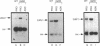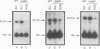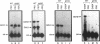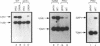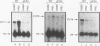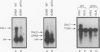Abstract
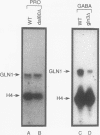
We demonstrate that expression of the UGA1, CAN1, GAP1, PUT1, PUT2, PUT4, and DAL4 genes is sensitive to nitrogen catabolite repression. The expression of all these genes, with the exception of UGA1 and PUT2, also required a functional GLN3 protein. In addition, GLN3 protein was required for expression of the DAL1, DAL2, DAL7, GDH1, and GDH2 genes. The UGA1, CAN1, GAP1, and DAL4 genes markedly increased their expression when the DAL80 locus, encoding a negative regulatory element, was disrupted. Expression of the GDH1, PUT1, PUT2, and PUT4 genes also responded to DAL80 disruption, but much more modestly. Expression of GLN1 and GDH2 exhibited parallel responses to the provision of asparagine and glutamine as nitrogen sources but did not follow the regulatory responses noted above for the nitrogen catabolic genes such as DAL5. Steady-state mRNA levels of both genes did not significantly decrease when glutamine was provided as nitrogen source but were lowered by the provision of asparagine. They also did not respond to disruption of DAL80.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André B., Jauniaux J. C. Nucleotide sequence of the yeast UGA1 gene encoding GABA transaminase. Nucleic Acids Res. 1990 May 25;18(10):3049–3049. doi: 10.1093/nar/18.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechet J., Greenson M., Wiame J. M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970 Jan;12(1):31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Benjamin P. M., Wu J. I., Mitchell A. P., Magasanik B. Three regulatory systems control expression of glutamine synthetase in Saccharomyces cerevisiae at the level of transcription. Mol Gen Genet. 1989 Jun;217(2-3):370–377. doi: 10.1007/BF02464906. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Boonchird C., Messenguy F., Dubois E. Characterization of the yeast ARG5,6 gene: determination of the nucleotide sequence, analysis of the control region and of ARG5,6 transcript. Mol Gen Genet. 1991 Apr;226(1-2):154–166. doi: 10.1007/BF00273599. [DOI] [PubMed] [Google Scholar]
- Brandriss M. C. Evidence for positive regulation of the proline utilization pathway in Saccharomyces cerevisiae. Genetics. 1987 Nov;117(3):429–435. doi: 10.1093/genetics/117.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckholz R. G., Cooper T. G. The allantoinase (DAL1) gene of Saccharomyces cerevisiae. Yeast. 1991 Dec;7(9):913–923. doi: 10.1002/yea.320070903. [DOI] [PubMed] [Google Scholar]
- Bysani N., Daugherty J. R., Cooper T. G. Saturation mutagenesis of the UASNTR (GATAA) responsible for nitrogen catabolite repression-sensitive transcriptional activation of the allantoin pathway genes in Saccharomyces cerevisiae. J Bacteriol. 1991 Aug;173(16):4977–4982. doi: 10.1128/jb.173.16.4977-4982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chisholm G., Cooper T. G. Isolation and characterization of mutants that produce the allantoin-degrading enzymes constitutively in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1088–1095. doi: 10.1128/mcb.2.9.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Ferguson D., Rai R., Bysani N. The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J Bacteriol. 1990 Feb;172(2):1014–1018. doi: 10.1128/jb.172.2.1014-1018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Kovari L., Sumrada R. A., Park H. D., Luche R. M., Kovari I. Nitrogen catabolite repression of arginase (CAR1) expression in Saccharomyces cerevisiae is derived from regulated inducer exclusion. J Bacteriol. 1992 Jan;174(1):48–55. doi: 10.1128/jb.174.1.48-55.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Rai R., Yoo H. S. Requirement of upstream activation sequences for nitrogen catabolite repression of the allantoin system genes in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5440–5444. doi: 10.1128/mcb.9.12.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert D., Vissers S., André B., Grenson M. The UGA43 negative regulatory gene of Saccharomyces cerevisiae contains both a GATA-1 type zinc finger and a putative leucine zipper. Curr Genet. 1992 Apr;21(4-5):301–307. doi: 10.1007/BF00351687. [DOI] [PubMed] [Google Scholar]
