Abstract
Stroke stimulates neurogenesis in the adult rodent brain. The molecules that mediate stroke-induced neurogenesis are not definitely known. Using microarrays containing approximately 400 known genes associated with stem cell and angiogenesis, we compared transcriptional profiles of subventricular zone (SVZ) tissue with cultured neural progenitor cells isolated from the SVZ 7 days after ischemic stroke in the adult mouse. In SVZ tissue, we found that stroke upregulated 58 genes which are involved in multiple signaling pathways during embryonic development, suggesting that stroke recaptures embryonic molecular signals. In neural progenitor cells cultured in growth medium, 23 gene expressions were increased after stroke and 8 out of 23 genes overlapped with upregulated genes in stroke SVZ tissue. Expression alterations of selected genes were confirmed by real-time RT-PCR and immunohistochemistry. These in vivo and in vitro data provide new insight into the genetic program of adult SVZ neural progenitor cells after stroke and demonstrate gene expression differences between SVZ tissue and cultured SVZ cells.
Keywords: Neurogenesis, subventricular zone (SVZ), microarray, progenitor cell, stroke
Introduction
Cerebral impairment caused by ischemic stroke prompts the proliferation, migration and differentiation of SVZ progenitor cells in the adult brain (Arvidsson et al., 2002; Parent et al., 2002; Jin et al., 2003; Zhang et al., 2004a; Zhang et al., 2004b). However the molecular basis underlying these neurogenic events remains unknown. The identification of molecular pathways and potential modulators of SVZ progenitor cells after stroke is not only critical for the development of stem cell based medical therapies, but also vital for the pharmacological promotion of neurogenesis.
Applying microarrays, several groups have shown complex gene profiles in cultured SVZ neurospheres and SVZ tissue under different conditions (Gurok et al., 2004; Pennartz et al., 2004; Lim et al., 2006). In cultured SVZ neurospheres, Gurok et al. measured gene expression changes during the course of neural progenitor cell differentiation (Gurok et al., 2004). Lim et al. measured in vivo gene expression profiles of adult SVZ tissue and demonstrated RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis (Lim et al., 2006). These in vivo and in vitro studies provide valuable information on gene profiling of SVZ cells from the adult animal.
In the present study, we analyzed the transcriptional patterns of SVZ neural progenitor cells in vivo and in vitro after middle cerebral artery occlusion (MCAo) by means of microarrays. Our data demonstrate that stroke significantly alters gene profile expression and there are distinct differences in gene expression between SVZ tissue and cultured SVZ cells.
Materials and Methods
Animal model of MCAo
Male C57BL/6J mice (3 to 4 months) were employed in this study. The right middle cerebral artery (MCA) was occluded by placement of an embolus at the origin of the MCA, as previously described (Zhang et al., 1997). MCAo evokes a peak increase of neurogenesis 7 days after stroke (Zhang et al., 2004b; Zhang et al., 2001). Therefore, all mice were sacrificed 7 days after MCAo.
Neurosphere culture and tissue samples
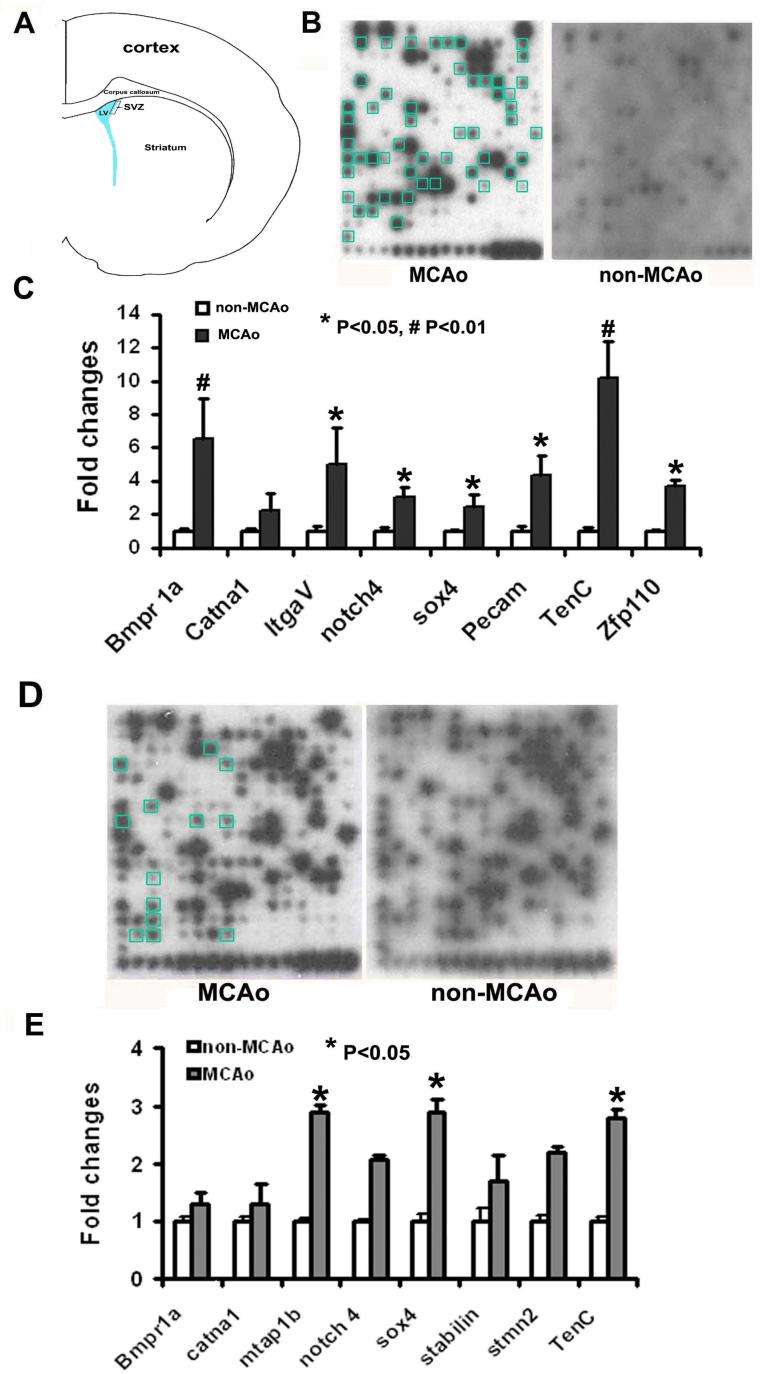
SVZ neural progenitor cells were dissociated from normal (n=3) and MCAo mice (n=3), as previously reported (Zhang et al., 2004a; Reynolds et al., 1992; Chiasson et al., 1999; Morshead et al., 2002). Briefly, brain localized to bregma 2.2 to −0.3 mm was cut into a coronal section and the SVZ (2 mm width) from the lateral wall of the lateral ventricle was aseptically removed (Fig. 1A). The ependymal cells were not included. The cells were plated at a density of 2×104 cells per milliliter in growth medium which includes DMEM/F-12 medium (Invitrogen Corporation, Carlsbad, CA, USA), epidermal growth factor (EGF, 20 ng/ml, R&D System, Minneapolis, MN, USA), and basic fibroblast growth factor (bFGF, 20 ng/ml, R&D System, Minneapolis, MN, USA). SVZ cells were retained in a CO2 incubator for 7 days and processed for microarray and RT-PCR analysis. SVZ tissue isolated from the ipsilateral hemispheres of rats subjected to MCAo (n=3) and hemispheres of normal rats (n=3), were pooled and stored at −80°C freezer until used in the microarray assay.
