Abstract
Staphylococci represent the most commonly encountered blood culture isolates. Differentiating Staphylococcus aureus from coagulase-negative staphylococci (CoNS) is important in guiding empirical therapy, especially since the majority of CoNS are contaminants. This study evaluated three rapid methods for the direct identification of S. aureus from blood cultures. A total of 157 patient blood cultures with gram stains showing gram-positive cocci in clusters were included. The following assays were evaluated: API RAPIDEC staph (API) (bioMerieux, Durham, N.C.), the tube coagulase test (TCT) read at 4 h, and peptide nucleic acid (PNA) fluorescence in situ hybridization (FISH) (AdvanDx, Woburn, Mass.). All assays yielded results of S. aureus or non-S. aureus. The direct rapid results were compared to results obtained with isolated colonies using the AccuProbe Staphylococcus aureus Culture Identification Test (Gen-Probe, San Diego, Calif.). API, TCT, and PNA FISH exhibited sensitivities of 96, 84, and 99% and specificities of 99, 100, and 100%, respectively. Direct identification testing by any of these three assays yielded acceptable performance and timely results. This ability to accurately detect S. aureus in blood cultures gives the physician information with which to initiate or tailor antimicrobial therapy. Coupled with direct susceptibility testing of positive blood culture broths, the patient and institution may experience improved outcomes.
Staphylococci are the most commonly isolated organisms from blood cultures. The majority of these are coagulase-negative staphylococci (CoNS), many of which are not clinically significant and are likely contaminants (5, 8, 13, 15). Thus, timely differentiation of staphylococci in blood cultures is instrumental in deciding on the use of empirical antimicrobial therapy.
Traditional identification of staphylococcal blood culture isolates requires subculture and overnight incubation to obtain isolated colonies. Rapid methods and variations of traditional methods have been developed that allow differentiation of staphylococci in a matter of hours, thus reducing the turnaround time (TAT) by more than 24 h compared to traditional testing.
The API RAPIDEC staph system (API) (bioMerieux, Durham, N.C.), direct tube coagulase test (TCT), and peptide nucleic acid (PNA) fluorescence in situ hybridization (FISH) (AdvanDx, Woburn, Mass.) have been evaluated for the direct identification of S. aureus in blood cultures, but not in parallel with each other. This prospective study sought to evaluate these rapid methods of differentiating S. aureus from CoNS and other staphylococci directly from blood cultures using the AccuProbe S. aureus Culture Identification kit (Gen-Probe, San Diego, Calif.) performed on isolated colonies as the reference method. In addition, a comparison of the work flow for each assay is presented as a practical component when considering applications in clinical microbiology.
MATERIALS AND METHODS
Specimens.
A total of 157 unique patient ESP (TREK Diagnostic Systems, Cleveland, Ohio) 80A aerobic blood cultures positive for gram-positive cocci (GPC) in clusters were included (Table 1). Blood cultures were collected from patients at Lahey Clinic, a 249-bed, suburban, teaching hospital, between April and September 2002. A greater number of clinically significant S. aureus isolates were preferred for testing in order to challenge the sensitivity of the assays while maximizing the number of API tests. API strips contain three tests but must be used within 2 days of opening the strip. Therefore, instances in which two bottles tested positive but only one API test remained led us to preferentially include S. aureus. CoNS were included without regard for clinical significance.
TABLE 1.
Isolates tested in this study
| Organism | No. of isolates tested |
|---|---|
| S. aureus | 69 |
| CoNS | 87 |
| Micrococcus sp. | 1 |
| Total | 157 |
API.
