Abstract
Aims
To determine the sensitivity and specificity of a multiplex PCR assay for the contemporary identification of major species involved in oral candidiasis, without extraction and purification of DNA from the samples under investigation; and evaluation of this method in comparison with routine phenotypic culture identification.
Methods
78 oral rinse solutions were collected. The concentrated oral rinse technique was used for a quantitative and qualitative study. Research and identification of Candida spp, with routine phenotypic culture identification (germ‐tube test in serum at 37°C for 3 hours and sugar assimilation strip analysis), were performed. Each sample was analysed with multiplex PCR directly on oral rinse solution. Samples giving discrepant results between routine phenotypic and PCR identification methods were resubcultured on CHROMagar Candida plates. The fungus‐specific primers ITS1, ITS2, CA3, and CA4 were used. For the identification of other species (C kefyr, C famata and C dubliniensis), ITS1F, ITS1K, and ITS2D primers were designed.
Results
Multiplex PCR correctly identified all samples, including those with single species, or with mixed species, negative samples and positive samples which appeared to be negative from routine phenotypic methods.
Conclusion
This multiplex PCR assay provides a rapid alternative to the conventional culture based technique for the identification and speciation of the most frequently isolated Candida species. The absence of an extraction method made identification of 10 species possible in a few hours.
Keywords: candida, PCR, rapid identification, molecular pathology
Oral candidosis is one of the most frequently observed pathologies in everyday practice.1 It is an opportunistic disease because of some promoting factors that alter the equilibrium in the oral cavity microbial ecosystem, helping the transformation of yeasts from commensals to pathogens.2 Although the transition from commensalism to disease may be associated with the virulence characteristics of the organism, it is widely accepted that host factors (iatrogenic and infective factors) are of paramount importance in the development of the infection.3
A number of relevant factors have been associated with oral carriage of yeast (denture wearing, periodontal disease, leucoplastic lesions, etc). They contribute to form the so‐called “mycotic count” and to cause the following clinical cases.4
Candida albicans has been by far the most common yeast species carried as a commensal by healthy individuals and the agent most frequently responsible for oral yeast infection. However, the emergence of non‐albicans Candida species (C parapsilosis, C tropicalis, C glabrata, etc) has also been observed and associated with long‐term treatment with amphotericin and fluconazole.5,6,7,8,9,10,11,12,13,14
More recently, oral candidosis has been associated with C dubliniensis in HIV‐infected individuals15,16 and patients with diabetes,17 and has been detected in the sputum of patients with cystic fibrosis.18
Since pathogenicity and antifungal susceptibility often vary among species, a rapid and accurate identification of the disease‐causing species of Candida is crucial for both clinical treatment and epidemiological studies of oral candidiasis.19 The conventional identification of pathogenic fungi in the clinical microbiology laboratory involves the examination of colony and microscopic morphologies and the assessment of various biochemical reactions.20,21 It often requires three or more days, and may be inaccurate. Moreover, the presence of more than one Candida species in the oral cavity of the same host is not infrequent.
In recent years, numerous DNA‐based methods such as DNA–DNA reassociation,22 DNA fingerprinting,23 and Southern hybridisation with appropriate DNA probes24,25 have been reported to recognise Candida species in culture or in clinical materials. However, these genotypic methods have the disadvantage of being laborious and time‐consuming, and also require specialised equipment.
The aims of the present study were to determine the sensitivity and specificity of a multiplex PCR method applied to oral rinse solution obtained in our laboratory, for the identification of Candida species that are frequently isolated from the oral cavity, and to evaluate this method in comparison with routine phenotypic culture identification. The method is based on the size variability of the ITS1 region in different species and on the amplification of a specific DNA fragment of the ITS2 region of C albicans. The test has allowed us to identify Candida species using seven species‐specific oligonucleotides in a single PCR tube.
Drawing from this strategy we studied the structure of other species of Candida and generated in the same region other primers that were able to amplify specific fragments. Indeed for the identification of other three species, C kefyr, C famata and C dubliniensis we designed the following primers: ITS1F (5′‐CCA GCG CTT AAT TGC G‐3′), ITS1K (5′‐ATC GTC TGA ACA AGG CCT GC‐3′), and ITS2D (5′‐GAG AAC CAA GAG ATC CGT TGT TG‐3′). Table 1 lists these primers and the generated PCR products.