Figure 1.
A schematic of a coronal brain section shows a box where SVZ tissue was harvested for microarray analysis and the ependymal cells were not included (A). Total RNAs obtained from SVZ tissue of normal mice or mice subjected to 7 day MCAo were extracted and microarray was performed by means of the non-radioactive GEArray Q series cDNA expression array filters. Panel B shows gene profiles of GE mouse stem cell cDNA chip from representative stroke and non-stroke SVZ tissues. Real-time RT-PCR analysis confirmed selective upregulated-genes detected on the microarrays in SVZ tissue (C). Panel D shows gene profiles of cultured SVZ cells harvested from normal mice and mice subjected from MCAo. Panel E shows real-time RT-PCR results of selective upregulated-genes in cultured SVZ cells. SVZ cells were cultured in the growth medium for 7 days. Blue boxes in panels B (MCAo) and D (MCAo) represent upregulated genes. *P<0.05, #P<0.01, n=5-6 /group, LV = lateral ventricle.
RNA isolation and gene expression microarray
Total RNAs from SVZ tissue and neurospheres were extracted using an RNeasy spin column purification kit (Qiagen, Valencia, CA, USA) in accordance with the manufacturer's procedure. To remove possible genomic DNA contamination, RNase-free DNase was used during the RNA purification steps.
The non-radioactive GEArray Q series cDNA expression array filters (MM-601.2N; SuperArray Incorporation, Frederick, MD, USA) were used and hybridization procedures were performed according to manufacturer's instructions. The biotin dUTP-labeled cDNA probes (biotin UTP-labeled oligo probes were applied in Oligo GEArray) were specifically generated in the presence of a designed set of gene-specific primers using total RNA (4 μg) and 1μl reverse transcriptase. The array filters were hybridized with biotin-labeled probes at 60°C for 17 hr. The filters were then washed twice with 2× saline sodium citrate buffer (SSC)/1% sodium dodecyl sulfate (SDS) and then twice with 0.1 x SSC/1% SDS at 60°C for 15 min each. Chemiluminescent detection steps were performed by subsequent incubation of the filters with alkaline phosphatase-conjugated streptavidin and CDP-Star substrate.
Data processing
For quantification, intensity of spots was measured using GEArray Expression Analysis Suite software (http://GEAsuite.superarray.com, SuperArray Incorporation, USA). Briefly, the lowest density spot on the array was found, and the average density across that spot was used as the background correction setting. Array spots were considered “present” only if genes were between the 25% and 75% quartile for normalization. To reduce the contamination by adjacent spots, “clover on” mode was used, which considers the four individual spots as a form of border for the capture of expression data. Total density was divided by the number of pixels to obtain average intensities that were used to compare gene expression levels between control and MCAo groups.
Reverse transcription and Quantitative real-time RT-PCR
Total RNAs isolated from SVZ tissue (n=6 rats) and cultured SVZ cells (n=4 rats) were processed by reverse-transcription. Real-time PCR was performed in ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) by using SYBR Green PCR Master Mix (Applied Biosystems) with 3-stage program parameters provided by the manufacturer, as follows: 2 min at 50°C to require optimal AmpErase uracil-N-glycosylase activity, 10 min at 95°C to activate AmpliTaq Gold DNA polymerase, and then each cycle 15 s at 95°C, 1 min at 60°C for 50 cycles. 2 min at 50°C, 10 min at 95°C, and then 40 cycles of 15 s at 95°C and 1 min at 60°C. Table 1 lists primers (Invitrogen Incorporation, Carlsbad, CA, USA) specific for the genes examined in the present study. Each sample was tested in triplicate and data obtained from three independent experiments were expressed as a subtraction of the quantity of specific transcripts to the quantity of the control gene (β-actin) in mean arbitrary units. CT values were quantified by the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Table 1.