The API RAPIDEC staph system detects the production of an S. aureus-specific enzyme, aurease. Aurease undergoes a reaction with prothrombin, and the resulting product cleaves a fluorescent peptide in the test well, thereby liberating fluorescence. When used directly with blood cultures, this assay yields results of S. aureus or non-S. aureus. (This assay is capable of identifying S. saprophyticus when using isolated colonies based on a β-galactosidase reaction.) A 10-ml aliquot of the blood culture broth was added to a Vacutainer serum separator tube (BD Vacutainer Systems, Franklin Lakes, N.J.) and was centrifuged at 1,300 × g for 10 min to obtain a bacterial pellet. This modification of the manufacturer's two-spin protocol is similar to that described by van Griethuysen et al. (18). The resulting pellet was added to 250 μl of demineralized water in the API strip and adjusted to match an internal 4.0 McFarland standard. Then 50 μl of the bacterial suspension was added to a control and test well on the strip. The strip was incubated at 35°C in non-CO2 for 2 h. The wells were examined for fluorescence using a Wood's lamp (365 nm). Positive results were subjectively determined by the presence of more fluorescence in the test well than in the control well.
TCT.
Free coagulase, produced by bacteria such as S. aureus, causes clotting when mixed with plasma. Five drops of blood culture broth was added to a tube containing 0.5 ml of rabbit plasma with EDTA (BBL, Cockeysville, Md.). Tubes were incubated for 4 h in a 35°C heat block. Clotting of any degree at any time indicated the presence S. aureus. For the purpose of this study, a positive TCT result was interpreted as S. aureus and a negative TCT result was interpreted as non-S. aureus. The TCT was evaluated at 2 h but not reported as negative until 4 h.
PNA FISH.
PNA FISH is available as a kit approved by the U.S. Food and Drug Administration containing all necessary reagents and uses a fluorescence-tagged PNA probe for S. aureus-specific 16S rRNA. This assay yields results of S. aureus or non-S. aureus. The protocol has been previously described (11, 12). Briefly, 1 drop of the blood culture broth was added to a slide containing a drop of fixation solution. This was flame fixed and ethanol (70%) fixed for 5 min before adding 1 drop of S. aureus PNA probe (AdvanDx) and a coverslip. The probe and specimen were incubated at for 90 min at 55°C. Coverslips were removed, and slides were immersed in a preheated wash solution for 30 min in a 55°C water bath. After air drying the slides, 1 drop of mounting medium followed by a coverslip was added. Slides were examined under a fluorescence microscope (Nikon, Tokyo, Japan) using a 100-W Hg bulb, a dual-band fluorescein isothiocyanate-Texas Red XF53 filter set (Omega Optical, Brattleboro, Vt.), and a 100× oil objective. The probe is specific for S. aureus; thus, bright green cocci in clusters seen in multiple fields were indicative of a positive S. aureus result.
AccuProbe.
The AccuProbe S. aureus Culture Identification Test is a DNA probe for S. aureus-specific 16S rRNA that yields results of S. aureus or non-S. aureus. AccuProbe results using isolated colonies were the reference standard used for definitive identification of all organisms. For each blood culture, a Trypticase soy agar plate with 5% sheep blood (BBL) was inoculated with a drop of broth for isolation. The plate was incubated overnight at 35°C, and the resulting colonies were used to perform the assay according to manufacturer instructions. A Leader 450i luminometer (Gen-Probe) was used to read the chemiluminescent signal. A result of ≥50,000 relative light units was considered positive for S. aureus.
Quality control.
For each lot and each shipment of API strips, isolated colonies of S. aureus (ATCC 29213) and a stock strain of CoNS were tested. The same quality control strains were tested by TCT, PNA FISH, and AccuProbe each day of testing.
