Abstract
At least one third of all cases of epithelial ovarian cancer are associated with the production of ascites, although its effect on tumor cell microenvironment remains poorly understood. This study addresses the effect of the heterologous acellular fraction of ovarian cancer-derived ascites on a cell line (OV-90) derived from the chemotherapy-naïve ovarian cancer patient. Ascites were assayed for their effect on cell invasion, growth, and spheroid formation. When compared to either no serum or 5% serum, ascites fell into one of two categories: stimulatory or inhibitory. RNA from OV-90 cells exposed to selected ascites were arrayed on an Affymetrix HG-U133A GeneChip. A supervised analysis identified a number of differentially expressed genes and quantitative polymerase chain reaction validation based on OV-90 cells exposed to 54 independent ascites demonstrated that stimulatory ascites affected the expression of ISGF3G, TRIB1, MKP1, RGS4, PLEC1, and MOSPD1 genes. In addition, TRIB1 expression was shown to independently correlate with prognosis when its expression was ascertained in an independent set of primary cultures established from ovarian ascites. The data support the validity of the strategy to uncover molecular events that are associated with tumor cell behavior and highlight the impact of ascites on the cellular and molecular parameters of ovarian cancer.
Keywords: Epithelial ovarian cancer, ovarian ascites, cell behavior, invasion, molecular profiling
Introduction
Ovarian cancer is the fifth leading cause of cancer-related deaths in the Western world, the second most common gynecological cancer, and the leading cause of death from gynecological malignancies. The most common form of cancer of the ovary is epithelial ovarian cancer (EOC). EOCs originate either from the normal ovarian surface epithelium itself or from the crypts and inclusion cysts on the surface epithelium [1]. Ovarian carcinomas can spread by local extension, lymphatic invasion, intraperitoneal implantation, hematogenous dissemination, and/or transdiaphragmatic passage. In the commonly observed intraperitoneal dissemination, malignant cells appear to implant anywhere over peritoneal surfaces though mainly in sites of stasis along the peritoneal fluid circulation.
At least one third of ovarian cancer patients present with ascites [2,3], a generally voluminous exudative fluid with a cellular fraction consisting mainly of ovarian cancer cells, lymphocytes, and mesothelial cells. The neoplastic cells in the ascites are present either as single cells, as aggregates, or as spheroids, and may contribute to the spread of cancer to secondary sites. Indeed, it has been demonstrated that ascites spheroids adhere to extracellular matrix through β1 integrins, indicating that this process may play a role in the dissemination of the disease [4]. The acellular fraction of ascites is known to harbor angiogenic factors such a vascular endothelial growth factor [5] and growth factors such as the epidermal growth factor [6], lysophosphatidic acid [7], and transforming growth factor (TGF) family members [8,9] among others. How this acellular fraction affects the tumor microenvironment, and specifically how it affects the cellular and molecular properties of tumor cells per se is still a matter of debate [10,11]. Indeed, the recent resurgence of intraperitoneal-based chemotherapies highlights the importance of understanding this important component of the disease [12–14]. Recently, a targeted approach demonstrated the ability of ascites to modulate the expression of urokinase plasminogen activator, its receptor, and integrins, and these modifications in expression were associated with changes in the cellular behavior of ovarian cancer cell lines [15].
In the present study, we conducted a comprehensive analysis of the effect of ascites on the growth characteristics of the OV-90 ovarian cancer cell line, which we have previously described [16]. This EOC cell line, derived from the cellular fraction of ascites from a chemotherapy-naïve patient, has been characterized by morphological, immunohistochemical, cytogenetic, and molecular analyses including gene expression profiles [16,17] and was shown to harbor mutations in genes implicated in ovarian cancer such as tumor protein p53 (TP53), cyclin-dependent kinase inhibitor 1A (CDNK2A), and TGFβRII. In addition to characterizing the cellular effects of ascites, a DNA microarray approach was used to assess differences in gene expression in OV-90 cell line grown in the presence or absence of serum, as well as in the presence of ascites without serum. Statistical analysis was used to identify differentially expressed genes that correlated with cellular invasion for the OV-90 cell line. Candidate genes were further validated on both arrayed RNA and an extended test set using quantitative polymerase chain reaction (Q-PCR), and their association with survival was tested in an independent set of primary cultures derived from patients with ovarian ascites.
Materials and Methods
Cell Culture, Clinical Material, and Patients
The OV-90 cell line was maintained in OSE media consisting of 50:50 medium 199:105 (Sigma-Aldrich, St. Louis, MO) supplemented with 10%fetal bovine serum(FBS), 2.5 µg/ml amphotericin B and 50 µg/ml gentamicin [18]. Following appropriate consent, ascites were collected at the time of clinical intervention at the Centre Hospitalier de l'Université de Montréal (Montreal, QC, Canada). Ascites were centrifuged at 2500 rpm for 5 minutes. The acellular fractions were stored at -20°C and tested within 6 months of reception. The protein concentration in ascites fluid was measured by the Bradford assay (Bio-Rad, Hercules, CA). Histopathology, grade, and stage of ovarian tumors were assigned according to the International Federation of Gynecology and Obstetrics criteria. Of the 54 ascites included in our study on invasion, two thirds were from patients diagnosed with papillary serous adenocarcinomas and most presented with stage IIIC and grade 3 disease (Table 1). One third of samples were from patients who had already received chemotherapy prior to surgery. The presence of neoplastic cells in ascites was determined from pathology reports. Table 2 describes the 28 ovarian cancer patients diagnosed with accompanying ascites that were included in the survival analysis.
Table 1.
Clinical Characteristics of Patients from Which Ascites Were Obtained.