Table 1 Lengths of the PCR products.
| Organism | Primers | PCR product (bp) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS1 | ITS1F | ITS1K | ITS2 | ITS2D | CA3 | CA4 | ||||
| C glabrata | + | − | − | + | + | − | − | 482–483 | 462–463 | – |
| C guilliermondii | + | − | − | + | + | − | − | 248 | 228 | – |
| C famata | − | + | − | + | + | − | − | 234 | 214 | – |
| C kefyr | − | − | + | + | + | − | − | 249 | 229 | – |
| C parapsilosis | + | − | − | + | + | − | − | 229 | 209 | – |
| C tropicalis | + | − | − | + | + | − | − | 218 | 199 | – |
| C albicans | + | − | − | + | + | + | + | 218–219 | 198–199 | 110 |
| C krusei | + | − | − | + | + | − | − | 182 | 166 | – |
| C lusitaniae | + | − | − | + | + | − | − | 148 | 128 | – |
| C dubliniensis | + | − | − | − | + | − | − | – | 198 | – |
Materials and methods
A total of 78 oral rinse solutions obtained from symptomatic patients with odontostomatological and maxillo‐facial diseases, with or without signs, attending the outpatients clinic of the Microbiology Laboratory of the Hygiene Unit of the Department of Public, Clinical and Preventive Medicine were collected during the period 2004 to 2005.
The concentrated oral rinse technique for the determination of bacterial and mycotic colony counts (a gargle with 9 ml of phosphate‐buffered saline) was used for a quantitative and qualitative study as previously described.26 From all positive cultures, Candida genus yeasts were identified presumptively (germ‐tube test in serum at 37°C for 3 hours) and definitively with API 20C AUX (bioMérieux, Rome, Italy) sugar assimilation strip analysis. Each sample was analysed with multiplex PCR directly on oral rinse solution. Samples giving discrepant results between routine phenotypic and PCR identification methods were resubcultured on CHROMagar Candida (Alfa Wassermann, Diagnostics SpA, Milan, Italy) plates to identify possible mixed yeast culture. PCR was performed directly on each oral rinse solution (without preliminary DNA extraction).
Patients gargled with 5 ml of phosphate‐buffered saline, and the resulting solution was centrifuged at 14000 rpm for 5 min. The supernatant was discarded. The residual pellet was resuspended with 100 μl sterile distilled water and heated at 90°C for 3 min; 20 μl samples were added to the mix for PCR.
The fungus‐specific primers ITS1 (5′‐TCC GTA GGT GAA CCT GCG G‐3′) and ITS2 (5′‐GCT GCG TTC TTC ATC GAT CG‐3′)27 were used to amplify a small conserved portion of the 18S rDNA region, the adjacent ITS1, and a small portion of the 28S rDNA region (fig 1), generating PCR products for C glabrata, C guilliermondii, C lusitaniae, C parapsilosis, C tropicalis and C krusei. In addition, C albicans‐specific primers CA3 (5′‐GGT TTG GAA AGA CGG TAG‐3′) and CA4 (5′‐AGT TTG AAG ATA TAC GTG GTA G‐3′)28 were also included in the PCR mixture to amplify a portion of the ITS2 region of C albicans.
Figure 1 Diagram of hybridisation sites of ITS1 and ITS2 primers.
Multiplex PCR was performed in duplicate with 20 μl of oral rinse solution in a total reaction volume of 50 μl, consisting of 10 mM Tris‐HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.8 mM deoxyribonucleoside triphosphates (0.2 mM each), 3.2 μM primers, Taq DNA polymerase (1.25 U), and 50 μl of a mineral oil overlay. PCR was carried out with a Perkin Elmer thermal cycler under the following condition: initial denaturation, 92°C, 2 min; 35 cycles of denaturation (95°C, 1 min), annealling (50°C, 1 min), and extension (72°C, 1 min); and final extension, 72°C, 10 min. A negative control run was performed with each test run by replacing the samples with sterilised water in the PCR mixture. A positive culture broth containing C albicans was run in parallel with unknown samples, and this culture broth was used as a positive control. Gel electrophoresis was conducted in TBE buffer (0.1 M Tris, 0.09 M boric acid, 1 mM EDTA, pH 8.4) at 100 V for 1–2 h using gel composed of 2% (w/v) agarose (Sigma‐Aldrich, Milan, Italy). Gel was stained with 0.5 μg ethidium bromide per ml distilled water.
