Abstract
Cbln1 and the orphan glutamate receptor GluRδ2 are pre- and postsynaptic components, respectively, of a novel transneuronal signaling pathway regulating synapse structure and function. We show here that Cbln1 is secreted from cerebellar granule cells in complex with a related protein, Cbln3. However, cbln1- and cbln3-null mice have different phenotypes and cbln1 cbln3 double-null mice have deficits identical to those of cbln1 knockout mice. The basis for these discordant phenotypes is that Cbln1 and Cbln3 reciprocally regulate each other's degradation and secretion such that cbln1-null mice lack both Cbln1 and Cbln3, whereas cbln3-null mice lack Cbln3 but have an approximately sixfold increase in Cbln1. Unlike Cbln1, Cbln3 cannot form homomeric complexes and is secreted only when bound to Cbln1. Structural modeling and mutation analysis reveal that, by constituting a steric clash that is masked upon binding Cbln1 in a “hide-and-run” mechanism of endoplasmic reticulum retention, a single arginine confers the unique properties of Cbln3.
Cbln1 is a secreted glycoprotein expressed in cerebellar granule neurons and is essential for synaptic integrity and function in this brain region (1, 9, 40). Mice lacking cbln1 are ataxic and exhibit multiple physiological and structural deficits in the synapses between granule cells and Purkinje cells (9). Although granule cell numbers are unaffected in young cbln1-null mice, Purkinje cells have numerous dendritic spines that lack presynaptic contributions from granule cells. This structural deficit is paralleled by impaired neurotransmission and a lack of cerebellar long-term depression (9). This complex phenotype is also seen in mice that lack the δ2 glutamate receptor (GluRδ2) (8, 10, 12, 15, 17), a Purkinje cell-specific protein that localizes to postsynaptic densities (18, 36, 44). An analysis of mice lacking both GluRδ2 and Cbln1 indicated a genetic interaction between the two, with GluRδ2 being the primary determinant of phenotype (9). Thus, Cbln1 secreted from presynaptic granule neurons and GluRδ2, a component of the Purkinje cell postsynapse, are components of a novel transneuronal signaling pathway regulating synapse structure and function (9).
Given the importance of mechanisms that control synaptic integrity and plasticity, we sought to identify additional components of the Cbln1 signaling pathway. Cbln1 is the prototype of a family of proteins (Cbln1 to Cbln4) that share a conserved C1q globular domain at their C termini (1, 27, 40, 41). This raises the possibility that other family members fulfill functions analogous to that of Cbln1. In addition, it raises the potential for the presynaptic component being a heteromeric complex containing multiple Cbln1 family members. The C1q motif is a protein-protein association domain that is present in a wide variety of secreted proteins and mediates the assembly of trimeric complexes (11, 13). In the case of Cbln1, two homotrimers assemble into hexamers via intermolecular disulfide bonds between conserved cysteine-containing motifs in the N termini of all family members (1). Although Cbln1 forms homomeric complexes, it also binds to Cbln3 in yeast two-hybrid assays (1, 27). As Cbln3 is coexpressed with Cbln1 in mature granule neurons (9, 27), this raises the question of whether Cbln3 can contribute to the maintenance of synaptic structure and function in cerebellum either by acting alone or by participating in heteromeric complexes with Cbln1.
To address the role of Cbln3 in synapse structure and function, we first determined whether Cbln3 existed in the same molecular complex as Cbln1. We then generated and characterized cbln3-null mice and crossed them to cbln1-null animals to obtain cbln1 cbln3 double-null mice and compared the phenotypes. Subsequently, we investigated the interrelationship between Cbln1 and Cbln3 with regard to their degradation, complex formation, and secretion. Finally, the structural basis of the functional differences between Cbln1 and Cbln3 was elucidated by mutational analysis.
MATERIALS AND METHODS
Antibodies and reagents.
Mouse monoclonal antibody to β-actin and Flag tag were from Sigma (St. Louis, MO). Mouse monoclonal anti-V5 tag was from Invitrogen (Carlsbad, CA). An antiserum to a synthetic peptide corresponding to amino acids 95 to 112 of mouse Cbln1 was generated and described previously (1) and is referred to as anti-Cbln1. Chloroquine was from Sigma. Lactacystin, MG132, and Z-Ile-Glu(OtBu)-Ala-Leucinal (ZAL) were from Calbiochem (La Jolla, CA).
Plasmids and constructs.
Standard molecular cloning and sequencing techniques were used to isolate and validate cDNA clones carrying full-length cbln1, cbln2, cbln3, and cbln4 from brain total RNA. These cDNAs were subcloned into the BamHI and XbaI sites of the pcDNA3.1 and pcDNA3.1V5-His vectors (Invitrogen). The cDNA of cbln1 was also inserted into the p3xFLAG-CMV-9 vector (Sigma). All truncation and mutation constructs were generated by standard PCR-based methods. All plasmids were purified with a Midi-Prep kit (QIAGEN, Valencia, CA) before use. Primer synthesis, DNA sequencing, and bioinformatics support were provided by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital.
Generation of cbln3−/− mice.
A cbln3 targeting vector was constructed by replacing all coding sequences, except for the first two codons of cbln3 plus the intervening sequences, with a fragment containing the LacZ coding sequence from pMC1847 and a neo cassette derived from the PGK.neo.TK plasmid. The LacZ was placed in frame with the start codon of cbln3. The left arm is a 2.28-kb XbaI-KpnI (KpnI was introduced by PCR following the second codon) fragment, and the right arm is a 5.98-kb ApaI fragment (see Fig. 2A). A thymidine kinase expression cassette was placed downstream to the right arm for negative selection. AB2.2 prime embryonic stem cells were transfected with targeting vector and screened for positive clones harboring disrupted cbln3 alleles using Southern blot analysis. The external probe for Southern blotting was a 0.57-kb XhoI-XbaI genomic DNA fragment upstream to the left arm, and the internal probe was a 1.45-kb XbaI-BamHI fragment cloned in pBlueScript. Positive clones were microinjected into blastocysts. Two chimeras underwent germ line transmission and generated founder strains.
FIG. 2.
Generation and characterization of cbln3-null mice. (A) The gene targeting strategy. The cbln3 targeting vector used to generate cbln3−/− embryonic stem cells is shown as well as the disrupted cbln3 allele. The positions of external probe A and internal probe B are shown. Restriction sites are shown as follows: A, ApaI; C, ClaI; N, NotI; X, XhoI; and Xb, XbaI. Technical details are provided in Materials and Methods. (B) DNA isolated from wild-type (WT) and heterozygous founder (cbln3+/−) mouse tails was digested with XhoI and ClaI and analyzed by Southern blotting. The wild-type and targeted alleles gave rise to bands of 13.3 kb (wild type) and 4.2 kb (targeted), respectively. (C) Total RNA isolated from cerebella of wild-type, cbln3+/−, and cbln3−/− mice was analyzed by Northern blotting using a probe for cbln3. cbln3−/− mice show loss of cbln3 mRNA, and cbln3+/− mice have intermediate levels of mRNA on Northern blots. (D) Cerebellar extracts of wild-type, cbln3+/− and cbln3−/− mice were immunoblotted with an anti-Cbln3 antiserum. Intermediate levels and complete loss of Cbln1 were evident in cbln3+/− and cbln3−/− mice, respectively. (E) Neuroanatomical analysis of cbln3−/− mice. Cresyl violet-stained sagittal sections of 1-month-old wild-type and cbln3−/− mice show no overt neuroanatomical anomalies. PEP-19 immunostaining of Purkinje cells in wild-type and cbln3−/− (lower panels) mice also revealed no overt differences in the positioning, number, and overall dendritic morphology between the two genotypes. Scale bars: upper panel, 1 mm; middle and lower panels, 50 μm. (F) Rota-rod test of cbln1−/− and cbln3−/− mice. Performance of wild-type (n = 6), cbln1−/− (n = 7), and cbln3−/− (n = 8) mice was assessed on a standardized accelerating Rota-rod. Motor performance was scored as the mean latency to fall (min) on the accelerating rod. In contrast to the low performance of cbln1−/− mice (P = 0.00006), cbln3−/− mice showed a level of performance similar to that of wild-type mice. Error bars represent standard error of the means.