| Ascites | Age | Histopathology | Grade | Stage | Neoplastic Cells in Ascites | Prior Chemotherapy | Clinical Intervention Associated with Ascites Collection |
| A1185(2)* | 82 | PSA | G3 | IIIC | Yes | Yes | Secondary cytoreduction |
| A1301 | 49 | SA | G3 | IIIC | Yes | No | Primary biopsy |
| A1317 | 60 | PSA | G3 | IV | No | Yes | Primary surgery |
| A1318 | 52 | PSA | G2 | IIIC | Yes | No | Primary surgery |
| A1322 | 71 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1322(2) | 71 | PSA | G3 | IIIC | Yes | Yes | Secondary cytoreduction |
| A1330 | 48 | SA | G3 | IIIC | Yes | No | Primary surgery |
| A1337 | 45 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1369 | 60 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1369(2) | 60 | PSA | G3 | IIIC | Yes | Yes | Secondary cytoreduction |
| A1396 | 54 | PSA | G2 | IIIC | Yes | No | Primary surgery |
| A1406 | 49 | PSA | G2 | IIIC | N/S | Yes | Secondary cytoreduction |
| A1464 | 63 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1483 | 74 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1519 | 73 | CCA | G3 | IV | N/S | No | Primary surgery |
| A1526 | 50 | PSA | G2 | IIIC | Yes | Yes | Secondary cytoreduction |
| A1526(2) | 50 | PSA | G2 | IIIC | Yes | Yes | Secondary cytoreduction |
| A1592 | 35 | MCA | G3 | IIIC | N/S | No | Primary surgery |
| A1607 | 59 | SA | GB | IB | No | No | Primary surgery |
| A1610 | 72 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1611 | 45 | MA | GB | IA | No | No | Primary surgery |
| A1613 | 76 | SA | G3 | IIIC | No | No | Primary surgery |
| A1642 | 79 | SA | G3 | IIB | Yes | No | Primary surgery |
| A1717 | 45 | PSA | G2 | IIIC | N/S | No | Primary surgery |
| A1739 | 64 | MA | N/S | N/S | N/S | No | Primary surgery |
| A1778 | 87 | PSA | G3 | IV | Yes | No | Primary surgery |
| A1793 | 55 | SA | G3 | IIIC | No | No | Primary surgery |
| A1801 | 65 | PSA | G2 | IIIC | Yes | No | Primary surgery |
| A1810 | 62 | PSA | G2 | IIIC | Yes | No | Primary surgery |
| A1813 | 55 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1830 | 56 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1835 | 69 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1876 | 44 | PSA | G3 | IIIC | No | No | Primary surgery |
| A1884 | 69 | PSA | G3 | IV | Yes | No | Primary surgery |
| A1891 | 62 | PSA | G3 | IIIC | No | No | Primary surgery |
| A1922 | 52 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1927 | 48 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1931 | 67 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1946 | 75 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A1998 | 70 | UA | N/S | N/S | Yes | Yes | N/S |
| A1998(2) | 70 | UA | N/S | N/S | Yes | Yes | N/S |
| A2069 | 63 | MCA | G3 | IIIC | Yes | No | Primary surgery |
| A2070 | 78 | SA | G3 | IIIC | Yes | No | Primary surgery |
| A2085 | 65 | PSA | G3 | IIIC | N/S | Yes | Secondary cytoreduction |
| A2085(2) | 65 | PSA | G3 | IIIC | N/S | Yes | Secondary cytoreduction |
| A2085(3) | 65 | PSA | G3 | IIIC | N/S | Yes | Secondary cytoreduction |
| A2090 | 76 | UA | N/S | IIIC | Yes | Yes | N/S |
| A2093 | 62 | PSA | G3 | IV | Yes | Yes | Secondary cytoreduction |
| A2093(2) | 62 | PSA | G3 | IV | Yes | Yes | Secondary cytoreduction |
| A2774 | 42 | EA | G3 | IB | No | No | Primary surgery |
| A2775 | 49 | PSA | G2 | IIIC | Yes | No | Primary surgery |
| A2834 | 63 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A2912 | 54 | PSA | G3 | IIIC | Yes | No | Primary surgery |
| A2965 | 71 | MCA | G3 | IIIC | Yes | No | Primary surgery |
PSA, papillary serous adenocarcinoma; SA, serous adenocarcinoma; MA, mucinous adenocarcinoma; MCA, mixed cell adenocarcinoma; CCA, clear cell adenocarcinoma; UA, undifferentiated adenocarcinoma; EA, endometrioid adenocarcinoma; N/S, not specified.
Ascites further characterized in this study are in bold.
Denotes second and third ascites collected from patient with the same number.
Table 2.
Patients' Cohort with Ovarian Cancer and Accompanying Ascites, for Survival Analyses.
| Patients | Death | Survival (months) | Histopathology |
| 90 | Yes | 0 | UA |
| 513 | No | 37 | SA |
| 595 | Yes | 11 | PSA |
| 665 | Yes | 32 | PSA |
| 747 | Yes | 9 | SA |
| 866 | No | 9 | PSA |
| 889 | Yes | 51 | SA |
| 892 | No | 67 | PSA |
| 893 | Yes | 46 | EA |
| 899 | No | 5 | SA |
| 908 | Yes | 11 | PSA |
| 926 | Yes | 54 | PSA |
| 944 | No | 60 | PSA |
| 960 | Yes | 16 | PSA |
| 962 | No | 72 | EA |
| 976 | Yes | 0 | CCA |
| 980 | No | 26 | PSA |
| 993 | No | 13 | SA |
| 999 | No | 43 | MA |
| 1005 | No | 12 | PSA |
| 1012 | Yes | 3 | EA |
| 1035 | No | 41 | CCA |
| 1127 | Yes | 45 | PSA |
| 1129 | Yes | 16 | CCA |
| 1193 | Yes | 6 | PSA |
| 1330 | No | 13 | SA |
| 1830 | No | 6 | PSA |
| 1946 | Yes | 0 | PSA |
UA, undifferentiated adenocarcinoma; SA, serous adenocarcinoma; PSA, papillary serous adenocarcinoma; EA: endometrioid adenocarcinoma; CCA, clear cell adenocarcinoma; MA, mucinous adenocarcinoma.
Patients died following the progression of the disease, except for patients 976 and 1946, death was due to myocardium infarction and digestive hemorrhage.
In Vitro Invasion Assay
Cellular invasion was assayed by determining the ability of cells to invade a synthetic basement membrane (Matrigel; Becton-Dickinson, Bedford, MA). Polycarbonate membranes (8-µm pore size) of the upper compartment of Transwell culture chambers were coated with 0.4 µg/ml Matrigel. The upper compartment was filled with OSE media containing 1% FBS and the lower compartment was filled with OSE media either with no serum, with 5% FBS, or with 5% of the indicated ascites. For inactivation, ascites were heated for 10 minutes at 100°C to denature proteins. Ovarian cancer cells were trypsinized and resuspended in OSE media containing 1% FBS. The cell suspension (20 × 103 cells/well) was placed in the upper compartment. Then, cells were incubated at 37°C and allowed to invade through the Matrigel barrier for 24 hours. Following incubation, membranes were fixed with methanol and stained (Giemsa; Sigma-Aldrich). Noninvading cells were removed using a cotton swab, whereas invading cells on the underside of the membrane were counted using an inverted microscope. All experiments were performed at least twice.
Cell Proliferation
Two thousand cells were plated either with no serum, with 5% FBS, or with 5% of the indicated ascites in six-well plates and incubated at 37°C. At defined intervals, cells were trypsinized and cell viability was assessed by a Trypan Blue exclusion assay. Cell numbers were evaluated using a hematocytometer. Each experiment was performed in triplicate.