Primer sequences of genes applied in real-time RT-PCR
| Genes | Sense | Anti-sense | Size |
|---|---|---|---|
| β-actin | 5′-CCATCATGAAGTGTGACGTTG | 5'-CAATGATCTTGATCTTCATGGTG | 150bp |
| Bmpr1a | 5′-ATGCAAGGATTCACCGAAAG | 5′-AACAACAGGGGGCAGTGTAG | 100bp |
| Catna1 | 5′-CTACGTGGCTTCCACCAAAT | 5′-TCTCTTCACCAACGGCTTCT | 109bp |
| CCL2 | 5′-CCCAATGAGTAGGCTGGAGA | 5′-TCTGGACCCATTCCTTCTTG | 125bp |
| CXCL 10 | 5′-AAGTGCTGCCGTCATTTTCT | 5′-CTATGGCCCTCATTCTCAC | 128bp |
| Edg1 | 5′-GGCCCCTCTCTTCATCCTAC | 5′-GATGATGGGGTTGGTACCTG | 124bp |
| Ephrin A1 | 5′-ATCAGGAATCCCAGTGC TG | 5′-CAATGCTGTGCAAAACCTGT | 134bp |
| Ephrin B2 | 5′-AATCTCCTGGGTTCCGAAGT | 5′-GTCTCCTGCGGTACTTGAGC | 111bp |
| Hgf | 5′-AGGAACAGGGGCTTTACGTT | 5′-GTCAAATTCATGGCCAAACC | 126bp |
| Hif1α | 5′-GAAATGGCCCAGTGAGAAAA | 5′-CTTCCACGTTGCTGACTTGA | 119bp |
| Mtap 1b | 5′-AACCTCATCTCCCCTGACCT | 5′-TGTTGGGTTCTGGGTCTTTC | 73bp |
| Notch4 | 5′-GCTCTTGCCACTCAATTTCC | 5′-GAGATAGCCTCAGGCAGGTG | 101bp |
| PECAM | 5′-TCACCATCAACAGCATCCAT | 5′-GGTGCTGAGACCTGCTTTTC | 120bp |
| Stmn2 | 5′-GCAATGGCCTACAAGGAAAA | 5′-TTCACCTCCATGTCGTCGTA | 116bp |
| Stabilin 1 | 5′-ACCTGTCATGGGAGAGTTGG | 5′-ACAGTGAAAGGTCCGTCACC | 110bp |
| Sox4 | 5′-AGTGAAGCGCGTCTACCTGT | 5′-TTCGTACAACCCCAGTGGAT | 102bp |
| TenC | 5′-TGGAGTACGAGCTGCATGAC | 5′-AAACTTGGTGGCGATGGTAG | 101bp |
| Thbs2 | 5′-GTGATGTCACCAGCAACACC | 5′-CAAGTCACGGAGCATGAAGA | 135bp |
| Timp 1 | 5′-CATGGAAAGCCTCTGTGGAT | 5′-AAGAAGCTGCAGGCATTGAT | 111bp |
| Zfp110 | 5′-CAGAAGGGCACTGGAAAGAG | 5′-TATGGAGGAGCCATGTGTCA | 141bp |
Immunohistochemistry and immunocytochemistry
For immunohistochemistry, frozen sections were air-dried for 30 seconds, washed with PBS three times (5 min each), followed by fixation with 4% paraformaldehyde or acetone for 20 min at room temperature, and then blocked with 1% bovine serum aluminum (BSA) in PBS for 30 min. The following primary antibodies were applied for 1 hr at room temperature: goat polyclonal antibody against, mouse monoclonal antibody against Tenascin C (1:500, Abcam, Cambridge, MA, USA). After three washes in PBS (5 min each), biotinylated secondary antibodies (1:200, Vector, Burlingame, CA, USA) were applied for 30 min at room temperature. The reaction product was detected using 3'-3'-diaminobenzidine- tetrahydrochloride (DAB, Sigma, St. Louis, MO, USA). Omitting primary antibodies was used as a negative control.
For double immunofluorescence, brain sections were incubated overnight in a mixture of two primary antibodies raised in different species (ie, rabbit anti-GFAP, mouse anti-nestin and mouse anti-Tuj1). Species-specific CY3-and FITC-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA) were used to visualize double-fluorescent immunostaining with Axioplan 2 fluorescent microscope (Carl Zeiss, Thornwood, NY, USA). Confocal microscopy was performed with a LSM 510 Meta (Carl Zeiss, Thornwood, NY, USA) and images were obtained at 2.2-μm optical sections. Specificity of immunostaining was confirmed by omitting the primary antibodies.
Quantification
Semi-quantitative measurements of immunoreactive cells were performed according to our previously published methods (Zhang et al., 1999; Zhang et al., 2005). Briefly, immunoreactive cells in the non-stroke and stroke SVZ were digitized under a 40x objective (BX40; Olympus Optical) using a 3-CCD color video camera (DXC-970MD; Sony, Tokyo, Japan) interfaced with an MCID image analysis system. The data are presented as a percentage of positive immunoreactivity area within the total SVZ area (pixel).
Statistical analysis
Student's t test was performed to compare immunoreactive cells in the SVZ between the non-stroke and stroke groups. The data are presented as mean ± SD; P < 0.05 was taken as a significant difference.
Results
General transcription profiles in SVZ neurospheres and SVZ tissue
To analyze genes involved in neurogenesis and angiogenesis after stroke, we employed the GEArray S Series Mouse Stem Cell Gene Array containing 258 known genes and the Oligo GEArray Mouse Angiogenesis Microarray containing 113 known genes (www.superarray.com). Figures 1 demonstrated gene profiles in MCAo and normal SVZ tissue from representative mouse stem cell gene arrays. To quantify the gene expression, all clones with more than 1.5 fold estimated differences were subjected to further evaluation. This threshold is based on a statistical analysis using online software provided by Superarray Inc, which has been widely employed (Campos et al, 2003).