Results
A total of 78 oral rinse solutions were analysed by routine phenotypic methods for species identification. From these samples, 63 strains of yeasts were isolated; 15 oral rinse solutions were negative. The most frequently isolated species was C albicans (48 strains, 76.2%), followed by C glabrata (6 strains, 9.5%), C tropicalis (5 strains, 7.9%), C krusei (2 strains, 3.2%), C parapsilosis (1 strain, 1.6%), and C famata (1 strain, 1.6%) (table 2).
Table 2 Comparative identification of Candida spp in oral rinse solutions by routine phenotypic identification methods versus multiplex PCR.
| Routine phenotypic identification | No of oral rinse solutions analysed | Multiplex PCR identification | No of oral rinse solutions analysed |
|---|---|---|---|
| C albicans | 48 | C albicans | 47 |
| C albicans + C glabrata | 3 | ||
| C glabrata | 6 | C glabrata | 3 |
| C glabrata + C guilliermondii | 1 | ||
| C tropicalis | 5 | C tropicalis | 1 |
| C tropicalis + C guilliermondii | 1 | ||
| C krusei | 2 | C krusei | 2 |
| C parapsilosis | 1 | C parapsilosis | 1 |
| C famata | 1 | C famata | 1 |
| C guilliermondii | 0 | C guilliermondii | 1 |
| C dubliniensis | 0 | C dubliniensis | 2 |
| C kefyr | 0 | C kefyr | 1 |
| Negative | 15 | Negative | 14 |
| Total | 78 | Total | 78 |
Multiplex PCR identification results matched phenotypic identification results for 45 of 64 positive samples (70.3%) (47 samples containing C albicans, 73.4%; 3 C glabrata, 4.7%; 2 C krusei, 3.1%; 2 C dubliniensis, 3.1%; 1 C tropicalis, 1.6%; 1 C parapsilosis, 1.6%; 1 C famata, 1.6%; 1 C gulliermondii, 1.6%; 1 C kefyr, 1.6%). Three samples that were found to contain C albicans only by phenotypic methods were identified by PCR to contain both C albicans and C glabrata. The subculture of the oral rinse solution on CHROMagar Candida medium indicated that this specimen contained a mixture of C albicans and C glabrata (table 3).
Table 3 Resolution of discrepancies between phenotypic and multiplex PCR methods for the identification of Candida species.
| Patient no | Identification by the following methods: | ||
|---|---|---|---|
| Routine phenotypic method | Multiplex PCR | Final identification* | |
| 293 | C albicans | C albicans + C glabrata | C albicans + C glabrata |
| 301 | C glabrata | C albicans + C glabrata | C albicans + C glabrata |
| 315 | C tropicalis | C albicans | C albicans |
| 330 | C tropicalis | C albicans | C albicans |
| 333 | C tropicalis | C albicans | C albicans |
| 348 | C albicans | C albicans + C glabrata | C albicans + C glabrata |
| 358 | C parapsilosis | C albicans | C albicans |
| 362 | C albicans | C glabrata + C guilliermondii | C glabrata + C guilliermondii |
| 374 | C glabrata | C albicans | C albicans |
| 392 | C glabrata | C albicans | C albicans |
| 420 | C albicans | C parapsilosis | C parapsilosis |
| 421 | C albicans | C tropicalis + C guilliermondii | C tropicalis + C guilliermondii |
| 424 | C albicans | C glabrata | C glabrata |
| 422 | C tropicalis | C guilliermondii | C guilliermondii |
| 427 | C albicans | C dubliniensis | C albicans |
| 433 | C albicans | C kefyr | C kefyr |
| 441 | C albicans | C dubliniensis | C albicans |
| 451 | C glabrata | C albicans | C albicans |
| Negative | Negative | C albicans | Negative |
*Final species identification was corroborated by colony morphology and colour on CHROMagar Candida medium and by reidentification by routine phenotypic methods.
Two samples that were found to contain C albicans only by phenotypic methods were identified by PCR to contain both C glabrata and C guilliermondii in one case and C tropicalis and C gulliermondii in another. In these two cases of mixed species, C glabrata and C guilliermondii and C tropicalis and C gulliermondii were also recovered on CHROMagar Candida plates (table 3).
