Abstract
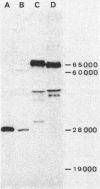
The nucleotide sequence of a 2.7-kilobase segment of DNA containing the sdhA and sdhB genes encoding the flavoprotein (Fp, sdhA) and iron-sulfur protein (Ip, sdhB) subunits of the succinate dehydrogenase of Bacillus subtilis was determined. This sequence extends the previously reported sequence encoding the cytochrome b558 subunit (sdhC) and completes the sequence of the sdh operon, sdhCAB. The predicted molecular weights for the Fp and Ip subunits, 65,186 (585 amino acids) and 28,285 (252 amino acids), agreed with the values determined independently for the labeled Fp and Ip antigens, although it appeared that the B. subtilis Fp was not functional after expression of the sdhA gene in Escherichia coli. Both subunits closely resembled the corresponding Fp and Ip subunits of the succinate dehydrogenase (SDH) and fumarate reductase of E. coli in size, composition, and amino acid sequence. The sequence homologies further indicated that the B. subtilis SDH subunits are equally related to the SDH and fumarate reductase subunits of E. coli but are less closely related than are the corresponding pairs of E. coli subunits. The regions of highest sequence conservation were identifiable as the catalytically significant flavin adenine dinucleotide-binding sites and cysteine clusters of the iron-sulfur centers.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. J., Blobel G. Immunoprecipitation of proteins from cell-free translations. Methods Enzymol. 1983;96:111–120. doi: 10.1016/s0076-6879(83)96012-3. [DOI] [PubMed] [Google Scholar]
- Beinert H., Thomson A. J. Three-iron clusters in iron-sulfur proteins. Arch Biochem Biophys. 1983 Apr 15;222(2):333–361. doi: 10.1016/0003-9861(83)90531-3. [DOI] [PubMed] [Google Scholar]
- Cammack R., Patil D. S., Weiner J. H. Evidence that centre 2 in Escherichia coli fumarate reductase is a [4Fe-4S]cluster. Biochim Biophys Acta. 1986 Apr 22;870(3):545–551. doi: 10.1016/0167-4838(86)90264-5. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Condon C., Lemire B. D., Weiner J. H. Molecular biology, biochemistry and bioenergetics of fumarate reductase, a complex membrane-bound iron-sulfur flavoenzyme of Escherichia coli. Biochim Biophys Acta. 1985 Dec;811(4):381–403. doi: 10.1016/0304-4173(85)90008-4. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Grundström T., Jaurin B., Robinson J. J., Weiner J. H. Location and nucleotide sequence of frdB, the gene coding for the iron-sulphur protein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982 Aug;126(1):211–216. doi: 10.1111/j.1432-1033.1982.tb06768.x. [DOI] [PubMed] [Google Scholar]
- Condon C., Cammack R., Patil D. S., Owen P. The succinate dehydrogenase of Escherichia coli. Immunochemical resolution and biophysical characterization of a 4-subunit enzyme complex. J Biol Chem. 1985 Aug 5;260(16):9427–9434. [PubMed] [Google Scholar]
- Cutting S., Mandelstam J. The nucleotide sequence and the transcription during sporulation of the gerE gene of Bacillus subtilis. J Gen Microbiol. 1986 Nov;132(11):3013–3024. doi: 10.1099/00221287-132-11-3013. [DOI] [PubMed] [Google Scholar]
- Darlison M. G., Guest J. R. Nucleotide sequence encoding the iron-sulphur protein subunit of the succinate dehydrogenase of Escherichia coli. Biochem J. 1984 Oct 15;223(2):507–517. doi: 10.1042/bj2230507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K. A., Hatefi Y., Crawford I. P., Baltscheffsky H. Purification, molecular properties, and amino acid composition of the subunits of Rhodospirillum rubrum succinate dehydrogenase. Arch Biochem Biophys. 1977 Apr 30;180(2):459–464. doi: 10.1016/0003-9861(77)90060-1. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Ghosh D., O'Donnell S., Furey W., Jr, Robbins A. H., Stout C. D. Iron-sulfur clusters and protein structure of Azotobacter ferredoxin at 2.0 A resolution. J Mol Biol. 1982 Jun 15;158(1):73–109. doi: 10.1016/0022-2836(82)90451-x. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Guest J. R., Roberts R. E., Stephens P. E. Hybrid plasmids containing the pyruvate dehydrogenase complex genes and gene-DNA relationships in the 2 to 3 minute region of the Escherichia coli chromosome. J Gen Microbiol. 1983 Mar;129(3):671–680. doi: 10.1099/00221287-129-3-671. [DOI] [PubMed] [Google Scholar]
- Hasnain S., Sammons R., Roberts I., Thomas C. M. Cloning and deletion analysis of a genomic segment of Bacillus subtilis coding for the sdhA, B, C (succinate dehydrogenase) and gerE (spore germination) loci. J Gen Microbiol. 1985 Sep;131(9):2269–2279. doi: 10.1099/00221287-131-9-2269. [DOI] [PubMed] [Google Scholar]
- Hederstedt L., Andersson K. K. Electron-paramagnetic-resonance spectroscopy of Bacillus subtilis cytochrome b558 in Escherichia coli membranes and in succinate dehydrogenase complex from Bacillus subtilis membranes. J Bacteriol. 1986 Aug;167(2):735–739. doi: 10.1128/jb.167.2.735-739.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hederstedt L., Hatefi Y. Modification of bovine heart succinate dehydrogenase with ethoxyformic anhydride and rose bengal: evidence for essential histidyl residues protectable by substrates. Arch Biochem Biophys. 1986 Jun;247(2):346–354. doi: 10.1016/0003-9861(86)90593-x. [DOI] [PubMed] [Google Scholar]
