Abstract
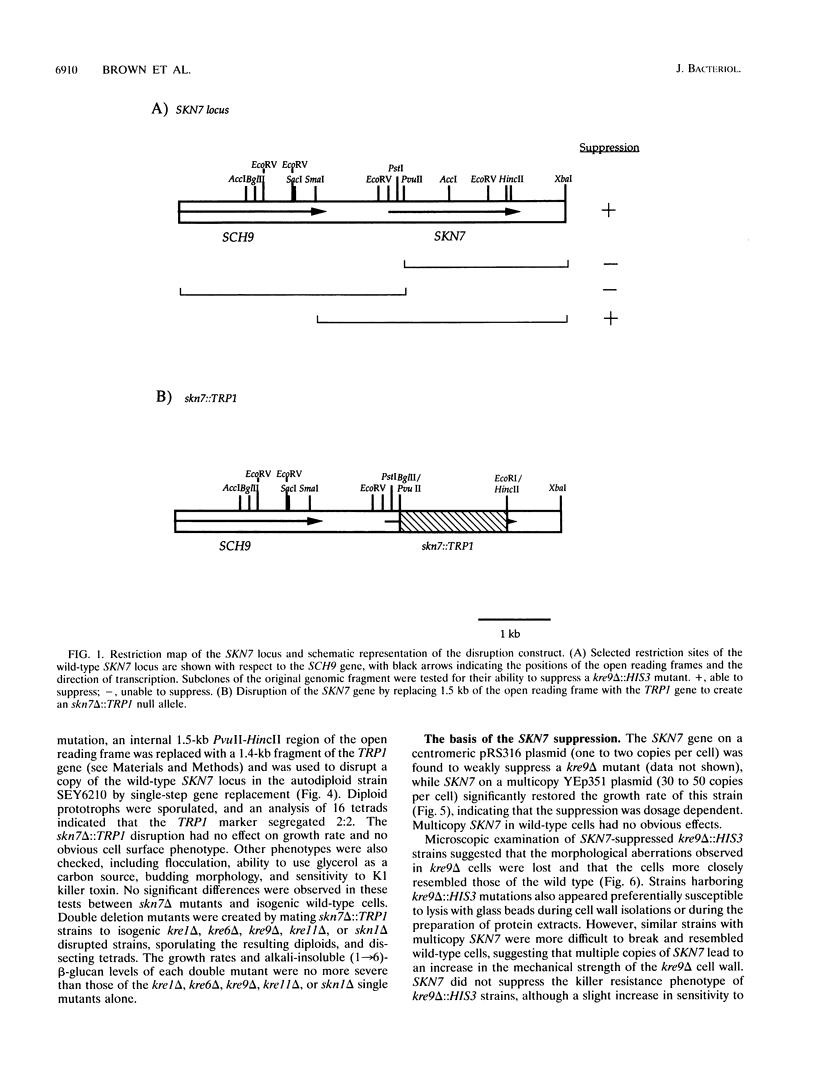
A search for genes which, at elevated copy number, could suppress the growth defect in a strain disrupted at the KRE9 locus has identified the SKN7 gene. SKN7 was mapped to the right arm of chromosome VIII and is predicted to encode a 70-kDa protein, Skn7p, with a region of homology to the DNA binding domain of the Saccharomyces cerevisiae heat shock transcription factor, Hsf1p. Skn7p also has a domain which shows similarity to the prokaryotic receiver modules found on an extensive family of two-component response regulators, including the products of the rcsC and barA genes. SKN7 did not suppress other mutations in the (1-->6)-beta-glucan biosynthetic pathway, suggesting that SKN7 does not act as a general bypass suppressor of this glucan.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- BADIN J., JACKSON C., SCHUBERT M. Improved method for determination of plasma polysaccharides with tryptophan. Proc Soc Exp Biol Med. 1953 Nov;84(2):289–291. [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Boone C., Sommer S. S., Hensel A., Bussey H. Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J Cell Biol. 1990 May;110(5):1833–1843. doi: 10.1083/jcb.110.5.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. L., Bussey H. The yeast KRE9 gene encodes an O glycoprotein involved in cell surface beta-glucan assembly. Mol Cell Biol. 1993 Oct;13(10):6346–6356. doi: 10.1128/mcb.13.10.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. L., Kossaczka Z., Jiang B., Bussey H. A mutational analysis of killer toxin resistance in Saccharomyces cerevisiae identifies new genes involved in cell wall (1-->6)-beta-glucan synthesis. Genetics. 1993 Apr;133(4):837–849. doi: 10.1093/genetics/133.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol Cell Biol. 1992 Apr;12(4):1764–1776. doi: 10.1128/mcb.12.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E., Slater M., Cabib E., Au-Young J., Sburlati A., Adair W. L., Jr, Robbins P. W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986 Jul 18;46(2):213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- Bussey H., Sacks W., Galley D., Saville D. Yeast killer plasmid mutations affecting toxin secretion and activity and toxin immunity function. Mol Cell Biol. 1982 Apr;2(4):346–354. doi: 10.1128/mcb.2.4.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E., Kang M. S. Fungal 1,3-beta-glucan synthase. Methods Enzymol. 1987;138:637–642. doi: 10.1016/0076-6879(87)38057-7. [DOI] [PubMed] [Google Scholar]
- Chiang G. G., Schaefer M. R., Grossman A. R. Complementation of a red-light-indifferent cyanobacterial mutant. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9415–9419. doi: 10.1073/pnas.89.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nobel J. G., Klis F. M., Ram A., Van Unen H., Priem J., Munnik T., Van Den Ende H. Cyclic variations in the permeability of the cell wall of Saccharomyces cerevisiae. Yeast. 1991 Aug-Sep;7(6):589–598. doi: 10.1002/yea.320070606. [DOI] [PubMed] [Google Scholar]
- Fujita A., Kikuchi Y., Kuhara S., Misumi Y., Matsumoto S., Kobayashi H. Domains of the SFL1 protein of yeasts are homologous to Myc oncoproteins or yeast heat-shock transcription factor. Gene. 1989 Dec 28;85(2):321–328. doi: 10.1016/0378-1119(89)90424-1. [DOI] [PubMed] [Google Scholar]
