b = 81.70 Å | a = b = 81.97 Å | a = 192.92 Å |
| c= 55.03 Å | c = 54.44 Å | b = 44.22 Å | |
| c = 69.17 Å | |||
| α = β = 90° | α = β= 90° | α = γ = 90° | |
| γ = 120° | γ = 120° | γ = 108.98° | |
| Complexes in the asymmetric unit | 1 | 1 | 3 |
| Resolution (Å) | 3.0 | 2.8 | 2.5 |
| Number of unique reflections | 4509 | 5005 | 19257 |
| I/σ | 27.6 (8.4) | 33.8 (3.8) | 24.2 (3.3) |
| Rsym a | 0.109 (0.423) | 0.073 (0.431) | 0.067 (0.439) |
| Data completeness (%) | 99.8 (100.0) | 95.7 (77.1) | 98.8 (95.6) |
| Phasing and Refinement | |||
| Mean F.O.M.(50 – 3.1 Å) | 0.36 (SOLVE) | ||
| Overall F.O.M. (50 – 3.0 Å) | 0.58 (RESOLVE) | 0.68 (RESOLVE) | |
| Solvent content for RESOLVE | 0.63 | 0.63 | |
| R b / Rfree c | 0.212 / 0.268 | 0.228 / 0.263 | |
| R.m.s. bond length (Å) | 0.007 | 0.007 | |
| R.m.s. bond angle (°) | 1.0 | 1.0 | |
| Average B value (Å2) d | 59.4 | 48.2 | |
| Wilson B value (Å2) | 72.9 | 59.4 | |
| Residues in most favored φ,ψ region (%) | 91.5 | 93.0 | |
The values in parentheses relate to highest resolution shells.
F.O.M., figure of merit.
Rsym = _h _i |Ii(h) - <I>| / _h_i Ii(h), where I is the observed intensity and <I> is the average intensity of multiple observations of symmetry-related reflections.
R = _||Fo| - k|Fc|| / _ |Fo|, where |Fo| and |Fc| are observed and calculated structure factor amplitude, respectively.
Rfree is calculated for a randomly chosen 10% (Crystal B) or 5% (Crystal C) of relections; the R factor is calculated for the remaining 90% (Crystal B) or 95% (Crystal C) of relections.
Average B value of all atoms in the asymmetric unit.
Surface plasmon resonance
The binding of wild-type and mutant Rabex-5 proteins to ubiquitin was monitored using SPR with a BIAcore T100 (BIAcore AB, Sweden), at 250C, with a flow rate of 20 ml min−1. Approximately 10,000 RU of GST, GST-ubiquitin, and GST-I44D and D58A ubiquitin mutants were immobilized on a CM5 surface that was stable for more than 60 experimental cycles. Briefly, a CM5 chip was activated using NHS/EDC (1:1) at a flow rate of 5 ml min−1 for 1200 s. GST (10 μM) and GST-Ubiquitin (10 μM) in 10 mM Acetate buffer (pH 5.5) was passed over separate flow cells at 5 μl min−1 for 2400 s, followed by a blocking step using ethanolamine (1M pH 8.5) at 5 μl min−1 for 1200 s. All binding experiments were performed in HPS-P. Binding of wild-type and mutant Rabex-5 proteins to ubiquitin was measured simultaneously by passing Rabex-5 over GST-, GST-ubiquitin, and GST-ubiquitin mutant- coupled flow cells with association and dissociation times of 100 s and 300 s, respectively. Between subsequent injections of Rabex-5 proteins, surfaces were regenerated with an injection of HPS-P supplemented with 500 mM NaCl for 15 s at 100 ml min−1.
We initially tried to fit our data assuming a 1:1 binding stoichiometry for a Rabex:Ubiquitin complex. However, poor fitting statistics and subsequent mutational data led us to define 2 independent binding sites on Rabex-5 for ubiquitin as follows:
Where R is Rabex-5 and U is ubiquitin. Corresponding data were fit to the following equation:
Where [Rabex] is the protein concentration of the flowing analyte, KD1 and KD2 are the dissociation constants for site 1 and site 2, respectively, Rmax1 and Rmax2 are the relative maximal change in response levels for site 1 and site 2, respectively, and RI is the residual or background signal. Fitting was performed using the BiaEvalution software, with globally floating KD, Rmax, and RI values.