RESULTS
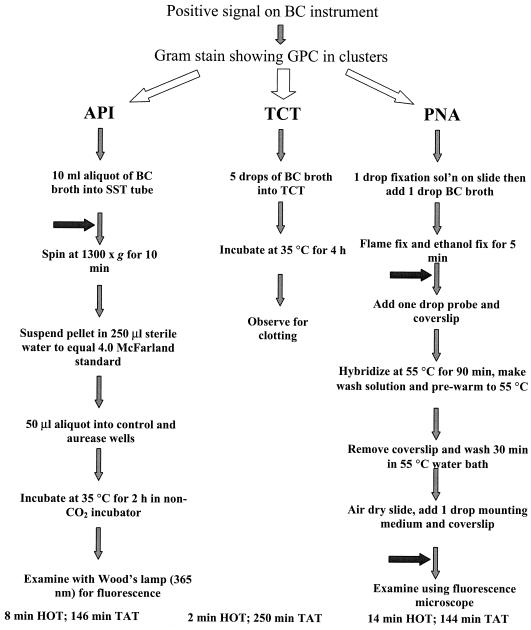
A total of 157 consecutive blood cultures from unique patients positive for GPC in clusters were evaluated. Of these cultures, 69 (44%) were positive for S. aureus as determined by AccuProbe. Specific work flow steps are detailed in Fig. 1. TATs, the time from positive blood culture instrument signal to result as measured once, were 146, 250, and 144 min, and hands-on times (HOTs) were 8, 2, and 14 min for API, TCT, and PNA FISH, respectively. The sensitivity, specificity, positive predictive value, and negative predictive value for each assay were as follows: 95.7, 98.9, 98.5, and 96.7% for API; 84.1, 100, 100, and 88.9% for TCT; and 98.6, 100, 100, and 98.6% for PNA FISH (Table 2). All assays yielded one or more false-negative (FN) result (Table 3). Of these combined 15 FN results, 9 (60%) were methicillin-resistant S. aureus isolates. For all S. aureus isolates tested, 43 of 69 (62%) were methicillin-resistant S. aureus isolates.
FIG. 1.
Flowchart for work with positive blood cultures. Horizontal arrows indicate points at which the assay can be stopped in order to batch specimens. Abbreviations: BC, blood culture; sol'n, solution.
TABLE 2.
Performance of three rapid assays to identify S. aureus directly from blood culturesa
| Assay | S (%) | SP (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|
| API | 95.7 | 98.9 | 98.5 | 96.7 |
| TCT | 84.1 | 100 | 100 | 88.9 |
| PNA FISH | 98.6 | 100 | 100 | 98.9 |
Abbreviations: S, sensitivity; SP, specificity; PPV, positive predictive value; NPV, negative predictive value.
TABLE 3.
Results of three rapid methods to identify S. aureus directly from blood cultures
| Assay and result | No. of isolates yielding AccuProbea result
|
|
|---|---|---|
| + | − | |
| API | ||
| + | 66 | 1 |
| − | 3 | 87 |
| TCTb | ||
| + | 58 | 0 |
| − | 11 | 88 |
| PNA FISH | ||
| + | 68 | 0 |
| − | 1 | 88 |
Definitive method.
TCT read at 4 h.
DISCUSSION
This study evaluated three rapid methods for the direct identification of S. aureus from positive blood cultures. API RAPIDEC staph was 95.7% sensitive and 98.9% specific with 3 FN and 1 false-positive result when ESP positive blood culture medium was used. Speers et al. reported similar performance (98% sensitivity; 100% specificity) using API in conjunction with BACTEC (Becton Dickinson, Sparks, Md.) nonresin bottles (16), which agreed with performance reported by others (9). A modified protocol by van Griethuysen and colleagues provided increased sensitivity (100 versus 91%) with slightly decreased specificity (97 versus 98%) when BACTEC nonresin bottles were used compared to when the protocol in the API package insert was used (18). This modified protocol resembles our procedure in that it requires only one centrifugation step from the positive blood culture broth. Performing the API test at the time the bottle is removed from the blood culture instrument yielded results in 146 min. Results, while subjective, were easily interpreted using a Wood's lamp (365 nm).
TCT read at 4 h was 84.1% sensitive and 100% specific. For the purposes of this evaluation of rapid methods, TCT was read at 4 h only. At 4 h, FN results were common, as 11 of 69 (16%) of S. aureus isolates were missed by reading the TCT after 4 h incubation. Of these 11 isolates giving FN at 4 h, 9 became positive within 24 h while 2 were still negative at 24 h. Other investigators report sensitivities of 79% (C. Tilley, T. Wakefield, and J. Dick, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. C-3, 2001) and 80% (7) for direct TCT at 2 and 3 h, respectively, whereas two reports show better sensitivity for the 2-h direct TCT (2, 3). Cooke et al. reported favorable performance of the 2-h direct TCT and showed no significant difference in performance if the test was read at 2, 6, or 24 h (2). Our in-house data show that direct TCT sensitivity increases to 97% at 24 h (data not shown), suggesting that reading the test at 4 h and again at 24 h may be appropriate and that negative results should not be reported until 24 h. The TCT has the longest TAT, as results are available 250 min after the bottle is removed from the instrument; however, this assay requires the least HOT and equipment.