The generation of cbln1−/− mice has been described previously (9). All mice were maintained at St. Jude Children's Research Hospital and had ad libitum access to food and water. All procedures involving animals were performed in accordance with the National Institutes of Health guidelines and were approved by the Animal Care and Use Committee of St. Jude Children's Research Hospital.
Production of polyclonal anti-Cbln3 antiserum.
An antiserum was raised to an epitope in Cbln3 located in the equivalent site to the cerebellin peptide motif in Cbln1. The synthetic peptide APPGRVAFAAVRSHHH, corresponding to amino acids 59 to 74 of mouse Cbln3, was coupled to keyhole limpet hemocyanin. Rabbits were immunized with 0.5 mg of keyhole limpet hemocyanin-peptide conjugate in complete Freund's adjuvant and given three booster immunizations with 0.25 mg of conjugate in incomplete Freund's adjuvant (Rockland, Gilbertsville, PA) at 1-week intervals. Antisera were further purified with an affinity column comprising Sepharose coupled to the peptide antigen. The specificity of the anti-Cbln3 antiserum was established by the immunoblotting of cerebellar extracts from cbln3−/− mice (Fig. 2D) and lysates of COS-7 cells transfected with recombinant vectors expressing Cbln1, Cbln2, Cbln3, or Cbln4 (see Fig. 5B). Further, the immunoreactive bands in lysates of cbln3-transfected cells were competed when the anti-Cbln3 antiserum was preadsorbed with excess immunogen peptide but not an irrelevant peptide (data not shown).
FIG. 5.
Cbln4 rescues secretion of Cbln3. (A) LexA and VP16 fusion constructs of Cbln1, Cbln2, Cbln3, and Cbln4 were expressed in yeast. Protein-protein interactions between all possible binary combinations of proteins were tested and scored. Methodological details of yeast two-hybrid assays are provided in Materials and Methods. ++, colonies turned blue within 1 h; −, colonies were negative at 2 h. (B) COS-7 cells were singly or doubly transfected with vectors expressing Cbln1, V5-tagged Cbln2 (Cbln2-V5), Cbln3, and Cbln4 (Cbln4-V5) as shown. Medium and cell lysates were immunoblotted with anti-Cbln3 and anti-V5 antibodies. −, absence of; +, presence of.
Immunoblotting.
Culture medium, cell lysates, and cerebellar extracts were run on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gels and electrotransferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). Subsequently, membranes were processed according to standard techniques and bound antibody was detected by the ECL chemiluminescence system (Amersham, Piscataway, NJ). For quantification, the intensity of each band was measured using FluorChem 8900 densitometric software (Alpha Innotech, San Leandro, CA) and normalized to β-actin level. Student's t test was used for statistical comparison.
Northern blotting.
The method and probes for Northern blotting were described previously (27). Briefly, total RNA from mouse cerebellum was extracted using RNAzol B (Tel-Test, Friendswood, TX) and hybridized to 32P-labeled probes from either cbln3 or cbln1. Blots were subsequently stripped and rehybridized to a 32P-labeled probe to β-actin.
Histological methods.
One-month-old mice were perfused transcardially with buffered 4% paraformaldehyde, and the cerebella were removed and postfixed as previously described (9). Frozen sections (16 μm) were stained using cresyl violet for standard histological analysis. For immunohistochemistry, a rabbit polyclonal antiserum to PEP-19 (the product of the pcp4 gene) was used to immunostain cerebellar Purkinje cells as described previously (45).
Rota-rod test.
Wild-type, cbln1−/− and cbln3−/− mice (gender and age matched; n = 6 to 8) were tested on an accelerating Rota-rod (San Diego Instruments, San Diego, CA). The Rota-rod was programmed to accelerate from 0 to 40 rpm in 4 min. Each mouse was tested over 4 consecutive days, with two 4-min trials each day. The latency of the mice to fall from the rod was scored as an index of their motor coordination.
Deglycosylation.
Cerebellar extracts were subjected to deglycosylation using endoglycosidase H (endo-H) (Roche, Indianapolis, IN) and N-glycosidase F (Calbiochem, La Jolla, CA). One hundred micrograms of extract was heated to 95°C for 5 min in reaction buffer (20 mM Tris-HCl, 50 mM sodium chloride, 0.2% SDS, 1% β-mercaptoethanol, pH 6.5), and subsequently, endoglycosidase H (10 mU) or N-glycosidase F (5 mU) was added and samples were incubated at 37°C for 4 h. The reaction was terminated by the addition of an equal volume of 2× sample buffer and then subjected to SDS-polyacrylamide gel electrophoresis and immunoblotting.
Cell cultures and transfection.
Primary dissociated cerebellar neuron cultures were prepared as previously described (1). After 3 weeks of culture, the medium and cell lysate were subjected to immunoblotting. African green monkey kidney COS-7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen) in 5% CO2 at 37°C. Cells were transfected using the FuGENE 6 transfection reagent (Roche, Indianapolis, IN) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were scraped into SDS sample-loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.01% bromophenol blue) and subjected to immunoblotting.
Coimmunoprecipitation.
Cerebellar extract or culture medium was precleared by adding 50 μl protein G-agarose beads (Roche) per 1 ml extract for 1 h at 4°C. Ten micrograms of immunoglobulin was added to supernatant samples, and the incubation continued overnight at 4°C. Subsequently, 50 μl protein G-agarose beads was added and the incubation continued for 1 h at 4°C. Protein G-agarose beads were then washed four times with a buffer containing 150 mM NaCl, 50 mM Tris-HCl, pH 6.8, 5 mM EDTA, 0.5% NP-40, 0.1% deoxycholate, and EDTA-free protease inhibitor cocktail (Roche). Beads were boiled for 5 min in 50 μl of 2× SDS sample-loading buffer, and supernatants were analyzed by immunoblotting as described above.
Yeast two-hybrid analysis.
The methods for yeast two-hybrid analysis were described previously (27). Briefly, cDNAs of interest were introduced into the yeast-Escherichia coli shuttle vector pSD10a and/or the Y.LexA vector. Saccharomyces cerevisiae strain S260 (lacking URA3 and TRP1), which contains a genomic lexA-operator-lacZ reporter gene integrated into the URA3 locus (16), was cotransformed with the LexA and VP16 fusion constructs. Transformants were selected on plates lacking Trp and Ura and transferred onto HybondN filters (Amersham, Piscataway, NJ). Filters were transferred to galactose medium to induce the expression of the fusion proteins, and LacZ-positive colonies were identified and scored in a β-galactosidase assay using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranosidase (X-Gal) from Promega (Madison, WI) as a substrate. Results of protein-protein interactions are shown as “++,” indicating that yeast colonies turned dark blue within 1 h; “+,”indicating that yeast colonies turned blue between 1 and 2 h; “± (trace activity),” indicating that yeast colonies are just above background at 2 h; and “− (no activity),” indicating that yeast colonies are at background at 2 h (27).