In SVZ tissue and cultured SVZ neural progenitor cells obtained from mice subjected to MCAo, 58 and 23, respectively, of 371 genes were upregulated compared with SVZ tissue and SVZ neural progenitor cells obtained from normal mice (Fig. 1, 2 and Table 2, 3). Many genes are associated with the development of neural progenitor cells. Eight additional novel genes upregulated by MCAo overlapped between SVZ tissue and cultured SVZ cells (Table 2, 3), which included Hif 1α, a transcription factor that is essential for adaptive responses of the cell to hypoxia (Ran et al., 2005; Sharp et al., 2004).
Figure 2.

Panels A and B are the results of real-time RT-PCR analysis showing mRNA levels of selective genes that were upregulated on the Oligo GEArray Mouse Angiogenesis microarray in SVZ tissue (A) and cultured SVZ neuropsheres (B). *P<0.05, #P<0.01, n=5-6 /group.
Table 2.
Gene expression profile in subventricular zone tissues after MCAo
| Symbol | Gene name | RefSeq number |
Fold change |
|---|---|---|---|
| Angptl4 | Angiopoietin-like 4 | NM_020581 | 1.53 |
| Bmp6 | Bone morphogenetic protein 6 | NM_007556 | 1.82 |
| Bmp8a | Bone morphogenetic protein 8a | NM_007558 | 1.88 |
| Bmpr1a | Bone morphogenetic protein receptor, type 1A | NM_009758 | 1.62 |
| Bmpr2 | Bone morphogenic protein receptor, type II (serine/threonine kinase) |
NM_007561 | 2.01 |
| Catna1 | Catenin alpha 1 | NM_009818 | 2.07 |
| Catna2 | Catenin alpha 2 | NM_009819 | 1.67 |
| CCL2 * | Chemokine (C-C motif) ligand 2 | NM_011333 | 2.99 |
| Cd34 | CD34 antigen | NM_133654 | 2.46 |
| CXCL10* | Chemokine (C-X-C motif) ligand 10 | NM_021274 | 9.92 |
| Cst3 | Cystatin C | NM_009976 | 2.26 |
| Erbb2ip | Erbb2 interacting protein | NM_021563 | 1.95 |
| Fgf12 | Fibroblast growth factor 12 | NM_010199 | 1.94 |
| Fgf22 | Fibroblast growth factor 22 | NM_023304 | 1.97 |
| Fgf23 | Fibroblast growth factor 23 | NM_022657 | 2.30 |
| Fgf3 | Fibroblast growth factor 3 | NM_008007 | 2.32 |
| Fgf5 | Fibroblast growth factor 5 | NM_010203 | 3.11 |
| Fgfr1 | Fibroblast growth factor receptor 1 | NM_010206 | 2.39 |
| Fzd1 | Frizzled homolog 1 (Drosophila) | NM_021457 | 2.58 |
| Fzd3 | Frizzled homolog 3 (Drosophila) | NM_021458 | 1.68 |
| Fzd8 | Frizzled homolog 8 (Drosophila) | NM_008058 | 2.09 |
| Gata2 * | GATA binding protein 2 | NM_008090 | 1.62 |
| Gcm2 | Glial cells missing homolog 2 (Drosophila) | NM_008104 | 1.59 |
| Gdf9 | Growth differentiation factor 9 | NM_008110 | 2.36 |
| Gjb1 * | Gap junction membrane channel protein beta 1 | NM_008124 | 1.96 |
| Hif1 α * | Hypoxia inducible factor 1, alpha subunit | NM_010431 | 2.29 |
| Icam5 * | Intercellular adhesion molecule 5, telencephalin | NM_008319 | 1.92 |
| Il6st | Interleukin 6 signal transducer | NM_010560 | 3.10 |
| Itga6 | Integrin alpha 6 | NM_008397 | 1.95 |
| Itga7 | Integrin alpha 7 | NM_008398 | 3.19 |
| Itgav | Integrin alpha V | NM_008402 | 4.23 |
| Itgax | Integrin alpha X | NM_021334 | 3.18 |
| Itgb4 | Integrin beta 4 | L04678 | 2.03 |
| Lifr | Leukemia inhibitory factor receptor | NM_013584 | 2.92 |
| Mmp2 | Matrix metalloproteinase 2 | NM_008610 | 1.63 |
| Mbp | Myelin basic protein | NM_010777 | 2.41 |
| Mtap2 | Microtubule-associated protein 2 | NM_008632 | 2.35 |
| Mtap1b | Microtubule-associated protein 1 B | NM_008634 | 1.57 |
| Myl4 | Myosin, light polypeptide 4, alkali; atrial, embryonic | NM_010858 | 1.84 |
| Ncam2 | Neural cell adhesion molecule 2 | NM_010954 | 2.44 |
| Nefl | Neurofilament, light polypeptide | NM_010910 | 3.05 |
| Nkx2-2 | NK2 transcription factor related, locus 2 (Drosophila) | NM_010919 | 3.38 |
| Nodal | Nodal | NM_013611 | 2.04 |
| Notch4 | Notch gene homolog 4 (Drosophila) | NM_010929 | 1.99 |
| Ntrk2 | Neurotrophic tyrosine kinase, receptor, type 2 | NM_008745 | 3.43 |
| Olig1 | Oligodendrocyte transcription factor 1 | NM_016968 | 2.86 |
| Plp | Proteolipid protein (myelin) | NM_011123 | 1.55 |
| Prdc | Protein related to DAN and cerberus | NM_011825 | 2.31 |
| Ptch | Patched homolog | NM_008957 | 1.75 |
| Stmn2 | Stathmin-like 2 | NM_025285 | 3.36 |
| Slc1a2 | Solute carrier family 1, member 2 | NM_011393 | 2.19 |
| Sox3 | SRY-box containing gene 3 | NM_009237 | 2.79 |
| Sox4 * | SRY-box containing gene 4 | NM_009238 | 2.39 |
| Sphk1 | Sphingosine kinase 1 | NM_025367 | 1.93 |
| Stab1 | Stabilin 1 | NM_138672 | 2.14 |
| Timp1 * | Tissue inhibitor of metalloproteinase 1 | NM_011593 | 2.85 |
| Utf1 | Undifferentiated embryonic cell transcription factor 1 | NM_009482 | 2.04 |
| Zfp110 | Zinc finger protein 110 | NM_022981 | 3.99 |
shows the upregulated-genes were detected on microarrays in both SVZ tissue and SVZ neuropsheres.