Four oral rinse solutions that were found to contain C tropicalis only by phenotypic methods were identified by PCR to contain 3 C albicans and 1 C guilliermondii; 1 sample containing C parapsilosis was identified by PCR to contain C albicans; 3 samples containing C glabrata were identified by molecular analysis to contain 3 C albicans; and 5 oral rinse solutions containing C albicans were identified by multiplex PCR to contain 1 C parapsilosis, 1 C glabrata, 1 C kefyr, and 2 C dubliniensis. The subculture of all these oral rinse solutions on CHROMagar Candida medium confirmed the identification of multiplex PCR except for C dubliniensis (table 3).
Finally, one of the oral rinse solutions which was negative with the phenotypic method was positive to the multiplex PCR (C albicans).
The time from the first oral rinse solution culture to species identification by routine phenotypic methods (subculture, germ tube and API 20CAUX) was 4 days (96 h). More slowly growing, individual isolates took as long as 6 days for growth and species identification by routine phenotypic methods (144 h, C guilliermondii isolate). For the multiplex PCR, on the other hand, no time was required to isolate and extract DNA from oral rinse solution, 3 h was required for PCR amplification and 1.5 h was required for agarose gel electrophoresis analysis. Therefore, species could be identified in as little as 5 h, including the time required to prepare PCR reagent mixture, in contrast to routine phenotypic methods which took several days.
The mycotic and bacterial colony count obtained from the oral rinse technique used for a quantitative study ranged from 10 CFU/ml to 5.46×106 CFU/ml for the mycotic count and from 9.8×105 CFU/ml to 15.3×107 CFU/ml for the bacterial colony count.
The limit of detection of the multiplex PCR was approximately 10 CFU/ml and was very close to that reported by Jaeger et al.29
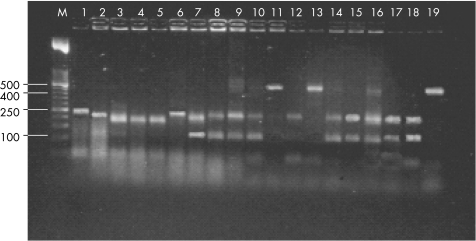
The high quantity of bacterial colony count did not interfere with the detection and identification of yeasts with multiplex PCR. In fact, coexisting bacteria in oral rinse solution specimens did not produce any detectable PCR products. Therefore, the multiplex PCR method could also detect mixed yeast cultures missed by routine subculturing methods, even when the oral rinse solution contained bacteria (fig 2).
Figure 2 Identification of yeasts present in oral rinse solutions by the multiplex PCR. M, 50 bp DNA ladder. Lane 1, C kefyr bp 249. Lane 2, C famata bp 234. Lane 3, C dubliniensis bp 198. Lane 4, C krusei bp 182. Lane 5, C parapsilosis bp 209. Lane 6, C guilliermondii bp 248. Lane 7, C albicans bp 110–218. Lane 8, C albicans bp 110–218. Lane 9, C albicans bp 110–218 and C glabrata bp 462–482. Lane 10, C albicans bp 110–218. Lane 11, C glabrata bp 462–482. Lane 12, C tropicalis bp 218. Lane 13, C glabrata bp 462–482; Lane 14, C albicans bp 110–218. Lane 15, C albicans bp 110–218. Lane 16, C albicans bp 110–218 and C glabrata bp 462–482. Lane 17, C albicans bp 110–218. Lane 18, C albicans bp 110–218. Lane 19, C glabrata bp 462–482.
Discussion
In order to improve the quality of our diagnostic research, we have set up a multiplex PCR assay for the contemporary identification of major species involved in oral candidiasis to avoid any time delay due to the extraction and purification of DNA from the samples under investigation.
To the best of our knowledge this is the first application of a PCR assay on oral rinse solutions obtained directly from the samples without DNA extraction.
A total of 78 oral rinse solutions were used for a PCR assay to simultaneously identify and type the Candida positivity directly from the clinical samples without any particular treatment. Multiplex PCR correctly identified all 78 samples, including 58 cultures from subjects with single species, 5 cultures of oral rinses from patients with colonisation of mixed species (identified to contain a single Candida species by routine phenotypic methods), 14 negative samples and 1 positive sample identified as negative from routine phenotypic methods.