Spheroid Formation
Spheroids were formed using a modification of the hanging droplet method [19]. Briefly, 4 × 103 cells were resuspended in 15 µl of OSE media supplemented either with 5% FBS, with 5% of the indicated ascites, or without serum, and then placed on the cover of a 150-mm tissue culture plate. The cover was placed over a plate that contained 15 ml of OSE to prevent dehydration of the hanging droplet. Spheroid formation was monitored after 4 days and representative spheroids were photographed.
RNA Extraction
Total RNA was extracted with a reagent (TRIzol; Gibco/BRL, Life Technologies, Inc., Grand Island, NY) as recommended by the manufacturer. RNA was extracted from tumor cells grown to 80% confluence in 100-mm Petri dishes. RNA quality was assessed using a 2100 Bioanalyzer with the RNA 6000 Nano LabChip kit (Agilent Technologies, Mississauga, ON, Canada) according to the manufacturer's protocol.
Microarray Analysis
Hybridization assays and data collection were performed at the McGill University and Genome Quebec Innovation Centre (Montreal, Canada). Briefly, 20 µg of total RNA from each sample was reverse-transcribed using an oligo-dT primer containing a T7 RNA polymerase binding site. In vitro transcription was performed on this cDNA and the resulting cRNA was biotinylated through incorporation of biotinylated dUTP and dCTP. Samples were fragmented in 40 mM Tris-acetate, 100 mM potassium acetate, and 30 mM MgCl2 (pH 8.1) at 95°C to reduce secondary structure. A total of 15 µg of cRNA was hybridized to an Affymetrix HG-U133A GeneChip array (Santa Clara, CA), washed, stained, and scanned with a Hewlett Packard Gene Array scanner (Palo Alto, CA) and .CEL files were normalized based on a quantile method.
Gene expression profiles were analyzed using R (www.r-project.org), a statistical programming language, and Bioconductor [20], an open source software library for the analyses of genomic data based on R. Background subtraction, normalization (quantile normalization), and expression value calculations were performed using the justGCrma function available as part of Bioconductor's gcrma package. Bioconductor's genefilter package was used to filter out genes with insufficient variation in expression across all samples tested. Expression values retained after this filtering process presented intensities greater than 100 U in at least two samples and a log base 2 scale of at least 0.2 for the interquartile range across all tested samples. Differentially expressed genes were identified using the limma package, which estimates the fold-change between predefined groups by fitting a linear model and using an empirical Bayes method to moderate standard errors of the estimated log-fold changes for expression values from each probe set.
Quantitative PCR
cDNA synthesis was prepared using the SuperScript First-Strand Synthesis System for reverse transcription-polymerase chain reaction (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instruction. Reverse transcription-polymerase chain reaction was performed on 2 µg of total RNA using 2.5 µl of the random hexamer solution. Samples were diluted 1:10 in water prior to Q-PCR. Positive and negative controls were included in all experiments.
Q-PCR was performed using the Rotor-gene 3000 (Corbett Research, Montreal Biotech Inc., Montreal, QC, Canada). Quantitect SYBR Green PCR (QIAGEN Inc., Mississauga, ON, Canada) was used for labeling in a final volume of 25 µl containing 5 µl of sample cDNA and 10 pg of the different primers and reactions performed as described by the manufacturer. Experiments were repeated at least twice. Serial dilutions (1:5) were performed to generate a standard curve for each gene tested to define the efficiency of the Q-PCR reaction and a melt curve was done to confirm the specificity of the reaction. We used the Pfaffl analysis method to measure the relative quantity of gene expression [21]. The algorithm is defined by R = (Etarget)ΔCp target(control - sample)/(Eref)ΔCp ref(control - sample), where R is the relative expression ratio, E is the efficiency of the PCR reaction, and ΔCp is the difference of the Ct (crossing point of the sample at a given threshold). The reference gene, ActinB, was selected based on its stable expression in all samples by microarray analysis. Moreover, Q-PCR confirmed its appropriateness because no significant statistical differences were noted among the samples. The first sample (with 5% FBS) served as the reference sample in each experiment. The mean value of the Ct from replicates was used to calculate R. Marker expression was evaluated in the OV-90 cell line either with no serum, with 5% FBS, or with 5% of the indicated ascites, under the same conditions used to evaluate the invasion potential of the OV-90 cell line. For each marker, a Pearson correlation was calculated between the scored invasion result (1 < 100% and 2 ≥ 100% of invasion) and the scored genes expression (1 < median and 2 ≥ median).
Statistical Analysis
Univariate Cox proportional hazard regression, Kaplan-Meier survival plots, and log-rank tests were performed to determine the significance of markers' ability to predict the survival of EOC patients (Table 2). The expression threshold used in the log-rank test that gave the best sensitivity-specificity values was established based on the receiver operating characteristics (ROC) curves. We used the ROC and SURVIVAL packages from R version 2.4.0 (Vienna, Austria).
The Spearman correlation coefficient test (two-tailed) was used to estimate the correlation between the invasion rates and clinical data and gene expression. Statistical analyses were performed with SPSS software 11.0 (SPSS Inc., Chicago, IL).
Results
Effect of Ascites on OV-90 Invasive Potential
To address the interactions among elements found within the peritoneal tumor environment, we characterized the effect of a large panel of ascites (Table 1) on the ability of an aggressive EOC cell line (OV-90) to stimulate invasion in an in vitro assay. Media containing 5% of ascites (acellular fraction) from patients with ovarian cancer was added to the lower chamber of Transwell plates containing micropore filters precoated with Matrigel and OV-90 cells were added to the upper chamber. The potential for ascites to affect OV-90 invasion was scored in comparison to media supplemented with 5% FBS (Figure 1A). OV-90 is capable of invasion in Matrigel assay in presence of 5% FBS (Figure 1). A large number of ascites led to an inhibition of OV-90 cell invasion compared to cells in the presence of FBS. A lower, but still important number of ascites was more stimulatory for invasion than the FBS control. We assessed the correlation between the invasion rates of ascites and clinical parameters available for patients: age, grade, stage, presence of neoplastic cells, and chemotherapy, but no significant correlation was observed (Table 1).
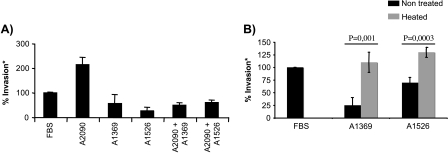
Figure 1.