Isothermal titration calorimetry
The Rabex-5 (9–73) fragments were dissolved in 50 mM Tris/HCl pH 7.4, 150 mM NaCl. The same buffer was used to prepare 0.3–0.75 mM solutions of bovine ubiquitin (Sigma), which was the injectant. Rabex-5 (9–73) was placed in the sample cell at 10–30 μM. The concentrations of the ubiquitin and Rabex-5 (9–73) were calculated by measuring absorbance at 280 nm and extinction coefficients of 1814 M−1 for ubiquitin and 21,175 M−1, 19,892 M−1, 19,892 M−1, 21,193 M−1, 21,141 M−1, 21,180 M−1, 21,127 M−1, 21,180 M−1, 21,185 M−1 and 19,759 M−1 for Rabex-5 (9–73) wild type and the Y26A, Y43A, R47A, D54A, L57D, A58D, L61D, E65A, Y25A, A58D mutants. Titrations (42 injections of 5 μl each) were performed at 30°C using a VP-ITC Microcalorimeter (MicroCal, LLC), and data were analyzed using Origin software (Origin Lab).
Interaction with E2 enzymes
UbcH5C was expressed using the plasmid pET15-UbcH5C 47. DNA coding for MmUbc7 was subcloned into the NcoI/BamHI sites of pET15. The human E3 gp78 encodes a specific MmUbc7 binding site between amino acid 574 – 643. A fragment coding for gp78 amino acid 574–643 was described previously 48, as was the GST-RNF25(86–363) construct 36. [35S]-Labeled UbcH5C and MmUbc7 were translated in vitro using the S30 T7 bacteria lysate system (Promega). For ubiquitin thioester formation, reactions contained 12 μL E2 translation mix, 100 nM murine E1 and 10 μg ubiquitin 80 μL reaction buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 4 mM ATP, 2 mM MgCl2). The reactions were carried out at 30o C for 10 min. To produce free E2, the same reaction conditions were used except that E1 was omitted. An aliquot containing 2 μg of GST fusion protein immobilized on glutathione-Sepharose 4B (Amersham) was incubated with 20 μL thioester reaction mix in 100 μL of binding buffer (PBS containing 0.2% Triton X-100 and 0.2 mM ZnCl2) for 2 h at 4°C with constant mixing. Equal loading of all samples was confirmed by SDS-PAGE and Coomassie blue staining (not shown). The immobilized proteins were collected by centrifugation and washed three times with 100 bed volumes of binding buffer. The mixture was quenched in SDS sample buffer (with or without DTT), separated by SDS-PAGE and processed for visualization on a Storm PhosphorImager (Amersham).
In vitro ubiquitination assays
Reaction mixtures contained 2 μg GST fusion proteins immobilized on glutathione-sepharose 4B (Amersham), 100nM recombinant murine E1 (expressed in Sf9 cells), 40 nM recombinant UbcH5C (expressed in E.coli), 2μMFlag-Ub, 0.5–1μM ubiquitin aldehyde in a final volume of 50μL reaction buffer (20mM Trisp H7.5, 100mM NaCl, 4mM ATP and 2mM MgCl2). Reactions were incubated at 30°C for 90m in with agitation, washed three times with 20 bed volumes of wash buffer (20mM Tris, pH7.5, 100mM NaCl, 1 mM DTT) and quenched with SDS sample buffer. Samples were subjected to SDS-PAGE and processed for immunoblot with anti-Flag antibody. The recombinant E2 enzymes were prepared as described49.