The advantages of this method are as follows: it does not require use of expensive or toxic chemical substances such as proteinase K or phenol‐chloroform; the total time from species identification is 5 h, compared to a mean of 5 days by routine phenotypic culture identification methods; identification of more than one species of yeast is possible in mixed cultures, with no cross‐reaction or interference with bacteria and viruses likely to be present in oral rinse solutions; and no modifications or changes in routine practice were necessary for the clinicians or the laboratory technicians, who were able to use the same samples used for conventional morphological and metabolic examination for the PCR assay.
Finally, considering the continuous decrease of the cost of purchasing a thermal cycler and the reagents necessary to use it, the estimated cost for this assay is similar to that of routine phenotypic culture identification.
Take‐home messages
A method for rapid identification by PCR of the presence of Candida spp has been developed (5 h versus a mean of 5 days by routine phenotypic culture identification methods).
The PCR assay is performed on the oral rinse solution obtained directly from the samples without DNA extraction.
The technique is accurate, and permits identification of more than one species of yeast in mixed cultures, with no cross‐reaction or interference with bacteria and viruses likely to be present in oral rinse solutions.
The method could be used routinely in clinical microbiology laboratories.
The method permits rapid identification of Candida isolates from high‐risk patients or patients with oral candidiasis, enabling prompt and appropriate treatment and thus reducing hospitalisation time.
In our experience, at least 10 species of Candida are frequently isolated from oral rinse solutions in laboratories: C albicans, C glabrata, C guilliermondii, C lusitaniae, C parapsilosis, C tropicalis, C krusei, C kefyr, C famata and C dubliniensis. In order to be able to detect the most common Candida species, we have found in the literature primers to detect C albicans, C glabrata, C guilliermondii, C lusitaniae, C parapsilosis, C tropicalis and C krusei.30 Moreover, in order to increase the number of primers to include all the medically important species of Candida, drawing from data in the literature, we have designed other primers in the region between the conserved portion of the 18S rDNA region, the adjacent ITS1, and a small portion of the 28S rDNA region in order to detect C kefyr, C famata and C dubliniensis.
The multiplex PCR is particularly suitable for use in a routine clinical microbiology laboratory because it can easily be automated; moreover it could be utilised for the rapid identification of Candida isolates from high‐risk patients or patients with oral candidiasis, enabling prompt and appropriate treatment and thus reducing hospitalisation time.
Acknowledgements
We thank Dr Pia Furno for editing the manuscript.
Footnotes
Funding: This work was supported in part by University of Naples “Parthenope” and Second University of Naples.
Competing interests: None declared.
References
- 1.Reichart P A, Samaranayake L P, Philipsen H P. Pathology and clinical correlates in oral candidiasis and its variants: a review. Oral Dis 2000685–91. [DOI] [PubMed] [Google Scholar]
- 2.Rogers T J, Balish E. Immunity to Candida albicans. Microbiol Rev 198044660–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saltarelli C G.Candida albicans. The pathogenic fungus. New York: Hemisphere Publishing, 1989
- 4.Odds F C.Candida and candidosis. London: Balliere Tindall, 1988
- 5.Nguyen M H, Peacock J E, Jr, Morris A J.et al The changing face of candidemia: emergence of non‐Candida albicans species and antifungal resistance. Am J Med 1996100617–623. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen H, Stenderup J. Invasive Candida norvegensis infection in immunocompromised patients. Scand J Infect Dis 199628311–312. [DOI] [PubMed] [Google Scholar]
- 7.Verduyn Lunel F M, Meis J F, Voss A. Nosocomial fungal infections: candidemia. Diagn Microbiol Infect Dis 199934213–220. [DOI] [PubMed] [Google Scholar]
- 8.Viscoli C, Girmenia C, Marinus A.et al Candidemia in cancer patients: a prospective, multicenter surveillance study by the Invasive Fungal Infection Group (IFIG) of the European Organization for Research and Treatment of Cancer (EORTC). Clin Infect Dis 1999281071–1079. [DOI] [PubMed] [Google Scholar]