Effect of ascites on the invasion of the ovarian cancer cell line OV-90. The invasive potential of OV-90 (solid bars) was determined by its ability to invade a synthetic basement membrane after 24 hours compared to 5% FBS (%Invasion*). (A) Effects of 54 ascites on the invasive potential of the cell line. (B) Invasion profile of OV-90 with OSE medium in the absence or presence of 5% FBS or with 5% of those ascites selected for gene expression analysis.
The acellular fraction of 10 independent EOC ascites (A1317, A1318, A1322, A1322(2), A1337, A1592, A1835, A1946, A2085, and A2090; Table 1) was selected for further characterization based on their ability to invade. The invasive potential was again compared to a 5% FBS control. In OV-90 cells, it should be noted that there is no difference in the invasion rate with or without 5% FBS and thus the classification of ascites as being either inhibitory or stimulatory is independent of the effect of serum. Inhibitory ascites reduce the invasion rate compared to either no serum or 5% FBS, whereas stimulatory ascites results from a higher number of cells able to cross a Matrigel barrier in comparison to controls. Ascites A1592, A1946, A2085, and A2090 induced an invasion rate greater to the one observed in the presence or the absence of FBS. A1317, A1318, A1322, A1322(2), A1337, and A1835 diminished OV-90 cell invasion potential (Figure 1B). These effects were not due to differences in protein concentration of each ascites because correcting for the amount of protein did not affect the overall patterns observed (data not shown).
Effect of Ascites on OV-90 Growth
To determine the effect of ascites on proliferation rates, 10 ascites were selected and tested for their ability to alter the growth potential of OV-90 cells compared to a 5% FBS control (Figure 2). OV-90 cells were incubated for 2 days in media supplemented either with no serum, with 5% FBS, or with 5% of the indicated ascites and cell growth was evaluated by a Trypan Blue exclusion assay. The highest growth rates were observed with exposure to FBS, as well as A1835, A1946 or A2085, whereas the remaining ascites samples conferred variable but lower growth rates. In particular, OV-90 exhibited lower growth rate in the presence of the A1337 and A1322(2) ascites compared to those observed with no serum. We noted that after 24 hours of incubation, the time used to monitor invasion, no statistically significant differences in the growth rate were observed (with one exception). This suggests that the monitored invasive effects are not a simple reflection of cell growth.
Figure 2.
Effect of ascites on the proliferation of the OV-90 ovarian cancer cell line. On day 0, 2 × 105 cells were incubated in media supplemented either with no serum, with 5% FBS, or with 5% ascites and the cell growth was evaluated with a Trypan Blue exclusion assay at 24 and 48 hours. Note that, with one exception, there were no statistically significant differences in growth after 24 hours in the different tested conditions. The asterisk denotes a statistical significance (P < .05).
Effect of Ascites on OV-90 Spheroid Formation
We have previously demonstrated that the OV-90 cells are able to grow as large compact spheroids [19]. Because the relation between these three-dimensional structures and invasive potential remains poorly defined, we determined the effect of ascites on the formation of in vitro spheroids. For this purpose, the OV-90 cell line was incubated either in the absence of serum, in 5% FBS, or in 5% of ascites. The formation of spheroids was monitored after 4 days. As shown in Figure 3, the OV-90 cell line formed multiple very small and nonreproducible spheroids from drop to drop in the absence of FBS, which is consistent with previous findings [19]. In the presence of FBS or ascites A1317 and A1592, however, OV-90 formed large (approximately 500 µm in diameter) and compact spheroids. This was reproducible from drop to drop with one unique spheroid of similar size formed in each drop. Although cell scattering around spheroids was observed, it was not significantly different between spheroids generated in the presence of different ascites. The ability for different ascites to induce large and compact spheroid formation was also confirmed with ascites A1318, A1322, A1322(2), A1337, A1835, A1946, A2085, and A2090, which gave similar results (Figure W1). Therefore, the ability of ascites to stimulate spheroid formation did not correlate with the invasion potential of the tested ascites.
Figure 3.
Effect of ascites on spheroid formation. Spheroids were formed using a modification of the hanging droplet method. Cells were incubated with OSE media supplemented either with no serum, with 5% FBS, or with 5% of the indicated ascites. Spheroid formation was monitored after 4 days. All pictures were taken at a magnification of × 100.
Ascites Invasive Phenotype Characterization
Ascites that stimulated or inhibited OV-90 cell invasion were combined to determine which effect is the most predominant (Figure 4A). We selected one stimulatory (A2090) and two inhibitory (A1369 and A1526) ascites. When stimulatory and inhibitory ascites were added together (5% A2090 + 5% A1369 or 5% A2090 + 5% A1526), we observed that the invasion rate was not significantly different compared to the presence of inhibitory ascites alone. This result suggests that the inhibitory ascites have a predominant impact on invasion of OV-90 cells compared to a stimulatory ascites. This effect was not due to an increased concentration of ascites in the assay as adjusting individually tested ascites to 10% had no effect on the invasive phenotype (data not shown).
Figure 4.
In vitro invasion assay with the ovarian cancer cell line OV-90. (A) Effect of simultaneous exposure to ascites that stimulate or inhibit the invasion of OV-90. Note that the inhibitory effect appears to be dominant. (B) Proteins of ascites A1369 and A1526 were inactivated by heating and their effect on the invasion of OV-90 was evaluated. Note that ascites no longer maintain their inhibitory effect.
To determine if this predominant effect is protein-dependent, inhibitory ascites were boiled at 100°C for 10 minutes before being tested on OV-90 cells (Figure 4B). The results showed that protein inactivation abolished the inhibitory effect of the two selected ascites (A1369 and A1526). These combined results suggest that inhibition of OV-90 cell invasion is protein-based and that the inhibitory effect is stronger than the stimulatory effect.
Modification of Gene Expression Induced By Ascites in OV-90 Cells
To identify potential molecular players in invasion regulated by ascites, gene profiling was performed using Affymetrix HG-U133A GeneChip arrays with RNA extracted from OV-90 cells after 24 hours of exposure to either no serum, to 5% FBS, or to 5% of 1 of 10 different ascites (A1317, A1318, A1322, A1322(2), A1337, A1592, A1835, A1946, A2085, and A2090).
For supervised analysis purposes, two groups were created for comparison. The group that stimulated invasion (referred here as GSTIMUL group) included samples with no FBS, with 5% FBS, or with 5% of the four ascites that stimulated OV-90 cell invasion (A1592, A1946, A2085, and A2090). An inhibitory group (referred here as GINHIB group) containing the six ascites that inhibited cell invasion (A1317, A1318, A1322, A1322(2), A1337, and A1835) was also defined. Expression analysis identified 243 probe sets to be differentially expressed (P ≤ .1) between the GSTIMUL and GINHIB groups (Table W1).
