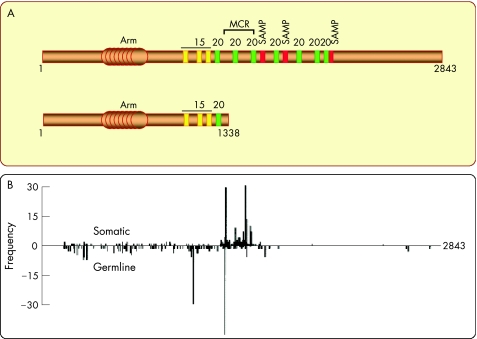
Figure 3 Schematic structure of the adenomatous polyposis coli (APC) protein and distribution of APC mutations found in patients with familial adenomatous polyposis. (A) The 2843‐amino acid sequence displays an armadillo domain near the N terminus. Approximately the central third of the molecule is sufficient to target β‐catenin for degradation. This region contains different motifs that are repeated and dispersed along the sequence. The 15‐amino‐acid repeats (yellow) can bind β‐catenin, but their functional significance is still obscure. The 20‐amino‐acid repeats (green) can bind β‐catenin with a very high affinity when they are phosphorylated. The SAMP (serine–alanine–methionine–proline) repeats (red) represent docking sites for axin/conductin. Both the latter motifs are crucial for efficient degradation of β‐catenin, because they bring β‐catenin and axin/conductin in close proximity. The mutation cluster region (MCR) delineates the region where most mutations associated with either familial polyposis or sporadic colorectal cancer have been found. These mutations interrupt the open reading frame and result in the production of truncated APC fragments lacking roughly the C‐terminal half. As an example, the structure below shows truncated APC in the SW480 colon cancer cell line. (B) The frequency of germ line and somatic mutations from patients with FAP is shown as a function of the position of the mutation in the APC sequence. The scheme was taken from.6

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.