- 9.Newman S L, Flanigan T P, Fisher A.et al Clinically significant mucosal candidiasis resistant to fluconazole treatment in patients with AIDS. Clin Infect Dis 199419684–686. [DOI] [PubMed] [Google Scholar]
- 10.Pfaller M A, Jones R N, Messer S A.et al National surveillance of nosocomial blood stream infection due to Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn Microbiol Infect Dis 199831327–332. [DOI] [PubMed] [Google Scholar]
- 11.Price M F, LaRocco M T, Gentry L O. Fluconazole susceptibilities of Candida species and distribution of species recovered from blood cultures over a 5‐year period. Antimicrob Agents Chemother 1994381422–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother 1995391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruhnke M, Eigler A, Tennagen I.et al Emergence of fluconazole‐resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J Clin Microbiol 1994322092–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wingard J R, Merz W G, Rinaldi M G.et al Increase in Candida krusei infection among patients with bone marrow transplantation and neutropenia treated prophylactically with fluconazole. N Engl J Med 19913251274–1277. [DOI] [PubMed] [Google Scholar]
- 15.Coleman D D, Sullivan B, Harrington K.et al Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV‐infected and AIDS patients. Oral Dis 19973S96–101. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan D, Coleman D. Candida dubliniensis: an emerging opportunistic pathogen. Curr Top Med Mycol 1997815–25. [PubMed] [Google Scholar]
- 17.Manfredi M, McCullough M J, Al‐Karaawi Z M.et al The isolation, identification and molecular analysis of Candida spp isolated from the oral cavities of patients with diabetes mellitus. Oral Microbiol Immunol 200217181–185. [DOI] [PubMed] [Google Scholar]
- 18.Peltroche‐Llacsahuanga H, Dohmen H, Haase G. Recovery of Candida dubliniensis from sputum of cystic fibrosis patients. Mycoses 20024515–18. [PubMed] [Google Scholar]
- 19.Williams D W, Lewis M A. Isolation and identification of Candida from the oral cavity. Oral Dis 200063–11. [DOI] [PubMed] [Google Scholar]
- 20.Warren N G, Hazen K C. Candida, Criptococcus, and other yeasts of medical importance. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, eds. Manual of clinical microbiology, 6th edn. Washington, DC: American Society for Microbiology, 1995723–737.
- 21.Wickes L B, Hicks J B, Merz W G.et al The molecular analysis of synonymy among medical important yeasts within the genus Candida. J Gen Microbiol 1992138901–907. [DOI] [PubMed] [Google Scholar]
- 22.Kurtzman C P, Phaff H J, Meyer S A. Nucleic acid relatedness among yeasts. In: Spencer JFT, Spencer DM, Smith ARW, eds. Yeast genetics: fundamental and applied aspects New York: Springer‐Verlag, 1983139–166.
- 23.Scherer S, Stevens D A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol 198725675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magee B B, D'Souza T M, Magee P T. Strain and species identification by restriction fragment length polymorphisms in the ribosomal DNA repeat of Candida species. J Bacteriol 19871639–1643. [DOI] [PMC free article] [PubMed]
- 25.Mason M M, Lasker B A, Riggsby W S. Molecular probe for identification of medically important Candida species and Torulopsis glabrata. J Clin Microbiol 198725563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samaranayake L P, MacFarlane T W, Lamey P J.et al A comparison of oral rinse and imprint sampling techniques for the detection of yeast, coliform and Staphylococcus aureus carriage in the oral cavity. J Oral Pathol 198615386–388. [DOI] [PubMed] [Google Scholar]
- 27.White T J, Bruns T, Lee S.et al Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gefland DH, Sninsky JJ, White TJ, eds. PCR protocols: a guide to methods and applications San Diego, CA: Academic Press, 1990315–322.
- 28.Huang C H. Specific identification of yeasts on the bases of PCR amplified ribosomal DNA immobilized by covalent bond on the piezoelectric quartz crystal. PhD thesis. National Taiwan University, Taipei, Taiwan 1996
- 29.Jaeger E E M, Carroll N M, Choudhury S.et al Rapid detection and identification of Candida, Aspergillus, and Fusarium species in ocular samples using PCR. J Clin Microbiol 2000382902–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang H C, Leaw S N, Huang A H.et al Rapid identification of yeasts in positive blood cultures by a multiplex PCR method. J Clin Microbiol 2001393466–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]