Differential Expression Validation of Selected Candidates By Q-PCR
As differences in expression are subtle but tended to be statistically significant, it was important to validate the value of the cutoff selected. For this purpose, we selected genes with different P values (Table 3) to test the robustness of their association with invasion. Table 3 describes the seven candidate genes upregulated in the GSTIMUL group [dickkopf homolog 1 (Xenopus) (DKK1), regulator of G-protein signaling 4 (RGS4), interferon-stimulated transcription factor 3, gamma 48 kDa (ISGF3G), tribbles homolog 1 (Drosophila) (TRIB1), MAP kinase phosphatase 1 (MKP1), cyclooxygenase 2 (COX2), and motile sperm domain containing 1 (MOSPD1)] and two candidate genes upregulated in the GINHIB group [plectin 1, intermediate filament binding protein 500 kDa (PLEC1) and myristoylated alanine-rich protein kinase C substrate (MARCKS)] that were selected for further validation. Q-PCR was used to validate the differential expression of selected candidates in RNA derived from OV-90 cells exposed individually to the entire panel of 54 ascites (Table 1). The relative expression ratio (R) of each gene, based on the Pfaffl method (see Materials and Methods section for details), was quantified and, for each experiment, the median ratio was calculated and the results scored. Pearson correlations were then calculated for each candidate correlating ascites invasion effects (stimulatory or inhibitory) and scored gene expression, as shown in Table 4. The scored gene expression of RGS4, ISGF3G, TRIB1, MKP1, MOSPD1, and PLEC1 correlated significantly with the ascites' invasion effects, but not the expression of DKK1, COX2, and MARCKS. These results also suggest that an extensive validation of candidates identified in the supervised analysis by Q-PCR is warranted to uncover the richness of genes implicated in the invasive process.
Table 3.
Selected Genes Differentially Expressed between the GSTIMUL Group and the GINHIB Group.
| P | Regulation in OV-90 Cells with Ascites That Inhibited the Invasion | Probe Set HG-U133a | UniGene | Description | Symbol | Cytoband | GO Biological Process Description | GO Molecular Function Description |
| .011 | Down | 204602_at | Hs.40499 | Dickkopf homolog 1 (Xenopus laevis) | DKK1 | 10q11.2 | Development | Signal transducer activity |
| Wnt receptor signaling pathway | Protein binding | |||||||
| Growth factor activity | ||||||||
| Low-density lipoprotein receptor binding | ||||||||
| .027/.082* | Down | 204337_at | Hs.386726 | Regulator of G-protein signaling 4 | RGS4 | 1q23.3 | Inactivation of MAPK activity | Signal transducer activity |
| 204339_s_at | G-protein-coupled receptor protein signaling pathway | GTPase activator activity | ||||||
| Calmodulin binding | ||||||||
| .044 | Down | 203882_at | Hs.1706 | Interferon-stimulated transcription factor 3, gamma 48 kDa | ISGF3G | 14q11.2 | Transcription | Transcription factor activity |
| Immune response | Protein binding | |||||||
| Cell surface receptor-linked signal transduction | Metal ion binding | |||||||
| Response to virus | ||||||||
| .052 | Down | 202241_at | Hs.444947 | Tribbles homolog 1 (Drosophila) | TRIB1 | 8q24.13 | Protein amino acid phosphorylation | Protein serine/threonine kinase activity |
| Cell proliferation | ||||||||
| Regulation of MAPK activity | ATP binding | |||||||
| .057 | Down | 201041_s_at | Hs.171695 | MAP kinase phosphatase 1 | MKP1 | 5q34 | Protein amino acid | Non-membrane-spanning protein |
| Dephosphorylation | Tyrosine phosphatase activity | |||||||
| Response to oxidative stress | Protein binding | |||||||
| Cell cycle | Hydrolase activity | |||||||
| MAP kinase phosphatase activity | ||||||||
| Protein tyrosine/serine/threonine phosphatase activity | ||||||||
| .066 | Down | 204748_at | Hs.196384 | Cyclooxygenase 2 | COX2 | 1q25.2 | Prostaglandin biosynthesis | Peroxidase activity |
| q25.3 | Cell motility | Prostaglandin-endoperoxide synthase activity | ||||||
| Physiological process | Metal ion binding | |||||||
| Blood pressure regulation | Oxidoreductase activity | |||||||
| Cyclooxygenase pathway | ||||||||
| Keratinocyte differentiation | ||||||||
| Anagen | ||||||||
| Lipid biosynthesis | ||||||||
| Inflammatory response | ||||||||
| .082 | Down | 218853_s_at | Hs.590789 | Motile sperm domain containing 1 | MOSPD1 | Xq26.3 | Structural molecule activity | |
| .011 | Up | 216971_s_at | Hs.434248 | Plectin 1, intermediate filament binding protein 500 kDa | PLEC1 | 8q24 | Cytoskeletal anchoring | Actin binding |
| Structural molecule activity | ||||||||
| .014 | Up | 213002_at | Hs.519909 | Myristoylated alanine-rich protein kinase C substrate | MARCKS | 6q22.2 | Cell motility | Calmodulin binding |
| Actin binding |
The GSTIMUL group consists of OV-90 samples with no FBS, with 5% FBS, or with 5% of the four ascites that stimulated cell invasion (A1592, A1946, A2085, and A2090). The GINHIB group consists of OV-90 samples with 5% of the six ascites that inhibited cell invasion (A1317, A1318, A1322, A1322(2), A1337, and A1835).
Corresponding to two different probeset on the Affymetrix HG-U133A GeneChip array.
Table 4.
Correlation between Invasion and Genes Expression.
| Genes | Pearson Correlation | P |
| DKK1 | 0.096 | .468 |
| RGS4 | 0.302* | .019 |
| ISGF3G | 0.377† | .003 |
| TRIB1 | 0.302* | .019 |
| MKP1 | 0.397† | .002 |
| COX2 | 0.206 | .114 |
| MOSPD1 | 0.357† | .005 |
| MARCKS | 0.226 | .082 |
| PLEC1 | 0.322* | .019 |
Pearson correlations were calculated between scored invasion results (1 < 100% of invasion and 2 ≥ 100% of invasion) and scored genes expression (1 < median and 2 ≥ median) for the six candidates quantified by Q-PCR.
Correlation is significant at the level 0.05.
Correlation is significant at the level 0.01.