Table 3.
Gene expression alteration in subventricular zone neurospheres after MCAo
| Symbol | Gene name | RefSeq number |
Fold change |
|---|---|---|---|
| CCL2 * | Chemokine (C-C motif) ligand 2 | NM_011333 | 2.21 |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A (P21) | NM_007669 | 1.65 |
| Ctgf | Connective tissue growth factor | NM_010217 | 1.77 |
| CXCL10* | Chemokine (C-X-C motif) ligand 10 | NM_021274 | 2.95 |
| Dnmt1 | DNA methyltransferase (cytosine-5) 1 | NM_010066 | 1.95 |
| Edg1 | Endothelial differentiation sphingolipid G-protein- coupled receptor 1 |
NM_007901 | 2.75 |
| Efnb2 | Ephrin B2 | NM_010111 | 3.27 |
| Egfr | Epidermal growth factor receptor | NM_007912 | 1.51 |
| Gata2 * | GATA binding protein 2 | NM_008090 | 2.49 |
| Gjb1 * | Gap junction membrane channel protein beta 1 | NM_008124 | 1.52 |
| Hgf | Hepatocyte growth factor | XM_131908 | 4.92 |
| Hif1α * | Hypoxia inducible factor 1, alpha subunit | NM_010431 | 3.47 |
| Icam5 * | Intercellular adhesion molecule 5, telencephalin | NM_008319 | 1.81 |
| Igf1r | Insulin-like growth factor I receptor | NM_010513 | 1.60 |
| Notch1 | Notch gene homolog 1 (Drosophila) | NM_008714 | 1.89 |
| Pofut1 | Protein O-fucosyltransferase 1 | NM_080463 | 1.67 |
| S100b | S100 protein, beta polypeptide, neural | NM_009115 | 1.68 |
| Sox4 * | SRY-box containing gene 4 | NM_009238 | 1.76 |
| Thy1 | Thymus cell antigen 1, theta | NM_009382 | 2.16 |
| Thbs2 | Thrombospondin 2 | NM_011581 | 1.81 |
| Timp1 * | Tissue inhibitor of metalloproteinase 1 | NM_011593 | 1.89 |
| Tenc | Tenascin C | NM_011607 | 3.46 |
| Vim | Vimentin | NM_011701 | 1.59 |
shows the upregulated-genes were detected on microarrays in both SVZ tissue and SVZ neuropsheres.
Comparison of expression profiles between SVZ tissue and cultured SVZ neurospheres induced by MCAo
To identify the difference in gene expression profiles after MCAo, specific gene subfamilies were compared. Fibroblast growth factors (FGFs) are multifunctional signaling proteins that regulate developmental processes and adult physiology. Compared with normal SVZ, we found that multiple FGF subtypes and FGF receptor were upregulated in MCAo SVZ tissue (Table 2). However, FGF subtype genes were not upregulated in cultured SVZ neural progenitor cells (Table 3).
Integrins are cell surface receptors involved in cell-cell and cell-extra cellular matrix interactions (Campos LS, 2005). Several integrins were upregulated in MCAo SVZ tissue (Table 2), whereas MCAo did not strikingly increase integrin expression in cultured neural progenitor cells (Table 3). In addition, several other matrix molecules, for example intercellular adhesion molecule 5 (ICAM 5) and Tenacin C (Tenc) were increased in in vivo SVZ tissue, and in cultured SVZ neurospheres (Table 2 and 3).
Compared with normal SVZ tissue, MCAo dramatically induced transforming growth factor-β signaling related molecules, including Bmp6, Bmp8a, Bmpr1a, Bmpr2 and Nodal expression (Table 2). In contrast, upregulation of TGF- β genes was not detected in MCAo derived neural progenitor cells cultured in the growth medium.
The wingless (Wnt) is a large family of secreted glycoproteins and plays a key role in cell fate specification and CNS patterning (Ciani and Salinas, 2005). Frizzled (Fzd) 1, Fzd3, Fzd8, Catna 1 and Catna 2 were upregulated in stroke SVZ tissue, but not in cultured stroke neural progenitor cells (Table 2).
Confirmation of gene expression profile by Real-Time RT-PCR and immunohistochemistry
To verify stroke-upregulated genes observed in microarrays, we performed real-time RT-PCR analysis for 21 selective genes (Table 1) in SVZ tissue and neurospheres. Upregulation of these genes was detected in MCAo SVZ tissues and cultured SVZ cells compared with that in normal SVZ cells (Fig. 1C, 1E, 2A and 2B), confirming our findings in the microarrays. Immunostaining analysis showed significant (P<0.05) increases of the ipsilateral SVZ cell immunoreactivity of Bmp8a (27.8 ± 5.1% vs 21.1 ± 4.3% for contralateral, n=8), integrin αV (37.5 ± 4.2% vs 18.1 ± 9.4% for contralateral, n=8), and sox 4 (15.0 ± 1.2% vs 9.4 ± 2.6% for contralateral, n=8), which are consistent with our previous findings (Liu et al., 2006). MCAo also resulted in a significant increase of tenascin C positive SVZ cells and double immunofluorescent staining revealed that these cells were GFAP immunoreactive (Fig. 3A to 3F).