Structure modeling.
The structural model of the C1q globular domains of Cbln1 and Cbln3 was generated with the Swiss-PdbViewer (Swiss Institute of Bioinformatics, Basel, Switzerland). We used the crystal structure of Acrp30 (32) as a template.
RESULTS
Cbln3 is a secreted glycoprotein that participates in heteromeric complexes with Cbln1.
As a prior study established that Cbln1 binds Cbln3 in yeast two-hybrid assays (27), experiments were conducted to establish whether Cbln3 is secreted and assembles into heteromeric complexes with Cbln1.
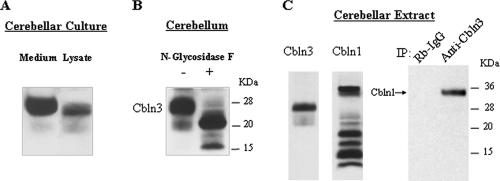
Cbln1 is secreted from cerebellar granule cells in primary dissociated cultures (1). Using the identical culture, the secretion of Cbln3 was assessed by immunoblotting using a Cbln3-specific antiserum. A major immunoreactive band of ∼28 kDa was detected in medium and cell lysate, along with several minor bands of lower molecular masses (Fig. 1A). The major immunoreactive band (28 kDa) has a molecular mass greater than that predicted for Cbln3 (∼20 kDa), suggesting posttranslational modification, possibly N-linked glycosylation. A cerebellar lysate was subjected to deglycosylation using N-glycosidase F to remove all asparagine-linked carbohydrates (1, 38). Following treatment, the major 28-kDa immunoreactive band was lost and a new major band of Cbln3-like immunoreactivity appeared at around 21 kDa (Fig. 1B), indicating that Cbln3 is an asparagine-linked glycoprotein.
FIG. 1.
Cbln3 is a secreted glycoprotein that participates in heteromeric complexes with Cbln1. (A) Medium or cell lysate of primary cultures of cerebellar neurons was immunoblotted with an anti-Cbln3 antiserum. (B) Cerebellar extracts were treated with (+) or without (−) N-glycosidase F and immunoblotted with anti-Cbln3 antiserum. Molecular mass markers are shown to the right of the panel. (C) Cerebellar extracts were immunoblotted with anti-Cbln1 and anti-Cbln3 antiserum. The same cerebellar extracts were also immunoprecipitated (IP) with either rabbit immunoglobulin G (Rb-IgG) or an anti-Cbln3 antiserum, and both were immunoblotted with anti-Cbln1 antiserum. Molecular mass markers are shown at right of the panel. The position of Cbln1 is shown.
To establish whether Cbln3 forms complexes with Cbln1, coimmunoprecipitation was performed on cerebellar lysates. Whereas Cbln3 exists as a single prominent species in cerebellum (Fig. 1B and C), multiple Cbln1-like immunoreactive bands, many of submonomeric size, are detected (Fig. 1C). As shown in Fig. 1C, complexes immunoprecipitated with a rabbit anti-Cbln3 antiserum (anti-Cbln3), but not an irrelevant rabbit antiserum (rabbit immunoglobulin G), contained a single Cbln1-like immunoreactive band at the correct molecular mass (∼35 kDa) (1). Although the anti-Cbln1 antiserum detects multiple Cbln1-like immunoreactive bands in cerebellar extracts, only the full-length, secreted form of Cbln1 coprecipitates with Cbln3, suggesting that the heteromeric complexes are composed of the secreted forms of the proteins. Thus, Cbln1 and Cbln3 are present in the same molecular complex in vivo.
Various technical and theoretical limitations preclude us from answering several important questions regarding the composition and relative abundance of Cbln-containing complexes in brain. Our lack of knowledge of the precise subunit stoichiometry of Cbln-containing complexes in vivo and technical limitations intrinsic to coimmunoprecipitation immunoblotting from whole tissues make it impossible to unambiguously determine the relative proportions of Cbln1 homomeric and Cbln1-Cbln3 heteromeric complexes in the cerebellum. Similarly, technical constraints involving the immunodepletion of whole-brain extracts make it impossible to assess unequivocally whether Cbln3 is found only in complex with Cbln1. Nevertheless, we established that full-length Cbln1 can participate in homomeric complexes as well as oligomeric complexes with mature Cbln3.
Phenotype of cbln3-deficient mice.
To study the function of Cbln3, we used homologous recombination to eliminate all three exons of cbln3 in the mouse genome (Fig. 2A). Successful targeting of cbln3 was confirmed by Southern blotting using external (probe A) and internal (probe B) probes (Fig. 2B). Northern blotting and Western blotting were performed on cerebella of cbln3+/+, cbln3+/−, and cbln3−/− mice. Homozygous cbln3−/− mice exhibited a complete loss of cbln3 mRNA (Fig. 2C) and protein (Fig. 2D), whereas heterozygous mice had intermediate levels of mRNA and protein (Fig. 2C and D).
The cbln3−/− mice are viable and fertile and have normal life spans and no obvious gross anatomical anomalies. As cbln3 is almost exclusively expressed in cerebellum (27), a more detailed study of this brain region was undertaken. The cerebella of cbln3−/− mice had normal foliation and laminated structures (Fig. 2E). All principal neuronal types were present in the cerebella of cbln3−/− mice (Fig. 2E), and Purkinje cells appeared normal when immunostained for PCP4/PEP-19 (Fig. 2E, lower panel), a Purkinje cell marker (45). In contrast to that with ataxic cbln1−/− mice (1), no behavioral deficit was observed in cbln3−/− mice based on general activity, gait analysis (data not shown), and performance on the accelerating Rota-rod (Fig. 2F).
We next tested whether there was a genetic interaction between cbln1 and cbln3 at the level of the cerebellar phenotype. cbln1+/− cbln3+/− and cbln1+/− cbln3−/− mice had normal phenotypes (data not shown). Thus, a partial or complete loss of Cbln3 does not produce a cbln1-null-like phenotype in a cbln1 heterozygous animal. Furthermore, the neuroanatomical and behavioral phenotypes of cbln1−/− cbln3−/− mice were indistinguishable from those of cbln1−/− cbln3+/+ mice (data not shown). Thus, although Cbln1 and Cbln3 are coexpressed and secreted from cerebellar granule cells in the same complex, null alleles of the two genes have distinct phenotypes, with no obvious genetic interaction between the two.
Inverse relationship between the levels of Cbln1 and Cbln3.