Survival
To evaluate the prognosis potential of the selected gene candidates RGS4, ISGF3G, TRIB1, MKP1, MOSPD1, and PLEC1, we sought to access the expression of these genes in samples derived from patients with ovarian ascites. Therefore, we extracted expression values from a microarray analysis of RNA extracted from 28 primary cultures derived from the cellular fraction of ascites from ovarian cancer patients (Table 2). Univariate Cox regression analysis showed a strong association only between TRIB1 gene expression and overall survival (P = .0007). Using a threshold determined by ROC analysis, a Kaplan-Meier curve coupled to a log-rank test identified the presence of two patient groups and high TRIB1 expression was associated with a poorer survival rate (log-rank P = .005) (Figure 5).
Figure 5.
Relation between TRIB1 expression and cumulative survival of patients with ovarian cancer in the context of concomitant ascites. The threshold was determined by ROC analysis. Kaplan-Meier graphical representation of survival curve illustrates the poor survival associated with a high expression of TRIB1 (P < .005).
Discussion
Although ascites is a common phenotype in ovarian cancer, the origin of malignant ascitic fluid and its relationship to the developing tumor is still poorly understood. The observation that ascites is often associated with the most invasive malignant tumors indirectly supports the notion that ascites is involved in the progression of ovarian cancer [22] presumably by favoring the dissemination of malignant cells within the peritoneal cavity. Contributing to the development of ascites are soluble factors produced by tumor cells that are known to increase vascular permeability and induce angiogenesis [5,23]. Although ascitic soluble factors such as chemokines, angiogenic factors, and growth factors have been implicated in ovarian cancer cell invasion [24–26], the combined effect of these factors in the progression of ovarian cancer has not been addressed. In this study, we assessed the effects of ascites on the invasion, proliferation, spheroid formation, and gene expression of the cell line OV-90 [16]. The overall aim was to determine how ascites alters its microenvironment and thus the biological characteristics of ovarian cancer cells.
Using a Matrigel-based invasion assay, 54 individual ascites showed varying effects on OV-90 cell invasion with some ascites being either poorly stimulatory or inhibitory compared to serum. This data is in contrast to other ascites that strongly stimulated the invasive capacity of this cell line. The varying invasive effects were not associated with total protein concentration in ascites, because adjusting for this factor did not affect the overall effect of invasion (data not shown). The inhibitory effect of ascites was lost when samples were heated, suggesting that the effect was due to protein inactivation rather than other soluble factors. Moreover, no correlation between the effect of ascites on invasion and chemotherapy, received prior to ascites collections, was noted.
Correlation of the invasion assay results with two other parameters, cellular proliferation and spheroid formation, was assessed because stimulation of cell growth could lead to bias in the number of cells counted in the upper chamber of the Transwell and because formation of compact spheroids may interfere with the ability to pass through the polycarbonate membrane pores. The initial characterization studies focused on 10 ascites possessing either stimulatory or inhibitory properties. Although cell growth and spheroid formation were influenced to some extent by ascites, neither parameter was strongly associated with OV-90 invasive capacity. In general, higher doubling times or saturation densities with ascites in comparison to serum were not observed. These findings are similar to results obtained by independent analyses of the PEO.36, OVHS, and SKOV-3 ovarian cancer cell lines [15,27], suggesting that although ascites might affect the in vitro biological characteristics (e.g., survival) of tumor cells, it does not necessarily contribute to the proliferation of tumor cells.
To investigate the molecular events associated with the invasive effect of ascites, global RNA gene expression from OV-90 cells in the presence or absence of serum or ascites was characterized. As expected, differential gene expression patterns occurred in treated cells from which we determined a subset of differentially expressed genes that correlated with invasion potential. Interestingly, most of the genes within this subset have not been previously linked to ovarian cancer invasion. Among these genes, DUSP1/MKP1, TRIB1, ISGF3g/IRF-9, RGS4, MOSPD1, and PLEC1 overexpression was validated by Q-PCR in a large set of samples confirming the potential role of these genes in the cellular invasion process.
The gene signature of ascites-stimulated OV-90 cells reinforces the role of the mitogen-activated protein kinase (MAPK) pathway in the invasion process. Extracellular signal -regulated protein kinase (ERK) MAPKs have already been shown to be involved in invasion through the activation of metalloproteinase [28–31] and Snail [32] promoters. Here we observed altered expression of two genes implicated in kinase regulation, namely TRIB1 and MKP1. TRIB1 is a serine/threonine kinase protein interacting with the mitogen-activated protein kinase 4. The effect of this interaction on MAPK pathway activity is unknown but TRIB1 overexpression inhibits Ras- and mitogen-activated protein kinase kinase/extracellular signal-regulated protein kinase kinase 1-mediated activator protein-1 activation whereas ERK activation is enhanced. On the other hand, MKP1, the MAP kinase phosphatase 1, is the target of the ERKs. This phosphorylation does not modify the intrinsic ability of MKP1 to dephosphorylate p44MAPK, but leads to the stabilization of the protein. Although these results suggest that ascites may regulate the invasion potential of OV-90 cells through TRIB1 overexpression that activates ERKs, the microarray data do not show Snail and metalloproteinase overexpression in OV-90 cells stimulated with invasive ascites. This does not rule out that other downstream targets of the ERK pathway could be affected. Indeed, RGS4, a member of the regulator of G protein signaling family, has been involved in the inhibition of MAPK and protein kinase B (PKB/AKT) activation in neuroblastoma cells as well as ERKs activation induced by angiotensin and endothelin but not by serum stimulation [33,34]. Moreover, RGS4 has also been associated with invasion and motility of glioma cells [35]. Our results are consistent with this latter observation because a direct correlation between RGS4 gene expression and invasion was determined.
The gene signature of ascites-stimulated OV-90 also revealed an altered regulation of the ISGF3g/IRF9 and MOSPD1 genes. ISGF3 is an interferon-dependent transcription factor involved in resistance to chemotherapeutic agents [36,37] but not in any known cellular invasion process. MOSPD1 belongs to the transmembrane MSP-containing protein family. Its role in mammals is largely unknown but it is thought to be involved in the formation of protein-protein networks [38]. These networks have not yet been associated with cellular invasion so far but our results suggest a new role for these proteins in cancer progression.
One explanation for the lack of association of the majority of the genes identified in this study with the invasive potential of neoplastic cells is perhaps not surprising because few studies have attempted to determine the direct effect of ascites on ovarian cancer cells. In addition, meta-analyses of gene profiling studies between normal and cancerous cells are difficult to compare with our study due to various factors such as differences in model systems, use of different platforms, and a dearth of clinical data accompanying the studies including whether tumor formation was accompanied with ascites formation. Consequently, over the long term, we favor a functional approach, taking advantage of the model systems we have developed coupled with in vitro and in vivo assays, to determine the precise role of promising candidate genes in the process of invasion.