- Crabeel M., Lavalle R., Glansdorff N. Arginine-specific repression in Saccharomyces cerevisiae: kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol Cell Biol. 1990 Mar;10(3):1226–1233. doi: 10.1128/mcb.10.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham T. S., Cooper T. G. Expression of the DAL80 gene, whose product is homologous to the GATA factors and is a negative regulator of multiple nitrogen catabolic genes in Saccharomyces cerevisiae, is sensitive to nitrogen catabolite repression. Mol Cell Biol. 1991 Dec;11(12):6205–6215. doi: 10.1128/mcb.11.12.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop P. C., Roon R. J. L-Asparaginase of Saccharomyces cerevisiae: an extracellular Enzyme. J Bacteriol. 1975 Jun;122(3):1017–1024. doi: 10.1128/jb.122.3.1017-1024.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A. D., Pabo C. O. Fingering too many proteins. Cell. 1988 Jun 3;53(5):675–675. doi: 10.1016/0092-8674(88)90083-9. [DOI] [PubMed] [Google Scholar]
- Genbauffe F. S., Cooper T. G. Induction and repression of the urea amidolyase gene in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Nov;6(11):3954–3964. doi: 10.1128/mcb.6.11.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992 Mar 20;68(6):1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Grenson M., Mousset M., Wiame J. M., Bechet J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim Biophys Acta. 1966 Oct 31;127(2):325–338. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- HIERHOLZER G., HOLZER H. REPRESSION DER SYNTHESE VON DPN-ABHAENGIGER GLUTAMINSAEUREDEHYDROGENASE IN SACCHAROMYCES CEREVISIAE DURCH AMMONIUMIONEN. Biochem Z. 1963 Dec 3;339:175–185. [PubMed] [Google Scholar]
- Hoffmann W. Molecular characterization of the CAN1 locus in Saccharomyces cerevisiae. A transmembrane protein without N-terminal hydrophobic signal sequence. J Biol Chem. 1985 Sep 25;260(21):11831–11837. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauniaux J. C., Grenson M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur J Biochem. 1990 May 31;190(1):39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x. [DOI] [PubMed] [Google Scholar]
- Jauniaux J. C., Vandenbol M., Vissers S., Broman K., Grenson M. Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae. Cloning of the PUT4 gene and study of PUT4 RNA levels in wild-type and mutant strains. Eur J Biochem. 1987 May 4;164(3):601–606. doi: 10.1111/j.1432-1033.1987.tb11169.x. [DOI] [PubMed] [Google Scholar]
- Ko L. J., Yamamoto M., Leonard M. W., George K. M., Ting P., Engel J. D. Murine and human T-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human T-cell receptor delta gene enhancer. Mol Cell Biol. 1991 May;11(5):2778–2784. doi: 10.1128/mcb.11.5.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywicki K. A., Brandriss M. C. Primary structure of the nuclear PUT2 gene involved in the mitochondrial pathway for proline utilization in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2837–2842. doi: 10.1128/mcb.4.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. I., Orkin S. H. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 1990 Nov;4(11):1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- Miller S. M., Magasanik B. Role of the complex upstream region of the GDH2 gene in nitrogen regulation of the NAD-linked glutamate dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Dec;11(12):6229–6247. doi: 10.1128/mcb.11.12.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minehart P. L., Magasanik B. Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1991 Dec;11(12):6216–6228. doi: 10.1128/mcb.11.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minehart P. L., Magasanik B. Sequence of the GLN1 gene of Saccharomyces cerevisiae: role of the upstream region in regulation of glutamine synthetase expression. J Bacteriol. 1992 Mar;174(6):1828–1836. doi: 10.1128/jb.174.6.1828-1836.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Three regulatory systems control production of glutamine synthetase in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2767–2773. doi: 10.1128/mcb.4.12.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasu T., Hall B. D. Nucleotide sequence of the GDH gene coding for the NADP-specific glutamate dehydrogenase of Saccharomyces cerevisiae. Gene. 1985;37(1-3):247–253. doi: 10.1016/0378-1119(85)90279-3. [DOI] [PubMed] [Google Scholar]
- Orkin S. H. Globin gene regulation and switching: circa 1990. Cell. 1990 Nov 16;63(4):665–672. doi: 10.1016/0092-8674(90)90133-y. [DOI] [PubMed] [Google Scholar]