One explanation for the discordance in phenotypes between cbln1 and cbln3 knockout mice is functional compensation at the transcriptional or posttranslational level. Therefore, Northern and Western blotting was performed on cbln1−/− and cbln3−/− cerebella. No compensatory changes in mRNA levels for cbln1 or cbln3 were observed in either knockout strain (Fig. 3A). In striking contrast, there were profound changes in the levels of the two proteins (Fig. 3B). In cbln1-null mice, the level of Cbln3 was barely detectable (Fig. 3B). In contrast, Cbln1 levels were greatly elevated in cbln3-null mice (Fig. 3B). Quantitative immunoblotting revealed a gene dosage-dependent effect. Cbln3 levels were reduced by ∼46% in cbln1+/− mice and ∼93% in cbln1−/− mice (Fig. 3C and D). In contrast, cbln3+/− mice had 170% the level of Cbln1 and cbln3−/− mice had a 640% increase (Fig. 3E and F). Thus, cbln1−/− mice essentially represent a double knockout of Cbln1 and Cbln3. This potentially explains why cbln1 cbln3 double-knockout mice are no more affected than cbln1-null mice.
FIG. 3.
Inverse relationship between the levels of Cbln1 and Cbln3. (A) Cerebellar RNA isolated from wild-type, cbln1−/−, and cbln3−/− mice was assessed by Northern blotting using cbln1, cbln3, and β-actin (loading control) probes. No overt compensatory responses were observed for either gene in the knockout strains. (B) Cerebellar extracts of wild-type, cbln1−/−, and cbln3−/− mice were immunoblotted with antisera to Cbln1, Cbln3, and β-actin (loading control). Note the marked changes in levels of Cbln1 and Cbln3. (C and D) Western blot (C) and quantification (D) of Cbln1 and Cbln3 protein levels in wild-type, cbln1+/−, and cbln1−/− cerebella (n = 3). Cbln3 levels are markedly reduced in cbln1+/− (P = 0.026) and cbln1−/− (P = 0.0009) mice. Error bars represent standard error of the means. (E and F) Western blot (E) and quantification of Cbln1 and Cbln3 protein levels (F) in wild-type, cbln3+/−, and cbln3−/− cerebella (n = 3). Cbln1 levels are significantly increased in cbln3+/− (P = 0.008) and cbln3−/− (P = 0.0001) cerebella. Error bars represent SEM.
Additional experiments elucidated the mechanisms underlying the interdependence between Cbln1 and Cbln3 protein levels.
Cbln1 is required for ER export and secretion of Cbln3.
The levels of many secreted glycoproteins are regulated by the quality control mechanism for endoplasmic reticulum (ER) exit (6, 39). Therefore, the influence of Cbln1 on ER retention and the secretion of Cbln3 was assessed.
A hallmark of proteins located or retained in the ER is the presence of asparagine-linked, high-mannose-containing carbohydrate chains that can be specifically cleaved by endo-H (14). Therefore, the glycosylation state of Cbln1 and Cbln3 was determined following endo-H digestion of cerebellar extracts (Fig. 4A). Cbln1 was insensitive to endo-H digestion in both wild-type and cbln3−/− mice (Fig. 4A, upper panel). Whereas Cbln3 was also endo-H insensitive in wild-type mice (Fig. 4A, left side of lower panel) the residual Cbln3 in cbln1−/− mice was completely endo-H sensitive (note the generation of the lower-molecular-weight band indicated in Fig. 4A). To confirm that the molecular weight shift was not due to another enzymatic activity in the endo-H, lysates were treated with N-glycosidase F (PNGaseF) that removes all asparagine-linked carbohydrates (38). Both Cbln1 and Cbln3 were sensitive to PNGaseF (Fig. 4A). The presence of immature Cbln3 in the cbln1-null cerebellum suggests that Cbln1 is essential for Cbln3 to exit the ER.
FIG. 4.
Cbln1 is required for ER export and secretion of Cbln3. (A) Cerebellar extracts were subjected to deglycosylation in the presence (+) of endo-H or N-glycosidase F (PNGase F) for 4 h. Subsequently, cerebellar extracts from wild-type (50 μg), cbln1−/− (100 μg), and cbln3−/− (20 μg) mice were immunoblotted with anti-Cbln1 and anti-Cbln3 antisera, respectively. The asterisk indicates the presence of endo-H-sensitive Cbln3 exclusively in the cbln1-null mouse cerebellum. (B) After 21 days in vitro, medium and cell lysates from primary dissociated cerebellar cultures from wild-type, cbln1−/−, and cbln3−/− mice were immunoblotted with an anti-Cbln3 antiserum. (C) COS-7 cells were transfected with vectors encoding Cbln1 and Cbln3 singly or in combination. Culture medium and cell lysates were immunoblotted with anti-Cbln1 or anti-Cbln3 antiserum. −, absence of; +, presence of. (D) COS-7 cells were transfected with vectors encoding Flag-tagged Cbln1 (Flag-Cbln1) and V5-tagged Cbln3 (Cbln3-V5). Medium was immunoprecipitated with anti-Flag (IP Flag) or anti-V5 (IP V5) antibodies, then immunoblotted with anti-Flag (IB Flag) or anti-V5 (IB V5) antibodies. −, absence of; +, presence of. (E) COS-7 cells were transfected with vectors encoding Cbln1 and Cbln3. At 24 h after transfection, cells were incubated for 8 h in the presence of dimethyl sulfoxide as a vehicle control (Con), the lysosome inhibitor 100 μM chloroquine (CQ), or a proteasome inhibitor, 10 μM lactacystin (LA), 10 μM MG132 (MG), or ZAL. Medium and cell lysates were immunoblotted with anti-Cbln1 or anti-Cbln3 antisera.
To assess whether Cbln1 influences the secretion of Cbln3, primary cultures of wild-type, cbln1−/−, and cbln3−/− cerebellar neurons were analyzed by Western blotting. As with Cbln1 (1), cultures from wild-type mice secrete large amounts of Cbln3 (Fig. 4B). When corrected for loading, approximately 95% of total Cbln3 is in the medium. Mirroring the situation in vivo, there is little Cbln3 present in cell lysates from cultures generated from cbln1-null mice (Fig. 4B). Moreover, only a trace of Cbln3 is present in the medium of these cultures (Fig. 4B). Based upon the ratio of Cbln3 in medium to lysate from wild-type (20:1) versus cbln1-null (3:1) cells, this result is consistent with impaired secretion of Cbln3 in cbln1-null granule cells. However, this result is ambiguous given the low levels of Cbln3.
To establish whether Cbln1 modifies the secretion of Cbln3, recombinant Cbln1 and Cbln3 were expressed singly or in combination in COS-7 cells and medium and cell lysate were analyzed by immunoblotting. Cbln1 and Cbln3 were detected in the lysates of transfected cells (Fig. 4C). Unlike primary granule cells, neither protein appeared to influence the level of the other. In marked contrast, whereas Cbln1 was robustly secreted into the medium, Cbln3 was secreted only when coexpressed with Cbln1 (Fig. 4C). Thus, despite equivalent levels of Cbln3 within the intracellular compartment, Cbln3 is secreted only when Cbln1 is present, potentially because Cbln1 binds to Cbln3.
Flag-tagged Cbln1 (Flag-Cbln1) and V5-tagged Cbln3 (Cbln3-V5) were expressed in COS-7 cells and heteromeric complexes characterized by sequential immunoprecipitation and immunoblotting with antibodies to the respective epitope tags (Fig. 4D). Cbln3-V5 was detected in the medium only when coexpressed with Flag-Cbln1 (Fig. 4D, bottom panel). Furthermore, the Cbln3-V5 present in the medium from cotransfected cells was coimmunoprecipitated with Flag-Cbln1 (Fig. 4D, first and third panels). Noticeably, whereas submonomeric fragments of Cbln1 were detected in transfected cells, only the mature form of the protein was present in the secreted complexes (Fig. 4C). Thus, full-length secreted Cbln1 and Cbln3 form heteromeric complexes both in vivo and in heterologous systems.