PNA FISH results showed the greatest correlation with AccuProbe results (98.6% sensitive and 100% specific). This may be due in part to the fact that PNA FISH and AccuProbe are both genetically based tests that target 16S rRNA. Only one FN result was observed. This performance is comparable to that previously reported with ESP bottles (12). Likewise, similar performance has been noted using BacT/Alert FAN media (bioMerieux) (11) and BACTEC bottles with and without resins (12). Although easy to perform, this assay required the most HOT and yielded results in 144 min. (Validation of a condensed PNA FISH protocol reducing TAT to 138 min and HOT to 13 min is currently being evaluated.) PNA FISH requires a fluorescence microscope with a unique dual-band fluorescein isothiocyanate-Texas Red filter set. To our knowledge, no other clinical microbiology applications currently use this filter set (R. Kinoshita, Omega Optical, personal communication). While currently limited to S. aureus detection, PNA FISH testing will offer a potentially flexible testing platform as further probes are implemented for other organisms. PNA probes for Candida albicans (14) and Enterococcus faecalis (K. Oliveira, K. Chapin, M. Musgnug, G. Haase, J. Weber-Heynemann, P. DeGirolami, J. Dakos, and H. Stender, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. D-2003, 2002) have been developed by AdvanDx.
API, TCT, and PNA FISH each have positive qualities, and all may have a place in a GPC algorithm for testing blood cultures depending on the laboratory setting, volume, and staffing. PNA FISH performs the best but requires specialized equipment and a trained technologist. TCT is the easiest to perform but lacks sensitivity. API yields satisfactory performance and is relatively easy to perform but more cumbersome due to a centrifugation step. TCT lends itself to testing as positive blood cultures are flagged; however, API and PNA FISH should be batched for optimal work flow.
Cost for each assay, while relevant, is not calculable given the uniqueness of each diagnostic test setting. Since patients with bloodstream infections are often treated empirically (10), the differentiation of CoNS and S. aureus isolates, especially from a single blood culture, would likely facilitate an appropriate change in therapy and an overall cost reduction (17). Kreger et al. reported that appropriate antimicrobial therapy is key in reducing mortality associated with bacteremia caused by gram-negative organisms but, more importantly, that early appropriate therapy was related to improved patient outcomes (6). Thus, it is important to be able to rapidly and accurately identify bacteria from positive blood cultures. Investigators have shown that rapid identification and susceptibility testing can lead to improved outcomes for the patient and reduced costs for the hospital (1, 4). These benefits may justify the added cost of definitively identifying S. aureus directly from blood cultures.
Acknowledgments
AdvanDx is acknowledged for providing PNA FISH kits.
This work was supported in part by the Robert E. Wise, M.D., Research and Education Institute, Lahey Clinic.
