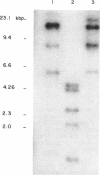
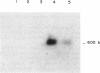
Abstract
Inserts which carried the CUP gene from Saccharomyces cerevisiae or Mucor racemosus were used as hybridization probes to measure the methylation state and expression of the CUP gene from Mucor rouxii at different stages of growth. It was observed that the fungus contains a CUP multigene family. All the CUP genes were present in a hypermethylated DNA region in nongrowing and isodiametrically growing spores and were not transcribed at these stages. After germ tube emergence, CUP genes became demethylated and transcriptionally active. Development, demethylation, and transcription of CUP genes were blocked by the ornithine decarboxylase inhibitor 1,4-diaminobutanone. These results suggest that genes that are activated during development became demethylated in this fungus.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aladjem M. I., Koltin Y., Lavi S. Enhancement of copper resistance and CupI amplification in carcinogen-treated yeast cells. Mol Gen Genet. 1988 Jan;211(1):88–94. doi: 10.1007/BF00338397. [DOI] [PubMed] [Google Scholar]
- Antequera F., Tamame M., Villanueva J. R., Santos T. DNA methylation in the fungi. J Biol Chem. 1984 Jul 10;259(13):8033–8036. [PubMed] [Google Scholar]
- BARTNICKI GARCIA S. SYMPOSIUM ON BIOCHEMICAL BASES OF MORPHOGENESIS IN FUNGI. III. MOLD-YEAST DIMORPHISM OF MUCOR. Bacteriol Rev. 1963 Sep;27:293–304. doi: 10.1128/br.27.3.293-304.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTNICKI-GARCIA S., NICKERSON W. J. Induction of yeast-like development in Mucor by carbon dioxide. J Bacteriol. 1962 Oct;84:829–840. doi: 10.1128/jb.84.4.829-840.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P., Greaves D. R. Programmed gene rearrangements altering gene expression. Science. 1987 Feb 6;235(4789):658–667. doi: 10.1126/science.3544215. [DOI] [PubMed] [Google Scholar]
- Butt T. R., Sternberg E. J., Gorman J. A., Clark P., Hamer D., Rosenberg M., Crooke S. T. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3332–3336. doi: 10.1073/pnas.81.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano C., Herrera-Estrella L., Ruiz-Herrera J. DNA methylation and polyamines in regulation of development of the fungus Mucor rouxii. J Bacteriol. 1988 Dec;170(12):5946–5948. doi: 10.1128/jb.170.12.5946-5948.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Compere S. J., Palmiter R. D. DNA methylation controls the inducibility of the mouse metallothionein-I gene lymphoid cells. Cell. 1981 Jul;25(1):233–240. doi: 10.1016/0092-8674(81)90248-8. [DOI] [PubMed] [Google Scholar]
- Dynan W. S. Understanding the molecular mechanism by which methylation influences gene expression. Trends Genet. 1989 Feb;5(2):35–36. doi: 10.1016/0168-9525(89)90016-4. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fogel S., Welch J. W. Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5342–5346. doi: 10.1073/pnas.79.17.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Faugeron G. Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol Cell Biol. 1989 Jul;9(7):2818–2827. doi: 10.1128/mcb.9.7.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer D. H. Metallothionein. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- Hamer D. H., Thiele D. J., Lemontt J. E. Function and autoregulation of yeast copperthionein. Science. 1985 May 10;228(4700):685–690. doi: 10.1126/science.3887570. [DOI] [PubMed] [Google Scholar]
- Heby O. Role of polyamines in the control of cell proliferation and differentiation. Differentiation. 1981;19(1):1–20. doi: 10.1111/j.1432-0436.1981.tb01123.x. [DOI] [PubMed] [Google Scholar]
- Inderlied C. B., Cihlar R. L., Sypherd P. S. Regulation of ornithine decarboxylase during morphogenesis of Mucor racemosus. J Bacteriol. 1980 Feb;141(2):699–706. doi: 10.1128/jb.141.2.699-706.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe E. R., Magill J. M., Magill C. W. Stage-specific DNA methylation in a fungal plant pathogen. J Bacteriol. 1986 Feb;165(2):420–423. doi: 10.1128/jb.165.2.420-423.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill J. M., Magill C. W. DNA methylation in fungi. Dev Genet. 1989;10(2):63–69. doi: 10.1002/dvg.1020100202. [DOI] [PubMed] [Google Scholar]
- Nemer M., Travaglini E. C., Rondinelli E., D'Alonzo J. Developmental regulation, induction, and embryonic tissue specificity of sea urchin metallothionein gene expression. Dev Biol. 1984 Apr;102(2):471–482. doi: 10.1016/0012-1606(84)90212-4. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Heguy A., Karin M. Structural and functional analysis of the human metallothionein-IA gene: differential induction by metal ions and glucocorticoids. Cell. 1984 May;37(1):263–272. doi: 10.1016/0092-8674(84)90322-2. [DOI] [PubMed] [Google Scholar]
- Russell P. J., Rodland K. D., Rachlin E. M., McCloskey J. A. Differential DNA methylation during the vegetative life cycle of Neurospora crassa. J Bacteriol. 1987 Jun;169(6):2902–2905. doi: 10.1128/jb.169.6.2902-2905.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. J., Welsch J. A., Rachlin E. M., McCloskey J. A. Different levels of DNA methylation in yeast and mycelial forms of Candida albicans. J Bacteriol. 1987 Sep;169(9):4393–4395. doi: 10.1128/jb.169.9.4393-4395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko-Bulgac E. P., Sherman F., Hicks J. B. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J Bacteriol. 1990 Mar;172(3):1276–1283. doi: 10.1128/jb.172.3.1276-1283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987 Dec 4;51(5):741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Jensen B. C., Richardson G. A. A portable signal causing faithful DNA methylation de novo in Neurospora crassa. Science. 1987 Oct 2;238(4823):48–53. doi: 10.1126/science.2958937. [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Yelton M. M., Hamer J. E., Timberlake W. E. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]