In summary, this study revealed novel candidate genes that may play an important role in ovarian cancer cell invasion and potentially affect clinical outcomes. This study has also begun to define the importance and subtleties of ascites in modulating the tumor microenvironment and suggests that both positive and negative regulators of tumor behavior may be present in ascites. Further functional assays are required to determine their exact role in this biological process. Continued evaluation of ascites on EOC behavior is warranted and a comprehensive systems biology approach is required to fully understand the complex interactions within the peritoneum that influence EOC progression and metastasis.
Supplementary Material
Acknowledgements
We are grateful to Louise Champoux, Lise Portelance, Manon de Ladurantaye, Jason Madore, and Marise Roy for technical assistance. We thank Pierre Drouin, Philippe Sauthier, and Philippe Gauthier for their assistance in tissue procurement. We are grateful to Luke Masson for reviewing the manuscript and Kate Morris for editing.
Footnotes
This research was supported by The Cancer Research Society, Inc., Strategic Grant Program in Genomics and Proteomics of Metastatic Cancer award to A.-M.M-M, P.N.T., M.C., and D.P. The ovarian tissue bank was supported by the Banque de tissus et de données of the Réseau de recherche sur le cancer of the Fonds de la Recherche en Santé du Québec, affiliated with the Canadian Tumor Repository Network.
This article refers to supplementary material, which is designated by “Figure W1” and “Table W1” and is available online at www.bcdecker.com.
V.O. was supported by studentships from the Canadian Institutes of Health Research, and V.O. and M.Z. by the Canderel fund of the Institut du cancer de Montréal.
References
- 1.Auersperg N, Wong AS, Choi KC, Kang SK, Leung PC. Ovarian surface epithelium: biology, endocrinology, and pathology. Endocr Rev. 2001;22:255–288. doi: 10.1210/edrv.22.2.0422. [DOI] [PubMed] [Google Scholar]
- 2.Auersperg N, Ota T, Mitchell GW. Early events in ovarian epithelial carcinogenesis: progress and problems in experimental approaches. Int J Gynecol Cancer. 2002;12:691–703. doi: 10.1046/j.1525-1438.2002.01152.x. [DOI] [PubMed] [Google Scholar]
- 3.Cvetkovic D. Early events in ovarian oncogenesis. Reprod Biol Endocrinol. 2003;1:68. doi: 10.1186/1477-7827-1-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burleson KM, Casey RC, Skubitz KM, Pambuccian SE, Oegema TR, Jr, Skubitz AP. Ovarian carcinoma ascites spheroids adhere to extracellular matrix components and mesothelial cell monolayers. Gynecol Oncol. 2004;93:170–181. doi: 10.1016/j.ygyno.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 5.Zebrowski BK, Liu W, Ramirez K, Akagi Y, Mills GB, Ellis LM. Markedly elevated levels of vascular endothelial growth factor in malignant ascites. Ann Surg Oncol. 1999;6:373–378. doi: 10.1007/s10434-999-0373-0. [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto S, Hirata M, Yamazaki A, Kageyama T, Hasuwa H, Mizushima H, Tanaka Y, Yagi H, Sonoda K, Kai M, et al. Heparin-binding EGF-like growth factor is a promising target for ovarian cancer therapy. Cancer Res. 2004;64:5720–5727. doi: 10.1158/0008-5472.CAN-04-0811. [DOI] [PubMed] [Google Scholar]
- 7.Mills GB, Eder A, Fang X, Hasegawa Y, Mao M, Lu Y, Tanyi J, Tabassam FH, Wiener J, Lapushin R, et al. Critical role of lysophospholipids in the pathophysiology, diagnosis, and management of ovarian cancer. Cancer Treat Res. 2002;107:259–283. doi: 10.1007/978-1-4757-3587-1_12. [DOI] [PubMed] [Google Scholar]
- 8.Saltzman AK, Hartenbach EM, Carter JR, Contreras DN, Twiggs LB, Carson LF, Ramakrishnan S. Transforming growth factor-alpha levels in the serum and ascites of patients with advanced epithelial ovarian cancer. Gynecol Obstet Invest. 1999;47:200–204. doi: 10.1159/000010095. [DOI] [PubMed] [Google Scholar]
- 9.Abendstein B, Stadlmann S, Knabbe C, Buck M, Muller-Holzner E, Zeimet AG, Marth C, Obrist P, Krugmann J, Offner FA. Regulation of transforming growth factor-beta secretion by human peritoneal mesothelial and ovarian carcinoma cells. Cytokine. 2000;12:1115–1119. doi: 10.1006/cyto.1999.0632. [DOI] [PubMed] [Google Scholar]
- 10.Jandu N, Richardson M, Singh G, Hirte H, Hatton MW. Human ovarian cancer ascites fluid contains a mixture of incompletely degraded soluble products of fibrin that collectively possess an antiangiogenic property. Int J Gynecol Cancer. 2006;16:1536–1544. doi: 10.1111/j.1525-1438.2006.00624.x. [DOI] [PubMed] [Google Scholar]
- 11.Said NA, Najwer I, Socha MJ, Fulton DJ, Mok SC, Motamed K. SPARC inhibits LPA-mediated mesothelial-ovarian cancer cell crosstalk. Neoplasia. 2007;9:23–35. doi: 10.1593/neo.06658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fung-Kee-Fung M, Provencher D, Rosen B, Hoskins P, Rambout L, Oliver T, Gotlieb W, Covens A. Intraperitoneal chemotherapy for patients with advanced ovarian cancer: a review of the evidence and standards for the delivery of care. Gynecol Oncol. 2007;105:747–756. doi: 10.1016/j.ygyno.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Petignat P, du Bois A, Bruchim I, Fink D, Provencher DM. Should intraperitoneal chemotherapy be considered as standard firstline treatment in advanced stage ovarian cancer? Crit Rev Oncol Hematol. 2007;62:137–147. doi: 10.1016/j.critrevonc.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, Copeland LJ, Walker JL, Burger RA. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed N, Riley C, Oliva K, Rice G, Quinn M. Ascites induces modulation of alpha6beta1 integrin and urokinase plasminogen activator receptor expression and associated functions in ovarian carcinoma. Br J Cancer. 2005;92:1475–1485. doi: 10.1038/sj.bjc.6602495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Provencher DM, Lounis H, Champoux L, Tetrault M, Manderson EN, Wang JC, Eydoux P, Savoie R, Tonin PN, Mes-Masson AM. Characterization of four novel epithelial ovarian cancer cell lines. In Vitro Cell Dev Biol Anim. 2000;36:357–361. doi: 10.1290/1071-2690(2000)036<0357:COFNEO>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 17.Tonin PN, Hudson TJ, Rodier F, Bossolasco M, Lee PD, Novak J, Manderson EN, Provencher D, Mes-Masson AM. Microarray analysis of gene expression mirrors the biology of an ovarian cancer model. Oncogene. 2001;20:6617–6626. doi: 10.1038/sj.onc.1204804. [DOI] [PubMed] [Google Scholar]