- Herscovics A., Orlean P. Glycoprotein biosynthesis in yeast. FASEB J. 1993 Apr 1;7(6):540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- Hill K., Boone C., Goebl M., Puccia R., Sdicu A. M., Bussey H. Yeast KRE2 defines a new gene family encoding probable secretory proteins, and is required for the correct N-glycosylation of proteins. Genetics. 1992 Feb;130(2):273–283. doi: 10.1093/genetics/130.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Martinac B., Zhu H., Kubalski A., Zhou X. L., Culbertson M., Bussey H., Kung C. Yeast K1 killer toxin forms ion channels in sensitive yeast spheroplasts and in artificial liposomes. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6228–6232. doi: 10.1073/pnas.87.16.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaden P., Hill K., Wagner J., Slipetz D., Sommer S. S., Bussey H. The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1----6)-beta-D-glucan synthesis and normal cell growth. Mol Cell Biol. 1990 Jun;10(6):3013–3019. doi: 10.1128/mcb.10.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa S., Tokishita S., Aiba H., Mizuno T. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol Microbiol. 1992 Mar;6(6):799–807. doi: 10.1111/j.1365-2958.1992.tb01530.x. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S., Kofoid E. C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S. Signal transduction schemes of bacteria. Cell. 1993 Jun 4;73(5):857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas J. C., Diaz M., Duran A., Perez P. Isolation and characterization of Schizosaccharomyces pombe mutants defective in cell wall (1-3)beta-D-glucan. J Bacteriol. 1991 Jun;173(11):3456–3462. doi: 10.1128/jb.173.11.3456-3462.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T., Bussey H. Yeast beta-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11295–11299. doi: 10.1073/pnas.88.24.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T., Delaney S., Bussey H. SKN1 and KRE6 define a pair of functional homologs encoding putative membrane proteins involved in beta-glucan synthesis. Mol Cell Biol. 1993 Jul;13(7):4039–4048. doi: 10.1128/mcb.13.7.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C., Valdivieso M. H., Ribas J. C., Durán A. Isolation and characterization of Saccharomyces cerevisiae mutants resistant to Calcofluor white. J Bacteriol. 1988 Apr;170(4):1950–1954. doi: 10.1128/jb.170.4.1950-1954.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. J. Similar and different domains of chitin synthases 1 and 2 of S. cerevisiae: two isozymes with distinct functions. Yeast. 1989 Nov-Dec;5(6):459–467. doi: 10.1002/yea.320050605. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988 Sep 9;54(6):855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Stock A. M., Mottonen J. M., Stock J. B., Schutt C. E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989 Feb 23;337(6209):745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- Stock A., Koshland D. E., Jr, Stock J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Gottesman S. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990 Feb;172(2):659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Wigler M. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 1988 May;2(5):517–527. doi: 10.1101/gad.2.5.517. [DOI] [PubMed] [Google Scholar]
- Valdivieso M. H., Mol P. C., Shaw J. A., Cabib E., Durán A. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J Cell Biol. 1991 Jul;114(1):101–109. doi: 10.1083/jcb.114.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederrecht G., Seto D., Parker C. S. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988 Sep 9;54(6):841–853. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- Wurtzel E. T., Chou M. Y., Inouye M. Osmoregulation of gene expression. I. DNA sequence of the ompR gene of the ompB operon of Escherichia coli and characterization of its gene product. J Biol Chem. 1982 Nov 25;257(22):13685–13691. [PubMed] [Google Scholar]