- Plumb M., Frampton J., Wainwright H., Walker M., Macleod K., Goodwin G., Harrison P. GATAAG; a cis-control region binding an erythroid-specific nuclear factor with a role in globin and non-globin gene expression. Nucleic Acids Res. 1989 Jan 11;17(1):73–92. doi: 10.1093/nar/17.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F. S., Cooper T. G. Structure and transcription of the allantoate permease gene (DAL5) from Saccharomyces cerevisiae. J Bacteriol. 1988 Jan;170(1):266–271. doi: 10.1128/jb.170.1.266-271.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F. S., Sumrada R. A., Cooper T. G. Identification of sequences responsible for transcriptional activation of the allantoate permease gene in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):602–608. doi: 10.1128/mcb.9.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Genbauffe F., Lea H. Z., Cooper T. G. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J Bacteriol. 1987 Aug;169(8):3521–3524. doi: 10.1128/jb.169.8.3521-3524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos F., Wiame J. M. Synthesis and activation of asparagine in asparagine auxotrophs of Saccharomyces cerevisiae. Eur J Biochem. 1979 Mar;94(2):409–417. doi: 10.1111/j.1432-1033.1979.tb12908.x. [DOI] [PubMed] [Google Scholar]
- Roon R. J., Even H. L. Regulation of the nicotinamide adenine dinucleotide- and nicotinamide adenine dinucleotide phosphate-dependent glutamate dehydrogenases of Saccharomyces cerevisiae. J Bacteriol. 1973 Oct;116(1):367–372. doi: 10.1128/jb.116.1.367-372.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Siddiqui A. H., Brandriss M. C. A regulatory region responsible for proline-specific induction of the yeast PUT2 gene is adjacent to its TATA box. Mol Cell Biol. 1988 Nov;8(11):4634–4641. doi: 10.1128/mcb.8.11.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. M., Andrésson O. S. DNA sequences of yeast H3 and H4 histone genes from two non-allelic gene sets encode identical H3 and H4 proteins. J Mol Biol. 1983 Sep 25;169(3):663–690. doi: 10.1016/s0022-2836(83)80164-8. [DOI] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomulka K. W., Moat A. G. Inorganic nitrogen assimilation in yeasts: alteration in enzyme activities associated with changes in cultural conditions and growth phase. J Bacteriol. 1972 Jan;109(1):25–33. doi: 10.1128/jb.109.1.25-33.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbol M., Jauniaux J. C., Grenson M. Nucleotide sequence of the Saccharomyces cerevisiae PUT4 proline-permease-encoding gene: similarities between CAN1, HIP1 and PUT4 permeases. Gene. 1989 Nov 15;83(1):153–159. doi: 10.1016/0378-1119(89)90413-7. [DOI] [PubMed] [Google Scholar]
- Vissers S., Andre B., Muyldermans F., Grenson M. Induction of the 4-aminobutyrate and urea-catabolic pathways in Saccharomyces cerevisiae. Specific and common transcriptional regulators. Eur J Biochem. 1990 Feb 14;187(3):611–616. doi: 10.1111/j.1432-1033.1990.tb15344.x. [DOI] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol Cell Biol. 1987 Dec;7(12):4431–4440. doi: 10.1128/mcb.7.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickerham L. J. A Critical Evaluation of the Nitrogen Assimilation Tests Commonly Used in the Classification of Yeasts. J Bacteriol. 1946 Sep;52(3):293–301. [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Ko L. J., Leonard M. W., Beug H., Orkin S. H., Engel J. D. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 1990 Oct;4(10):1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- Yoo H. S., Cooper T. G. Sequences of two adjacent genes, one (DAL2) encoding allantoicase and another (DCG1) sensitive to nitrogen-catabolite repression in Saccharomyces cerevisiae. Gene. 1991 Jul 31;104(1):55–62. doi: 10.1016/0378-1119(91)90464-m. [DOI] [PubMed] [Google Scholar]
- Yoo H. S., Cooper T. G. The DAL7 promoter consists of multiple elements that cooperatively mediate regulation of the gene's expression. Mol Cell Biol. 1989 Aug;9(8):3231–3243. doi: 10.1128/mcb.9.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Cooper T. G. The ureidoglycollate hydrolase (DAL3) gene in Saccharomyces cerevisiae. Yeast. 1991 Oct;7(7):693–698. doi: 10.1002/yea.320070705. [DOI] [PubMed] [Google Scholar]
- Yoo H. S., Genbauffe F. S., Cooper T. G. Identification of the ureidoglycolate hydrolase gene in the DAL gene cluster of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2279–2288. doi: 10.1128/mcb.5.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon L. I., Youssoufian H., Mather C., Lodish H. F., Orkin S. H. Activation of the erythropoietin receptor promoter by transcription factor GATA-1. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10638–10641. doi: 10.1073/pnas.88.23.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]