- 18.Kruk PA, Maines-Bandiera SL, Auersperg N. A simplified method to culture human ovarian surface epithelium. Lab Invest. 1990;63:132–136. [PubMed] [Google Scholar]
- 19.Zietarska M, Maugard CM, Filali-Mouhim A, Alam-Fahmy M, Tonin PN, Provencher DM, Mes-Masson AM. Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC) Mol Carcinog. 2007;48:872–885. doi: 10.1002/mc.20315. [DOI] [PubMed] [Google Scholar]
- 20.Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;(Suppl):45–51. [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen-Gunther J, Mannel RS. Ascites as a predictor of ovarian malignancy. Gynecol Oncol. 2002;87:77–83. doi: 10.1006/gyno.2002.6800. [DOI] [PubMed] [Google Scholar]
- 23.Richardson M, Gunawan J, Hatton MW, Seidlitz E, Hirte HW, Singh G. Malignant ascites fluid (MAF), including ovarian-cancer-associated MAF, contains angiostatin and other factor(s) which inhibit angiogenesis. Gynecol Oncol. 2002;86:279–287. doi: 10.1006/gyno.2002.6760. [DOI] [PubMed] [Google Scholar]
- 24.Westermann AM, Beijnen JH, Moolenaar WH, Rodenhuis S. Growth factors in human ovarian cancer. Cancer Treat Rev. 1997;23:113–131. doi: 10.1016/s0305-7372(97)90024-4. [DOI] [PubMed] [Google Scholar]
- 25.Brown MR, Blanchette JO, Kohn EC. Angiogenesis in ovarian cancer. Baillieres Best Pract Res Clin Obstet Gynaecol. 2000;14:901–918. doi: 10.1053/beog.2000.0134. [DOI] [PubMed] [Google Scholar]
- 26.Schutyser E, Struyf S, Proost P, Opdenakker G, Laureys G, Verhasselt B, Peperstraete L, Van de Putte I, Saccani A, Allavena P, et al. Identification of biologically active chemokine isoforms from ascitic fluid and elevated levels of CCL18/pulmonary and activation-regulated chemokine in ovarian carcinoma. J Biol Chem. 2002;277:24584–24593. doi: 10.1074/jbc.M112275200. [DOI] [PubMed] [Google Scholar]
- 27.Yabushita H, Shimazu M, Noguchi M, Kishida T, Narumiya H, Sawaguchi K. Vascular endothelial growth factor activating matrix metalloproteinase in ascitic fluid during peritoneal dissemination of ovarian cancer. Oncol Rep. 2003;10:89–95. [PubMed] [Google Scholar]
- 28.Montesano R, Soriano JV, Hosseini G, Pepper MS, Schramek H. Constitutively active mitogen-activated protein kinase kinase MEK1 disrupts morphogenesis and induces an invasive phenotype in Madin-Darby canine kidney epithelial cells. Cell Growth Differ. 1999;10:317–332. [PubMed] [Google Scholar]
- 29.Lakka SS, Jasti SL, Kyritsis AP, Yung WK, Ali-Osman F, Nicolson GL, Rao JS. Regulation of MMP-9 (type IV collagenase) production and invasiveness in gliomas by the extracellular signal-regulated kinase and jun amino-terminal kinase signaling cascades. Clin Exp Metastasis. 2000;18:245–252. doi: 10.1023/a:1006724826083. [DOI] [PubMed] [Google Scholar]
- 30.Tan X, Egami H, Abe M, Nozawa F, Hirota M, Ogawa M. Involvement of MMP-7 in invasion of pancreatic cancer cells through activation of the EGFR mediated MEK-ERK signal transduction pathway. J Clin Pathol. 2005;58:1242–1248. doi: 10.1136/jcp.2004.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed N, Oliva K, Wang Y, Quinn M, Rice G. Downregulation of urokinase plasminogen activator receptor expression inhibits Erk signalling with concomitant suppression of invasiveness due to loss of uPAR- beta1 integrin complex in colon cancer cells. Br J Cancer. 2003;89:374–384. doi: 10.1038/sj.bjc.6601098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbera MJ, Puig I, Dominguez D, Julien-Grille S, Guaita-Esteruelas S, Peiro S, Baulida J, Franci C, Dedhar S, Larue L, et al. Regulation of Snail transcription during epithelial to mesenchymal transition of tumor cells. Oncogene. 2004;23:7345–7354. doi: 10.1038/sj.onc.1207990. [DOI] [PubMed] [Google Scholar]
- 33.Leone AM, Errico M, Lin SL, Cowen DS. Activation of extracellular signal-regulated kinase (ERK) and Akt by human serotonin 5-HT(1B) receptors in transfected BE(2)-C neuroblastoma cells is inhibited by RGS4. J Neurochem. 2000;75:934–938. doi: 10.1046/j.1471-4159.2000.0750934.x. [DOI] [PubMed] [Google Scholar]
- 34.Albig AR, Schiemann WP. Identification and characterization of regulator of G protein signaling 4 (RGS4) as a novel inhibitor of tubulogenesis: RGS4 inhibits mitogen-activated protein kinases and vascular endothelial growth factor signaling. Mol Biol Cell. 2005;16:609–625. doi: 10.1091/mbc.E04-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatenhorst L, Senner V, Puttmann S, Paulus W. Regulators of G-protein signaling 3 and 4 (RGS3, RGS4) are associated with glioma cell motility. J Neuropathol Exp Neurol. 2004;63:210–222. doi: 10.1093/jnen/63.3.210. [DOI] [PubMed] [Google Scholar]
- 36.Luker KE, Pica CM, Schreiber RD, Piwnica-Worms D. Overexpression of IRF9 confers resistance to antimicrotubule agents in breast cancer cells. Cancer Res. 2001;61:6540–6547. [PubMed] [Google Scholar]
- 37.Schmidt M, Schler G, Gruensfelder P, Hoppe F. Differential gene expression in a paclitaxel-resistant clone of a head and neck cancer cell line. Eur Arch Otorhinolaryngol. 2006;263:127–134. doi: 10.1007/s00405-005-0936-z. [DOI] [PubMed] [Google Scholar]
- 38.Laurent F, Labesse G, de Wit P. Molecular cloning and partial characterization of a plant VAP33 homologue with a major sperm protein domain. Biochem Biophys Res Commun. 2000;270:286–292. doi: 10.1006/bbrc.2000.2387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.