REFERENCES
- 1.Barenfanger, J., C. Drake, and G. Kacich. 1999. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J. Clin. Microbiol. 37:1415-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooke, R. P., and C. T. Jenkins. 1997. Comparison of commercial slide agglutination kits with a tube coagulase test for the rapid identification of Staphylococcus aureus from blood culture. J. Clin. Pathol. 50:164-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis, T. E., D. D. Fuller, and E. C. Aeschleman. 1992. Rapid, direct identification of Staphylococcus aureus and Streptococcus pneumoniae from blood cultures using commercial immunologic kits and modified conventional tests. Diagn. Microbiol. Infect. Dis. 15:295-300. [DOI] [PubMed] [Google Scholar]
- 4.Doern, G. V., R. Vautour, M. Gaudet, and B. Levy. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 32:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirchoff, L. V., and J. N. Sheagren. 1985. Epidemiology and clinical significance of blood cultures positive for coagulase-negative staphylococcus. Infect. Control 12:479-486. [DOI] [PubMed] [Google Scholar]
- 6.Kreger, B. E., D. E. Craven, and W. R. McCabe. 1980. Gram-negative bacteremia IV. Am. J. Med. 68:344-355. [DOI] [PubMed] [Google Scholar]
- 7.McDonald, C. L., and K. C. Chapin. 1995. Rapid identification of Staphylococcus aureus from blood culture bottles by a classic 2-hour tube coagulase test. J. Clin. Microbiol. 33:50-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirrett, S., M. P. Weinstein, L. G. Reimer, M. L. Wilson, and L. B. Reller. 2001. Relevance of the number of positive bottles in determining clinical significance of coagulase-negative staphylococci in blood cultures. J. Clin. Microbiol. 39:3279-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell, C. J., C. Geary, and M. Stevens. 1991. Detection of Staphylococcus aureus in blood cultures: evaluation of a two-hour method. Med. Lab. Sci. 48:106-109. [PubMed] [Google Scholar]
- 10.Munson, E. L., D. J. Diekema, S. E. Beekman, K. C. Chapin, and G. V. Doern. 2003. Detection and treatment of bloodstream infection: laboratory reporting and antimicrobial management. J. Clin. Microbiol. 41:495-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira, K., G. W. Procop, D. Wilson, J. Coull, and H. Stender. 2002. Rapid identification of Staphylococcus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol. 40:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira, K., S. M. Brecher, A. Durbin, D. S. Shapiro, D. R. Schwartz, P. C. De Girolami, J. Dakos, G. W. Procop, D. Wilson, C. S. Hanna, G. Haase, H. Peltroche-Llacsahuanga, K. C. Chapin, M. C. Musgnug, M. H. Levi, C. Shoemaker, and H. Stender. 2003. Direct identification of Staphylococcus aureus from positive blood cultures. J. Clin. Microbiol. 41:889-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter, S. S., S. E. Beekman, J. L. Croco, D. J. Diekema, F. P. Koontz, M. A. Pfaller, and G. V. Doern. 2002. Minimizing the workup of blood culture contaminants: implementation and evaluation of a laboratory-based algorithm. J. Clin. Microbiol. 40:2437-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigby, S., G. W. Procop, G. Haase, D. Wilson, G. Hall, C. Kurtzman, K. Oliveira, S. Von Oy, J. J. Hyldig-Neilsen, J. Coull, and H. Stender. 2002. Fluorescence in situ hybridization with peptide nucleic acid probes for rapid identification of Candida albicans directly from blood culture bottles. J. Clin. Microbiol. 40:2182-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Souvenir, D., D. E. Anderson, Jr., S. Palpant, H. Mroch, S. Askin, J. Anderson, J. Claridge, J. Eiland, C. Malone, M. W. Garrison, P. Watson, and D. M. Campbell. 1998. Blood cultures positive for coagulase-negative staphylococci: antisepsis, pseudobacteremia, and therapy of patients. J. Clin. Microbiol. 36:1923-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speers, D. J., T. R. Olma, and G. L. Gilbert. 1998. Evaluation of four methods for rapid identification of Staphylococcus aureus from blood cultures. J. Clin. Microbiol. 36:1032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trenholme, G. M., R. L. Kaplan, P. H. Karakusis, T. Stine, J. Fuhrer, W. Landau, and S. Levin. 1989. Clinical impact of rapid identification and susceptibility testing of bacterial blood culture isolates. J. Clin. Microbiol. 27:1342-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Griethuysen, A., A. Buiting, W. Goessens, P. van Keulen, R. Wintermans, and J. Kluytmans. 1998. Multicenter evaluation of a modified protocol for the RAPIDEC Staph system for direct identification of Staphylococcus aureus in blood cultures. J. Clin. Microbiol. 36:3707-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]