Figure 3.

Microphotographs of tenascin C immunoreactive cells in the SVZ. MCAo induced an increase in tenascin C immunoreactive cells in the ipsilateral SVZ (A, ipsi), although tenascin C immunoreactive cells were presented in the contralateral SVZ (A, contral). Panel B shows semi-quantitative data of tenascin C immunoreactivity. Double immunofluorescent staining revealed that tenascin C (TENC) immunoreactive cells (C, D and F, arrows) were GFAP positive (E and F, arrows). LV = lateral ventricle.
Discussion
Using a customized microarray approach, genes that mediate neurogenesis and angiogenesis were screened in adult SVZ progenitor cells in vivo and in vitro after MCAo. Gene profiles in normal SVZ progenitor cells are generally consistent with previous in vivo and in vitro findings (Gurok et al., 2004; Pennartz et al., 2004; Lim et al., 2006). Ischemic stroke triggered many genes in the adult SVZ cells, which are involved in multiple signaling pathways during embryogenesis. These genes included: 1) TGF-beta1 and Bmp superfamily, 2) Wnt family, 3) Notch signals, suggesting that embryonic molecular morphogenes and signals might mediate stroke-induced neurogenesis in the adult SVZ niche (Alvarez-Buylla and Lim., 2004; Van et al., 2005; Sun et al., 2005; James et al., 2005; Bonnert et al., 2006). These gene profiles triggered by stroke may be specific to SVZ cells because in the ischemic cortex and subcortex, stroke upregulates many genes associated with stress, inflammation, apoptosis, trophic factors and immediate early genes (Soriano et al., 2000; Raghavendra et al., 2002; Lu et al., 2004; Vikman and Edvinsson., 2006).
To examine gene profile changes in vivo and in vitro after stroke, we employed SVZ tissue and a neurosphere assay that has been commonly used for studying neurogenesis in vitro (Zhang et al., 2004a; Reynolds and Weiss., 1992; Chiasson et al., 1999; Morshead et al., 2002). The gene profiles of SVZ tissue dissected 7 days after stroke have much in common with the profiles of stroke SVZ cells isolated by laser capture microdissection (Liu et al., 2006). The 7 day time point is the peak stage of neurogenesis in the SVZ cell population after stroke (Zhang et al., 2004a). Stroke upregulated more genes in SVZ tissue than that in the SVZ neurosphere cells cultured in the growth medium, which is expected because the cell population of SVZ tissue is different from that of neurosphere cells which only contain type A, B, and C cells (Alvarez-Buylla and Lim., 2004) where SVZ tissue contains SVZ neural progenitor cells, endothelial cells and microglial cells. Furthermore, neural progenitor cells in SVZ tissue are in different stages of proliferation, differentiation, and migration, which are regulated by a variety of genes, whereas neurospheres employed in the present study were cultured in the growth medium containing high levels of exogenous EGF and bFGF that propagate neurospheres, likely leading to remarkable alterations in their transcriptional profiles (Hack et al., 1993; Gabay et al., 2003; Bonnert et al., 2006).
Our observation that stroke-increased expression of MMP2 and Timp1 is intriguing, based on the emerging data that MMPs and TIMPs are involved in neurogenesis and neuroblast migration (Tsukatani et al., 2003; Yong VW, 2005; Lee et al., 2006; Zhang et al., 2006). MMPs regulate cell migration and invasion (Hu et al., 2006). Neuroblasts in the SVZ of adult rodent travel the rostral migratory stream (RMS), as chains through tunnels formed by astrocytes, to the olfactory bulb where they differentiate into granule and periglomerular neurons throughout adult life (Garcia-Verdugo et al., 1998). Stroke increases the migration speed of neuroblasts and induces neuroblast migration towards the ischemic boundary region (Jin et al., 2003; Zhang et al., 2004a). Thus, increased expression of MMP2 and Timp1 in SVZ cells might promote neuroblast migration. In addition, stroke upregulated tenascin C expression, which mediates neuroblast migration (Peretto et al., 2005).
Although there were discrepancies of gene profile changes in neurospheres and their in vivo counterparts, eight genes upregulated by MCAo overlapped in SVZ tissue and neurospheres, suggesting that cultured SVZ cells retain signals initiated by stroke. Among them, Hif 1 α was strongly upregulated, suggesting that SVZ cells are subjected to hypoxia which could trigger changes of gene profiles (Zhu et al., 2005). Interestingly, when SVZ cells derived from MCAo mice were incubated under normal oxygen conditions, Hif 1α expression was still upregulated in these cells. Our findings are consistent with recent reports that dynamic gene profile changes were found in neurospheres cultured in different media and that SVZ cells remain stable in vitro after brain injury (Gurok et al., 2004, Dizon et al., 2006; Dictuset al., 2007). Therefore, in vivo and in vitro gene profile changes after stroke are complementary, but not identical.
In summary, we provide a dataset in comparison of in vivo and vitro gene expression changes in the adult neural progenitor cells after stroke. Our data show that stroke induces complex gene profile changes in the SVZ niche, which could serve as an initial step for future analysis of the molecular signals that mediate adult neurogenesis after stroke.