Proteins retained in the ER by the quality control mechanism are subsequently translocated to the cytosol for degradation, predominantly in the proteasome (39). To determine whether Cbln3 is subject to similar processing, COS-7 cells expressing Cbln1 or Cbln3 were treated with a proteasome inhibitor, lactacystin, MG132, or ZAL, or the lysosome inhibitor chloroquine. Whereas the levels of Cbln1 increased modestly in the presence of proteasome inhibitors, the levels of Cbln3 increased dramatically (Fig. 4E). Despite the marked increase in Cbln3, it was still not secreted (Fig. 4E). In the case of Cbln3, the proteasome inhibitors increased the intensity of multiple bands that were also present in untreated cells and, in addition, generated new bands of submonomeric masses (Fig. 4E). Whereas proteasome inhibitors also increased the intensity of existing bands in Cbln1-transfected cells, they did not generate any prominent new bands. Chloroquine caused a small increase in Cbln3 levels (Fig. 4E). In contrast, although not as potent as the proteasome inhibitors, chloroquine caused a relatively large increase in Cbln1-like immunoreactive bands (Fig. 4E). These data suggest that Cbln3 is degraded predominantly via the proteasome, whereas Cbln1 is eliminated in both the proteasome and the lysosome.
Specificity of Cbln1 family member interactions.
All Cbln1 family members share a C1q globular domain in their C termini, raising the possibility of additional homomeric and heteromeric complexes. Therefore, protein-protein interactions among the four family members were assessed using the yeast two-hybrid assay. All family members interacted with all other family members (Fig. 5A). In addition, all family members, with the striking exception of Cbln3 (Fig. 5A), interacted with themselves. Thus, Cbln3 is unique in being unable to undergo homomeric association.
The data also raise the question of whether Cbln2 or Cbln4 can rescue the secretion of Cbln3. Therefore, we examined the secretion of Cbln3 in COS-7 cells cotransfected with cbln2 or cbln4. Cbln4, a secreted protein, rescued the secretion of Cbln3 (Fig. 5B). In contrast, Cbln2, a nonsecreted transmembrane protein (41), did not (Fig. 5B). Thus, heteromeric binding of Cbln3 to a family member is essential for secretion but is insufficient if the interacting partner is not itself secreted.
The C1q domain of Cbln3 is essential for heteromeric interactions.
The C1q globular domain in Cbln1 is essential for mediating homomeric protein-protein interactions (1). However, the structural bases of heteromeric interactions among Cbln1 family members and the lack of homomeric interactions of Cbln3 are undetermined. Therefore, structure-binding analyses were performed to assess whether the C1q domains in Cbln1 and Cbln3 were required for heteromeric association and whether sequences outside of the C1q domain in Cbln3 inhibited its homomeric binding.
N-terminal truncations of Cbln1 and Cbln3 were generated with either LexA or VP16 fused to their N termini (Fig. 6A) in yeast expression plasmids. The choice of truncations is based upon a structure-binding analysis of homomeric interactions in Cbln1 (1). Cbln1Δ21N and Cbln3Δ24N are the secreted mature forms of Cbln1 and Cbln3, respectively. Cbln1Δ54N is the product generated by cleavage at a putative tumor necrosis factor (TNF) alpha-cleaving enzyme-like site in Cbln1 responsible for liberating the peptide, cerebellin (1, 33, 34, 40), and Cbln3Δ57N is the equivalent fragment in Cbln3. Cbln1Δ61N and Cbln3Δ63N constitute just the C1q domains of the two family members. Cbln1Δ72N is the predicted product of cleavage at the C terminus of the cerebellin peptide sequence in Cbln1 (1), and Cbln3Δ74N is the equivalent fragment of Cbln3; these products lack the first beta strand (designated A) of the C1q motif and abolish homomeric interactions in Cbln1 (1).
FIG. 6.
The C1q domains of Cbln1 and Cbln3 are essential for heteromeric interactions. Schematic diagrams of the truncation mutants of Cbln1 (A) and Cbln3 (B) used in yeast two-hybrid analysis are shown. The signal sequence is shown in blue, two cysteines are red, and the C1q domain is orange. The location of cerebellin, a naturally occurring peptide fragment of Cbln1, is shown by a black box. The various Mt forms of Cbln1 and Cbln3 were assayed in combination with full-length proteins as well as with themselves. All constructs scored as negative when expressed alone (data not shown). ++, colonies turned blue within 1 h; +, colonies turned blue by 2 h; ±, marginal coloration at 2 h; −, no coloration at 2 h.
For the assays summarized in Fig. 6A and B, we assessed the binding of mutant (Mt) forms of Cbln1 or Cbln3 to wild-type Cbln1 or Cbln3 in both LexA and VP16 configurations. In addition, homomeric binding of mutant forms of Cbln1 and Cbln3 was also assessed. Full-length Cbln1, Cbln1Δ21N, Cbln1Δ54N, and Cbln1Δ61N bound to Cbln3, whereas Cbln1Δ72N, which deletes the A strand of the C1q globular domain, did not (Fig. 6A). In assays of Cbln3 mutants, full-length Cbln3, Cbln3Δ24N, and Cbln3Δ57N bound to Cbln1 and Cbln3Δ63N showed weaker or marginal binding, whereas Cbln3Δ74N did not bind to Cbln1 (Fig. 6B). Thus, the C1q globular domains in both Cbln1 and Cbln3 are essential for heteromeric interactions. In addition, eliminating all sequences N terminal to the C1q domain of Cbln3 (mt-Cbln3Δ63N with mt-Cbln3Δ63N) does not confer the ability to undergo self-interaction (Fig. 6B). Therefore, the structural basis for the inability of Cbln3 to form a homomeric complex must lie within the C1q domain.
Arg119 in Cbln3 determines homomeric binding and secretion.
To identify amino acids in Cbln3 that influence homomeric assembly, two assumptions were made. First, amino acids that preclude protein-protein association should lie at or near subunit interfaces and, second, they should differ between Cbln3 and other family members. To identify interfaces, structural models of the trimeric C1q domains of Cbln1 and Cbln3 were generated using Swiss-PdbViewer software (Fig. 7A). The C1q domain contains 10 beta strands (designated A to H) oriented in a so-called 10-fold beta sandwich configuration, the structural hallmark of the C1q/TNF superfamily of proteins (32). The C strand lies at the interface of the three subunits (Fig. 7A) and is the most highly conserved region in terms of amino acid sequence in not only the Cbln1 family but also the entire C1q/TNF superfamily (1, 11, 13). Surveying a range of C1q/TNF superfamily members revealed that Cbln3 was unique in possessing a charged arginine (R119) residue within the C strand (1, 11, 13) (Fig. 7A). Within the predicted model of Cbln3 homotrimers, the side chains of arginine 119 converge at the center of the structure (Fig. 7A and B). Charged residues can provide intersubunit electrostatic repulsion, thereby regulating the assembly and stability of some proteins (4, 5). Noticeably, the equivalent amino acid in Cbln1 (asparagine 115) does not constitute a steric clash in a predicted Cbln1 homotrimer, and it can also potentially accommodate the arginine residue in a Cbln1+Cbln3 heterotrimer (Fig. 7B).