Acknowledgments
We thank Michelle Horning and Xiaolin Zhu for outstanding technical assistance, and Anand Saxena and the staff of National Synchrotron Light Source (NSLS), Brookhaven National Laboratory (BNL) beamline X12C for assistance with data collection, and Jaewon Kim for discussions. This research was supported by the intramural program of the National Insitutes of Health, NIDDK (to JHH), NCI (to AMW), and NICHD (JSB). MSK is a PRAT fellow of the NIGMS. Research carried out at the NSLS, BNL, was supported by the U.S. Department of Energy (DOE), Division of Materials Sciences and Division of Chemical Sciences, under Contract No. DE-AC02-98CH10886.
Footnotes
Coordinates. The crystallographic coordinates have been deposited in the protein data bank with accession code ____ (to be provided).
References
- 1.Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol. 2001;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 2.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 3.Pfeffer S, Aivazian D. Targeting RAB GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 4.Seabra MC, Wasmeier C. Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol. 2004;16:451–457. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Burd CG, Mustol PA, Schu PV, Emr SD. A yeast protein related to a mammalian ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol Cell Biol. 1996;16:2369–2377. doi: 10.1128/mcb.16.5.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delprato A, Merithew E, Lambright DG. Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell. 2004;118:607–617. doi: 10.1016/j.cell.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 7.McBride HM, et al. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 8.Gournier H, Stenmark H, Rybin V, Lippe R, Zerial M. Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. Embo J. 1998;17:1930–1940. doi: 10.1093/emboj/17.7.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horiuchi H, et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 10.Lippe R, Miaczynska M, Rybin V, Runge A, Zerial M. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–72. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 12.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 13.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 14.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 15.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 16.Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends BiochemSci. 2003;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- 18.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 19.Shih SC, et al. A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. Embo J. 2003;22:1273–1281. doi: 10.1093/emboj/cdg140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donaldson KM, Yin HW, Gekakis N, Supek F, Joazeiro CAP. Ubiquitin signals protein trafficking via interaction with a novel ubiquitin binding domain in the membrane fusion regulator, Vps9p. Curr Biol. 2003;13:258–262. doi: 10.1016/s0960-9822(03)00043-5. [DOI] [PubMed] [Google Scholar]
- 21.Davies BA, et al. Vps9p CUE domain ubiquitin binding is required for efficient endocytic protein traffic. J Biol Chem. 2003;278:19826–19833. doi: 10.1074/jbc.M301059200. [DOI] [PubMed] [Google Scholar]
- 22.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappa B signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 23.Huang L, et al. Structure of an E6AP-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 24.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence MC, Colman PM. Shape Complementarity at Protein-Protein Interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 26.Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. Embo J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends BiochemSci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- 28.Fisher RD, et al. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;278:28976–28984. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- 29.Polo S, et al. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 30.Shih SC, et al. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- 31.Raiborg C, et al. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 32.Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4:534–9. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- 33.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 34.Alam SL, et al. Ubiquitin interactions of NZF zinc fingers. Embo J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang SY, et al. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorick KL, et al. RING fingers mediate ubiquitin-conjugating enzymes (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prag G, et al. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–620. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- 38.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of "parallel" expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 40.Vagin AA, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 41.Terwilliger TC. Using prime-and-switch phasing to reduce model bias in molecular replacement. Acta Crystallogr Sect D-Biol Crystallogr. 2004;60:2144–2149. doi: 10.1107/S0907444904019535. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr Sect D-Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron- Density Maps and the Location of Errors in These Models. Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 44.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr Sect D-Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum- likelihood method. Acta Crystallogr Sect D-Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 46.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr Sect D-Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 47.Jensen JP, Bates PW, Yang M, Vierstra RD, Weissman AM. Identification of a family of closely related human ubiquitin conjugating enzymes. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- 48.Chen B, et al. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its CUE domain, RING fingers, and an E2-binding site. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0506618103. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lorick KL, Jensen JP, Weissman AM. Expression, purification, and properties of the Ubc4/5 family of E2 enzymes. Methods Enzymol. 2005;398:54–68. doi: 10.1016/S0076-6879(05)98006-3. [DOI] [PubMed] [Google Scholar]