Acknowledgements
We are grateful to Cindi Roberts who provided the technical support for immunohistochemistry. This work was supported by NINDS grants PO1 NS23393, PO1 NS42345, RO1NS38292 and RO1HL 6476.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41:683–686. doi: 10.1016/s0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Arvidsson A, Collin T, Kirik D, Kokaia Z, Lindvall O. Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat Med. 2002;8:963–970. doi: 10.1038/nm747. [DOI] [PubMed] [Google Scholar]
- Bonnert TP, Bilsland JG, Guest PC, Heavens R, McLaren D, Dale C, Thakur M, McAllister G, Munoz-Sanjuan I. Molecular characterization of adult mouse subventricular zone progenitor cells during the onset of differentiation. Eur J Neurosci. 2006;24:661–675. doi: 10.1111/j.1460-9568.2006.04912.x. [DOI] [PubMed] [Google Scholar]
- Chiasson BJ, Tropepe V, Morshead CM, van der Kooy D. Adult mammalian forebrain ependymal and subependymal cells demonstrate proliferative potential, but only subependymal cells have neural stem cell characteristics. J Neurosci. 1999;19:4462–4471. doi: 10.1523/JNEUROSCI.19-11-04462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AH, Zhao Y, Pollman MJ, Gibbons GH. DNA microarray profiling to identify angiotensin-responsive genes in vascular smooth muscle cells: potential mediators of vascular disease. Circ Res. 2003;92:111–118. doi: 10.1161/01.res.0000049100.22673.f6. [DOI] [PubMed] [Google Scholar]
- Campos LS. Beta1 integrins and neural stem cells: making sense of the extracellular environment. Bioessays. 2005;27:698–707. doi: 10.1002/bies.20256. [DOI] [PubMed] [Google Scholar]
- Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- Dictus C, Tronnier V, Unterberg A, Herold-Mende C. Comparative analysis of in vitro conditions for rat adult neural progenitor cells. J Neurosci Methods. 2007 Jan 4; doi: 10.1016/j.jneumeth.2006.11.012. online. [DOI] [PubMed] [Google Scholar]
- Dizon ML, Shin L, Sundholm-Peters NL, Kang E, Szele FG. Subventricular zone cells remain stable in vitro after brain injury. Neuroscience. 2006;142:717–725. doi: 10.1016/j.neuroscience.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Gabay L, Lowell S, Rubin LL, Anderson DJ. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–499. doi: 10.1016/s0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- Garcia-Verdugo JM, Doetsch F, Wichterle H, Lim DA, Alvarez-Buylla A. Architecture and cell types of the adult subventricular zone: in search of the stem cells. J Neurobiol. 1998;36:234–248. doi: 10.1002/(sici)1097-4695(199808)36:2<234::aid-neu10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Gurok U, Steinhoff C, Lipkowitz B, Ropers HH, Scharff C, Nuber UA. Gene expression changes in the course of neural progenitor cell differentiation. J Neurosci. 2004;24:5982–6002. doi: 10.1523/JNEUROSCI.0809-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack N, Sue-A-Quan A, Mills GB, Skorecki KL. Expression of human tyrosine kinase-negative epidermal growth factor receptor amplifies signaling through endogenous murine epidermal growth factor receptor. J Biol Chem. 1993;268:26441–26446. [PubMed] [Google Scholar]
- Hu B, Jarzynka MJ, Guo P, Imanishi Y, Schlaepfer DD, Cheng SY. Angiopoietin 2 induces glioma cell invasion by stimulating matrix metalloprotease 2 expression through the alphavbeta1 integrin and focal adhesion kinase signaling pathway. Cancer Res. 2006;66:775–783. doi: 10.1158/0008-5472.CAN-05-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Jin K, Sun Y, Xie L, Peel A, Mao XO, Batteur S, Greenberg DA. Directed migration of neuronal precursors into the ischemic cerebral cortex and striatum. Mol. Cell. Neurosci. 2003;24:171–189. doi: 10.1016/s1044-7431(03)00159-3. [DOI] [PubMed] [Google Scholar]
- Lee SR, Kim HY, Rogowska J, Zhao BQ, Bhide P, Parent JM, Lo EH. Involvement of matrix metalloproteinase in neuroblast cell migration from the subventricular zone after stroke. J Neurosci. 2006;26:3491–3495. doi: 10.1523/JNEUROSCI.4085-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Suarez-Farinas M, Naef F, Hacker CR, Menn B, Takebayashi H, Magnasco M, Patil N, Alvarez-Buylla A. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol Cell Neurosci. 2006;31:131–148. doi: 10.1016/j.mcn.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Liu XS, Zhang ZG, Zhang RL, Gregg S, Morris DC, Wang Y, Chopp M. Stroke induces gene profile changes associated with neurogenesis and angiogenesis in adult subventricular zone progenitor cells. J Cereb Blood Flow Metab. 2006 doi: 10.1038/sj.jcbfm.9600371. In press. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu XC, Williams AJ, Yao C, Berti R, Hartings JA, Whipple R, Vahey MT, Polavarapu RG, Woller KL, Tortella FC, Dave JR. Microarray analysis of acute and delayed gene expression profile in rats after focal ischemic brain injury and reperfusion. J Neurosci Res. 2004;77:843–857. doi: 10.1002/jnr.20218. [DOI] [PubMed] [Google Scholar]