FIG. 7.
Arg119 determines the inability of Cbln3 to undergo self-assembly and secretion. (A) The amino acid sequences of the C β strands of Cbln1 to Cbln4 are aligned as shown (left panel). Residues in red are identical, those in blue are conservative substitutions, and dissimilar residues are black. Note the critical arginine residue (italicized) in Cbln3. A trimeric structural model of the C1q globular domain of Cbln3 was built using the Swiss-PdbViewer. Top and side views of predicted Cbln3 trimers are shown in the right and middle panels. The C strands are white, and the arrows point to Arg119 of Cbln3. (B) Structural models of homo- and heterotrimeric complexes of Cbln1 and Cbln3. Left panel, homomeric Cbln1 model; middle panel, homomeric Cbln3 model; right panel, heteromeric Cbln1 and Cbln3 model. Cbln1 subunits are red, and Cbln3 subunits are green. The arrows point to Asn115 in Cbln1 and Arg119 in Cbln3. (C) LexA and VP16 fusions of Cbln1, Cbln3, and Cbln3(R119A) were expressed in yeast for interaction analyses. Protein-protein interactions were scored as described for Fig. 5 and 6. Dotted circles indicate the altered properties of Cbln3R119A compared to those of Cbln3. (D) COS-7 cells were transfected with combinations of vectors expressing Cbln1, Cbln3, and Cbln3(R119A). Medium and cell lysate were immunoblotted with anti-Cbln1 and anti-Cbln3 antibodies. Note the secretion of Cbln3R119A (R119A) in the absence of Cbln1 (left panel).
Arginine- and lysine-containing motifs have been implicated in ER retention and retrieval, especially in the immune and nervous systems (7, 20, 29, 30, 37). We therefore used mutational analysis to establish whether arginine 119 prevents Cbln3 from assembling into homotrimers and ultimately undergoing secretion. Cbln1, Cbln3, and the point mutant, Cbln3(R119A) were expressed in LexA and VP16 fusion vectors and subjected to yeast two-hybrid analysis (Fig. 7C). Cbln3 did not undergo self-association but did bind to Cbln1. However, Cbln3(R119A) not only bound to Cbln1 but also interacted with itself and Cbln3 (Fig. 7C), indicating that Arg119 is a determinant for assembly. To establish whether this was also true for secretion, Cbln3 and Cbln3(R119A) were expressed in COS-7 cells in the presence or absence of Cbln1 and medium and cell lysates were analyzed by Western blotting. Whereas Cbln3 was not secreted into the medium in the absence of Cbln1, Cbln3(R119A) was robustly secreted (Fig. 7D, left panel). Neither Cbln3 nor Cbln3(R119A) markedly influenced the secretion of Cbln1 in cotransfected cells (Fig. 7D, right panel). Thus, a single arginine residue in Cbln3 appears responsible for its inability to undergo homomeric association and secretion.
DISCUSSION
Previously, we identified Cbln1 and GluRδ2 as components of a transneuronal signaling pathway that contributes to the maintenance of synaptic structure and function in the cerebellum (9). Cbln1 is secreted from presynaptic granule cells, and its genetic elimination results in a number of structural and physiological changes in postsynaptic Purkinje cells. The primary deficit in cbln1-null mice is the presence of numerous naked postsynaptic spines on Purkinje cell dendrites despite the fact that normal numbers of parallel fibers and presynaptic nerve endings are present in these animals (9). Furthermore, there is an apparent compensatory hypertrophy of the postsynaptic densities associated with the remaining intact synapses (9). These ultrastructural alterations are associated with impaired neurotransmission between granule cells and Purkinje cells and also the loss of cerebellar long-term depression in Purkinje cells (9). The elimination of Cbln1 also has consequences on the interaction of other neurons with Purkinje cells. For example, climbing fibers that in adults have a one-to-one relationship with Purkinje cells fail to be pruned back during development in cbln1-null mice, resulting in multiple innervations. In addition, the supernumerary climbing fibers penetrate to a greater depth through the molecular layer and make synaptic connections with the distal dendrites of Purkinje cells, a territory normally innervated by granule cell parallel fibers (9). Remarkably, the same spectrum of deficits are seen when GluRδ2, an orphan glutamate receptor concentrated in the postsynaptic density of Purkinje cells (18, 36, 44), is knocked out (8, 10, 12, 15, 17). As there is also a genetic interaction between cbln1 and GluRδ2, it is proposed that secreted presynaptic Cbln1 and membrane-associated postsynaptic GluRδ2 are components of a common signaling pathway involved in the maintenance of synaptic integrity and function (9). The present study was aimed at identifying additional components and characteristics of this pathway.
We now extend our understanding of this transneuronal regulatory pathway by showing that Cbln1 is secreted from presynaptic granule cells in a heteromeric complex containing the related family member Cbln3. Moreover, Cbln1 and Cbln3 each regulate the level of the other through a mechanism involving the ER quality control system and ER-associated degradation (ERAD). Because of this interrelationship, cbln1-null mice are functionally deficient in both Cbln1 and Cbln3, raising the possibility that these mice have a compound phenotype attributable to the loss of both family members. As cbln3-null mice have no overt phenotype, we must conclude either that the greatly enhanced level of Cbln1 in these mice confers functional redundancy or that Cbln1/Cbln3 heteromeric complexes serve a function distinct from that of Cbln1 homomeric complexes and that it was not detected in our analyses. Nevertheless, viewing this interaction from the perspective of the presynaptic compartment, a complex mechanism is in place in cerebellar granule cells to regulate the stoichiometry of Cbln1 and Cbln3. Indeed, even haploinsufficiency for either family member, as occurs in heterozygous mice, results in compensatory changes in the level of the other protein.
Prominent submonomeric fragments of Cbln1 are present in brain (1, 9) (Fig. 1C), suggesting that Cbln1 undergoes substantial proteolytic processing in vivo. Indeed, these fragments are more abundant than the mature, secreted form of Cbln1 and they appear to be generated in the presynaptic compartment, as they never participate in either the heteromeric complexes isolated in vivo (Fig. 1C) or the homomeric or heteromeric complexes secreted in vitro (1) (Fig. 4D) but are detected in lysates of cultured granule cells and transfected cells (1). This processing appears to be the mechanism by which the naturally occurring peptide, cerebellin, is generated from Cbln1 (33, 40), and it may also serve to modify the conformation of the Cbln1 hexamer (1). In striking contrast, the great majority of Cbln3 present in cerebellum is the full-length, mature protein, which is also the only form detected in the medium of cultured granule neurons or transfected cell lines. Thus, despite the fact that the loss of Cbln1 results in a near disappearance of Cbln3, it is Cbln1 that in wild-type mice is most overtly subject to proteolysis.
The fate of many secreted and membrane glycoproteins is determined by the quality control mechanism for ER exit (6, 39) that ensures that unfolded proteins do not progress through the secretion pathway. Our data show that Cbln1 is essential for Cbln3 to escape elimination by ERAD and undergo subsequent secretion through the formation of heteromeric complexes. The molecular mechanism mediating this process involves a critical arginine residue in Cbln3 and is reminiscent of the process controlling the assembly of many heteromultimeric membrane proteins. Indeed there are a number of cases in which heteromeric interactions ensure the forward trafficking and surface expression of membrane proteins through the secretory pathway by masking specific ER retention motifs that frequently contain arginine or lysine residues (2, 7, 20, 21, 29-31, 35, 43). This “hide and run” mechanism helps ensure not only protein integrity but also appropriate subunit stoichiometry and correct cellular localization of the final functional complex (21). Examples of this in the nervous system include the B1 and B2 subunits of the GABAB receptor (3, 20, 26), the GluR2 subunit of the amino-3-hydroxy-5-methyl-4-isoazole propionate receptor (7), the NR1 subunit of the N-methyl-d-aspartate receptor (31), and the KA2 and GluR5-2b kainate receptors (29, 30). Indeed, alternative splicing in the NR1 receptor generates an RXR-type ER retention motif and the functional role of RNA editing of the Q/R site in the GluR2 subunit of the amino-3-hydroxy-5-methyl-4-isoazole propionate receptor is to generate an ER retention sequence (7), indicating that there is a dynamic regulation of protein sorting and trafficking in the nervous system.
In the case of Cbln3, an arginine residue critical for ER retention is predicted to lie in the core of the hypothetical Cbln3 trimer in a location that would provide for charge repulsion or steric clash between the subunits. Electrostatic repulsion has been implicated in regulating the assembly and stability of some protein complexes (4, 5). The key arginine residue in Cbln3 lies in the middle of the C strand of the C1q domain that lies along the interior interface of the predicted trimer. This is the most conserved region of the C1q domain and is the only part of the 10-fold beta-sandwich structure that harbors primary sequence identity beyond the immediate C1q family into the more distantly related TNF family (13). Remarkably, of the entire 10-fold beta-sandwich superfamily of proteins, only Cbln3 and one of the multimerins have charged amino acids within their C strands, indicating the uniqueness of this configuration. Structural analysis suggests that this arginine residue can be accommodated in a Cbln1-Cbln3 heteromeric complex, where an asparagine lies at the equivalent position.
Whereas arginine-based ER retention motifs are well documented for membrane proteins, there is a paucity of studies reporting regulated trafficking of secreted protein using this mechanism. Soluble ER proteins, such as BiP and protein disulfide isomerases, contain C-terminal KDEL sequences that are responsible for the recognition and retrieval of these proteins from post-ER compartments (22, 28). ER retention of these proteins is mediated by an integral membrane protein, ERD2 or KDEL receptor, that promotes uptake into vesicles coated with COPI complexes (19, 25). However, much less is known about the molecular mechanisms mediating the retention of proteins with arginine-containing motifs (21, 23), although COPI and 14-3-3 proteins have been implicated (21, 23, 24, 42). Therefore, we cannot distinguish whether the critical arginine in Cbln3 is a formal ER retention signal, possibly being recognized by a resident ER transmembrane protein, or whether it functions to preclude homomeric subunit interactions, thereby resulting in unfolded or incompletely folded Cbln3 monomers. These unfolded Cbln3 monomers could then bind to ER chaperones, which possess their own ER retention (KDEL) motifs and subsequently undergo elimination by ERAD.
As cbln1-null mice lack both Cbln1 and Cbln3 and because Cbln1 protein levels increase dramatically in cbln3-null mice, the respective contributions of Cbln1 and Cbln3 to the cbln1 knockout phenotype are ambiguous. As the Cbln3-R119A mutant is secreted in the absence of Cbln1, it may be possible to establish whether Cbln3 homomeric complexes can rescue the cbln1-null phenotype through genetic knockin or transgenic mouse strategies.
Acknowledgments
We thank Linda Hendershot and Jie Zheng for helpful discussion.
This work was supported in part by the NIH Cancer Center CORE grant CA 21765, the American Lebanese Syrian Associated Charities, and NIH grants NS042828, NS040361, and NS040749 to J.I.M.
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Bao, D., Z. Pang, and J. I. Morgan. 2005. The structure and proteolytic processing of Cbln1 complexes. J. Neurochem. 95:618-629. [DOI] [PubMed] [Google Scholar]
- 2.Bichet, D., V. Cornet, S. Geib, E. Carlier, S. Volsen, T. Hoshi, Y. Mori, and W. M. De. 2000. The I-II loop of the Ca2+ channel alpha1 subunit contains an endoplasmic reticulum retention signal antagonized by the beta subunit. Neuron 25:177-190. [DOI] [PubMed] [Google Scholar]
- 3.Calver, A. R., M. J. Robbins, C. Cosio, S. Q. Rice, A. J. Babbs, W. D. Hirst, I. Boyfield, M. D. Wood, R. B. Russell, G. W. Price, A. Couve, S. J. Moss, and M. N. Pangalos. 2001. The C-terminal domains of the GABAB receptor subunits mediate intracellular trafficking but are not required for receptor signaling. J. Neurosci. 21:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.del Alamo, M., and M. G. Mateu. 2005. Electrostatic repulsion, compensatory mutations, and long-range non-additive effects at the dimerization interface of the HIV capsid protein. J. Mol. Biol. 345:893-906. [DOI] [PubMed] [Google Scholar]
- 5.Douglas, C. C., D. Thomas, J. Lanman, and P. E. Prevelige, Jr. 2004. Investigation of N-terminal domain charged residues on the assembly and stability of HIV-1 CA. Biochemistry 43:10435-10441. [DOI] [PubMed] [Google Scholar]
- 6.Ellgaard, L., and A. Helenius. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181-191. [DOI] [PubMed] [Google Scholar]
- 7.Greger, I. H., L. Khatri, and E. B. Ziff. 2002. RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron 34:759-772. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto, K., R. Ichikawa, H. Takechi, Y. Inoue, A. Aiba, K. Sakimura, M. Mishina, T. Hashikawa, A. Konnerth, M. Watanabe, and M. Kano. 2001. Roles of glutamate receptor δ2 subunit (GluRδ2) and metabotropic glutamate receptor subtype 1 (mGluR1) in climbing fiber synapse elimination during postnatal cerebellar development. J. Neurosci. 21:9701-9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirai, H., Z. Pang, D. Bao, T. Miyazaki, L. Li, E. Miura, J. Parris, Y. Rong, M. Watanabe, M. Yuzaki, and J. I. Morgan. 2005. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat. Neurosci. 8:1534-1541. [DOI] [PubMed] [Google Scholar]