- Morshead CM, Benveniste P, Iscove NN, van der Kooy D. Hematopoietic competence is a rare property of neural stem cells that may depend on genetic and epigenetic alterations. Nat Med. 2002;8:268–273. doi: 10.1038/nm0302-268. [DOI] [PubMed] [Google Scholar]
- Parent JM, Vexler ZS, Gong C, Derugin N, Ferriero DM. Rat forebrain neurogenesis and striatal neuron replacement after focal stroke. Ann. Neurol. 2002:802–813. doi: 10.1002/ana.10393. [DOI] [PubMed] [Google Scholar]
- Pennartz S, Belvindrah R, Tomiuk S, Zimmer C, Hofmann K, Conradt M, Bosio A, Cremer H. Purification of neuronal precursors from the adult mouse brain: comprehensive gene expression analysis provides new insights into the control of cell migration, differentiation, and homeostasis. Mol Cell Neurosci. 2004;25:692–706. doi: 10.1016/j.mcn.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Peretto P, Giachino C, Aimar P, Fasolo A, Bonfanti L. Chain formation and glial tube assembly in the shift from neonatal to adult subventricular zone of the rodent forebrain. J Comp Neurol. 2005;487:407–427. doi: 10.1002/cne.20576. [DOI] [PubMed] [Google Scholar]
- Raghavendra Rao VL, Bowen KK, Dhodda VK, Song G, Franklin JL, Gavva NR, Dempsey RJ. Gene expression analysis of spontaneously hypertensive rat cerebral cortex following transient focal cerebral ischemia. J Neurochem. 2002;83:1072–1086. doi: 10.1046/j.1471-4159.2002.01208.x. [DOI] [PubMed] [Google Scholar]
- Ran R, Xu H, Lu A, Bernaudin M, Sharp FR. Hypoxia preconditioning in the brain. Dev Neurosci. 2005;27:87–92. doi: 10.1159/000085979. [DOI] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Sharp FR, Ran R, Lu A, Tang Y, Strauss KI, Glass T, Ardizzone T, Bernaudin M. Hypoxic preconditioning protects against ischemic brain injury. NeuroRx. 2004;1:26–35. doi: 10.1602/neurorx.1.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano MA, Tessier M, Certa U, Gill R. Parallel gene expression monitoring using oligonucleotide probe arrays of multiple transcripts with an animal model of focal ischemia. J Cereb Blood Flow Metab. 2000;20:1045–1055. doi: 10.1097/00004647-200007000-00004. [DOI] [PubMed] [Google Scholar]
- Sun Y, Lowther W, Kato K, Bianco C, Kenney N, Strizzi L, Raafat D, Hirota M, Khan NI, Bargo S, Jones B, Salomon D, Callahan R. Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-beta signaling. Oncogene. 2005;24:5365–5374. doi: 10.1038/sj.onc.1208528. [DOI] [PubMed] [Google Scholar]
- Tsukatani T, Fillmore HL, Hamilton HR, Holbrook EH, Costanzo RM. Matrix metalloproteinase expression in the olfactory epithelium. Neuroreport. 2003;14(8):1135–1140. doi: 10.1097/01.wnr.0000075306.76650.bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Vikman P, Edvinsson L. Gene expression profiling in the human middle cerebral artery after cerebral ischemia. Eur J Neurol. 2006;13:1324–1332. doi: 10.1111/j.1468-1331.2006.01496.x. [DOI] [PubMed] [Google Scholar]
- Yong VW. Metalloproteinases: mediators of pathology and regeneration in the CNS. Nat Rev Neurosci. 2005;6:931–944. doi: 10.1038/nrn1807. [DOI] [PubMed] [Google Scholar]
- Zhang R, Zhang Z, Wang L, Wang Y, Gousev A, Zhang L, Ho KL, Morshead C, Chopp M. Activated neural stem cells contribute to stroke-induced neurogenesis and neuroblast migration toward the infarct boundary in adult rats. J. Cereb. Blood. Flow. Metab. 2004a;24:441–448. doi: 10.1097/00004647-200404000-00009. [DOI] [PubMed] [Google Scholar]
- Zhang R, Zhang Z, Zhang C, Zhang L, Robin A, Wang Y, Lu M, Chopp M. Stroke transiently increases subventricular zone cell division from asymmetric to symmetric and increases neuronal differentiation in the adult rat. J. Neurosci. 2004b;24:5810–5815. doi: 10.1523/JNEUROSCI.1109-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RL, Chopp M, Zhang ZG, Jiang Q, Ewing JR. A rat model of embolic focal cerebral ischemia. Brain Research. 1997;766:83–92. doi: 10.1016/s0006-8993(97)00580-5. [DOI] [PubMed] [Google Scholar]
- Zhang RL, Zhang ZG, Zhang L, Chopp M. Proliferation and differentiation of progenitor cells in the cortex and the subventricular zone in the adult rat after focal cerebral ischemia. Neuroscience. 2001;105:33–41. doi: 10.1016/s0306-4522(01)00117-8. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Davies K, Prostak J, Fenstermacher J, Chopp M. Quantitation of microvascular plasma perfusion and neuronal microtubule-associated protein in ischemic mouse brain by laser-scanning confocal microscopy. J. Cereb. Blood. Flow. Metab. 1999;19:68–78. doi: 10.1097/00004647-199901000-00008. [DOI] [PubMed] [Google Scholar]
- Zhang RL, Zhang ZG, Chopp M. Neurogenesis in the adult ischemic brain: generation, migration, survival, and restorative therapy. Neuroscientist. 2005;11:408–416. doi: 10.1177/1073858405278865. [DOI] [PubMed] [Google Scholar]
- Zhang ZG, Letourneau Y, Zhang RL, Gregg SR, Hozeska A, Wang Y, Morris D, Chopp M. Atypical PKC ζ mediates neuroblast migration from the subventricular zone toward the ischemic boundary regions. Stroke. 2006;37:629. [Google Scholar]
- Zhu LL, Wu LY, Yew DT, Fan M. Effects of hypoxia on the proliferation and differentiation of NSCs. Mol Neurobiol. 2005;31:231–242. doi: 10.1385/MN:31:1-3:231. [DOI] [PubMed] [Google Scholar]