- 10.Ichikawa, R., T. Miyazaki, M. Kano, T. Hashikawa, H. Tatsumi, K. Sakimura, M. Mishina, Y. Inoue, and M. Watanabe. 2002. Distal extension of climbing fiber territory and multiple innervation caused by aberrant wiring to adjacent spiny branchlets in cerebellar Purkinje cells lacking glutamate receptor delta 2. J. Neurosci. 22:8487-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Innamorati, G., E. Bianchi, and M. I. Whang. 2006. An intracellular role for the C1q-globular domain. Cell. Signal. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 12.Kashiwabuchi, N., K. Ikeda, K. Araki, T. Hirano, K. Shibuki, C. Takayama, Y. Inoue, T. Kutsuwada, T. Yagi, and Y. Kang. 1995. Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR delta 2 mutant mice. Cell 81:245-252. [DOI] [PubMed] [Google Scholar]
- 13.Kishore, U., C. Gaboriaud, P. Waters, A. K. Shrive, T. J. Greenhough, K. B. Reid, R. B. Sim, and G. J. Arlaud. 2004. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 25:551-561. [DOI] [PubMed] [Google Scholar]
- 14.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-664. [DOI] [PubMed] [Google Scholar]
- 15.Kurihara, H., K. Hashimoto, M. Kano, C. Takayama, K. Sakimura, M. Mishina, Y. Inoue, and M. Watanabe. 1997. Impaired parallel fiber→Purkinje cell synapse stabilization during cerebellar development of mutant mice lacking the glutamate receptor δ2 subunit. J. Neurosci. 17:9613-9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurschner, C., and J. I. Morgan. 1996. Analysis of interaction sites in homo- and heteromeric complexes containing Bcl-2 family members and the cellular prion protein. Brain Res. Mol. Brain Res. 37:249-258. [DOI] [PubMed] [Google Scholar]
- 17.Lalouette, A., A. Lohof, C. Sotelo, J. Guenet, and J. Mariani. 2001. Neurobiological effects of a null mutation depend on genetic context: comparison between two hotfoot alleles of the delta-2 ionotropic glutamate receptor. Neuroscience 105:443-455. [DOI] [PubMed] [Google Scholar]
- 18.Landsend, A. S., M. Amiry-Moghaddam, A. Matsubara, L. Bergersen, S. Usami, R. J. Wenthold, and O. P. Ottersen. 1997. Differential localization of delta glutamate receptors in the rat cerebellum: coexpression with AMPA receptors in parallel fiber-spine synapses and absence from climbing fiber-spine synapses. J. Neurosci. 17:834-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, M. J., and H. R. Pelham. 1992. Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell 68:353-364. [DOI] [PubMed] [Google Scholar]
- 20.Margeta-Mitrovic, M., Y. N. Jan, and L. Y. Jan. 2000. A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron 27:97-106. [DOI] [PubMed] [Google Scholar]
- 21.Michelsen, K., H. Yuan, and B. Schwappach. 2005. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 6:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munro, S., and H. R. Pelham. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899-907. [DOI] [PubMed] [Google Scholar]
- 23.Nufer, O., and H. P. Hauri. 2003. ER export: call 14-3-3. Curr. Biol. 13:R391-R393. [DOI] [PubMed] [Google Scholar]
- 24.O'Kelly, I., M. H. Butler, N. Zilberberg, and S. A. Goldstein. 2002. Forward transport. 14-3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell 111:577-588. [DOI] [PubMed] [Google Scholar]
- 25.Orci, L., M. Stamnes, M. Ravazzola, M. Amherdt, A. Perrelet, T. H. Sollner, and J. E. Rothman. 1997. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell 90:335-349. [DOI] [PubMed] [Google Scholar]
- 26.Pagano, A., G. Rovelli, J. Mosbacher, T. Lohmann, B. Duthey, D. Stauffer, D. Ristig, V. Schuler, I. Meigel, C. Lampert, T. Stein, L. Prezeau, J. Blahos, J. Pin, W. Froestl, R. Kuhn, J. Heid, K. Kaupmann, and B. Bettler. 2001. C-terminal interaction is essential for surface trafficking but not for heteromeric assembly of GABAB receptors. J. Neurosci. 21:1189-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang, Z., J. Zuo, and J. I. Morgan. 2000. Cbln3, a novel member of the precerebellin family that binds specifically to Cbln1. J. Neurosci. 20:6333-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelham, H. R. 1988. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 7:913-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren, Z., N. J. Riley, E. P. Garcia, J. M. Sanders, G. T. Swanson, and J. Marshall. 2003. Multiple trafficking signals regulate kainate receptor KA2 subunit surface expression. J. Neurosci. 23:6608-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren, Z., N. J. Riley, L. A. Needleman, J. M. Sanders, G. T. Swanson, and J. Marshall. 2003. Cell surface expression of GluR5 kainate receptors is regulated by an endoplasmic reticulum retention signal. J. Biol. Chem. 278:52700-52709. [DOI] [PubMed] [Google Scholar]
- 31.Scott, D. B., T. A. Blanpied, G. T. Swanson, C. Zhang, and M. D. Ehlers. 2001. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 21:3063-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro, L., and P. E. Scherer. 1998. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 8:335-338. [DOI] [PubMed] [Google Scholar]
- 33.Slemmon, J. R., R. Blacher, W. Danho, J. L. Hempstead, and J. I. Morgan. 1984. Isolation and sequencing of two cerebellum-specific peptides. Proc. Natl. Acad. Sci. USA 81:6866-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slemmon, J. R., W. Danho, J. L. Hempstead, and J. I. Morgan. 1985. Cerebellin: a quantifiable marker for Purkinje cell maturation. Proc. Natl. Acad. Sci. USA 82:7145-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Standley, S., K. W. Roche, J. McCallum, N. Sans, and R. J. Wenthold. 2000. PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron 28:887-898. [DOI] [PubMed] [Google Scholar]
- 36.Takayama, C., S. Nakagawa, M. Watanabe, M. Mishina, and Y. Inoue. 1996. Developmental changes in expression and distribution of the glutamate receptor channel delta 2 subunit according to the Purkinje cell maturation. Brain Res. Dev. Brain Res. 92:147-155. [DOI] [PubMed] [Google Scholar]
- 37.Teasdale, R. D., and M. R. Jackson. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 12:27-54. [DOI] [PubMed] [Google Scholar]
- 38.Trimble, R. B., and A. L. Tarentino. 1991. Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: endo F1, endo F2, and endo F3. Endo F1 and endo H hydrolyze only high mannose and hybrid glycans. J. Biol. Chem. 266:1646-1651. [PubMed] [Google Scholar]
- 39.Trombetta, E. S., and A. J. Parodi. 2003. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19:649-676. [DOI] [PubMed] [Google Scholar]
- 40.Urade, Y., J. Oberdick, R. Molinar-Rode, and J. I. Morgan. 1991. Precerebellin is a cerebellum-specific protein with similarity to the globular domain of complement C1q B chain. Proc. Natl. Acad. Sci. USA 88:1069-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wada, C., and H. Ohtani. 1991. Molecular cloning of rat cerebellin-like protein cDNA which encodes a novel membrane-associated glycoprotein. Brain Res. Mol. Brain Res. 9:71-77. [DOI] [PubMed] [Google Scholar]
- 42.Yuan, H., K. Michelsen, and B. Schwappach. 2003. 14-3-3 dimers probe the assembly status of multimeric membrane proteins. Curr. Biol. 13:638-646. [DOI] [PubMed] [Google Scholar]
- 43.Zerangue, N., B. Schwappach, Y. N. Jan, and L. Y. Jan. 1999. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22:537-548. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, H. M., R. J. Wenthold, Y. X. Wang, and R. S. Petralia. 1997. Delta-glutamate receptors are differentially distributed at parallel and climbing fiber synapses on Purkinje cells. J. Neurochem. 68:1041-1052. [DOI] [PubMed] [Google Scholar]
- 45.Ziai, M. R., L. Sangameswaran, J. L. Hempstead, W. Danho, and J. I. Morgan. 1988. An immunochemical analysis of the distribution of a brain-specific polypeptide, PEP-19. J. Neurochem. 51:1771-1776. [DOI] [PubMed] [Google Scholar]