- Hederstedt L., Maguire J. J., Waring A. J., Ohnishi T. Characterization by electron paramagnetic resonance and studies on subunit location and assembly of the iron-sulfur clusters of Bacillus subtilis succinate dehydrogenase. J Biol Chem. 1985 May 10;260(9):5554–5562. [PubMed] [Google Scholar]
- Hederstedt L. Molecular properties, genetics, and biosynthesis of Bacillus subtilis succinate dehydrogenase complex. Methods Enzymol. 1986;126:399–414. doi: 10.1016/s0076-6879(86)26040-1. [DOI] [PubMed] [Google Scholar]
- Hederstedt L., Rutberg L. Biosynthesis and membrane binding of succinate dehydrogenase in Bacillus subtilis. J Bacteriol. 1980 Dec;144(3):941–951. doi: 10.1128/jb.144.3.941-951.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hederstedt L., Rutberg L. Succinate dehydrogenase--a comparative review. Microbiol Rev. 1981 Dec;45(4):542–555. doi: 10.1128/mr.45.4.542-555.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson C. E., Perham R. N., Finch J. T. Structure and symmetry of B. stearothermophilus pyruvate dehydrogenase multienzyme complex and implications for eucaryote evolution. Cell. 1979 May;17(1):85–93. doi: 10.1016/0092-8674(79)90297-6. [DOI] [PubMed] [Google Scholar]
- Johnson M. K., Morningstar J. E., Bennett D. E., Ackrell B. A., Kearney E. B. Magnetic circular dichroism studies of succinate dehydrogenase. Evidence for [2Fe-2S], [3Fe-xS], and [4Fe-4S] centers in reconstitutively active enzyme. J Biol Chem. 1985 Jun 25;260(12):7368–7378. [PubMed] [Google Scholar]
- Johnson M. K., Morningstar J. E., Cecchini G., Ackrell B. A. Detection of a tetranuclear iron-sulfur center in fumarate reductase from Escherichia coli by electron paramagnetic resonance spectroscopy. Biochem Biophys Res Commun. 1985 Sep 16;131(2):756–762. doi: 10.1016/0006-291x(85)91303-8. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Moran C. P., Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983 Apr 28;302(5911):800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- Kenney W. C., Mowery P. C., Seng R. L., Singer T. P. Localization of the substrate and oxalacetate binding site of succinate dehydrogenase. J Biol Chem. 1976 Apr 25;251(8):2369–2373. [PubMed] [Google Scholar]
- Magnusson K., Hederstedt L., Rutberg L. Cloning and expression in Escherichia coli of sdhA, the structural gene for cytochrome b558 of the Bacillus subtilis succinate dehydrogenase complex. J Bacteriol. 1985 Jun;162(3):1180–1185. doi: 10.1128/jb.162.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson K., Philips M. K., Guest J. R., Rutberg L. Nucleotide sequence of the gene for cytochrome b558 of the Bacillus subtilis succinate dehydrogenase complex. J Bacteriol. 1986 Jun;166(3):1067–1071. doi: 10.1128/jb.166.3.1067-1071.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson K., Rutberg B., Hederstedt L., Rutberg L. Characterization of a pleiotropic succinate dehydrogenase-negative mutant of Bacillus subtilis. J Gen Microbiol. 1983 Apr;129(4):917–922. doi: 10.1099/00221287-129-4-917. [DOI] [PubMed] [Google Scholar]
- Maguire J. J., Magnusson K., Hederstedt L. Bacillus subtilis mutant succinate dehydrogenase lacking covalently bound flavin: identification of the primary defect and studies on the iron-sulfur clusters in mutated and wild-type enzyme. Biochemistry. 1986 Sep 9;25(18):5202–5208. doi: 10.1021/bi00366a033. [DOI] [PubMed] [Google Scholar]
- Makaroff C. A., Zalkin H., Switzer R. L., Vollmer S. J. Cloning of the Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase gene in Escherichia coli. Nucleotide sequence determination and properties of the plasmid-encoded enzyme. J Biol Chem. 1983 Sep 10;258(17):10586–10593. [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Miles J. S., Guest J. R. Complete nucleotide sequence of the fumarase gene (citG) of Bacillus subtilis 168. Nucleic Acids Res. 1985 Jan 11;13(1):131–140. doi: 10.1093/nar/13.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morningstar J. E., Johnson M. K., Cecchini G., Ackrell B. A., Kearney E. B. The high potential iron-sulfur center in Escherichia coli fumarate reductase is a three-iron cluster. J Biol Chem. 1985 Nov 5;260(25):13631–13638. [PubMed] [Google Scholar]
- Neville D. M., Jr Molecular weight determination of protein-dodecyl sulfate complexes by gel electrophoresis in a discontinuous buffer system. J Biol Chem. 1971 Oct 25;246(20):6328–6334. [PubMed] [Google Scholar]
- Piggot P. J., Hoch J. A. Revised genetic linkage map of Bacillus subtilis. Microbiol Rev. 1985 Jun;49(2):158–179. doi: 10.1128/mr.49.2.158-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D. W., Schulz G. E., Guest J. R. Structural relationship between glutathione reductase and lipoamide dehydrogenase. J Mol Biol. 1984 Apr 15;174(3):483–496. doi: 10.1016/0022-2836(84)90332-2. [DOI] [PubMed] [Google Scholar]
- Robinson J. J., Weiner J. H. Molecular properties of fumarate reductase isolated from the cytoplasmic membrane of Escherichia coli. Can J Biochem. 1982 Aug;60(8):811–816. doi: 10.1139/o82-101. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Singer T. P., Johnson M. K. The prosthetic groups of succinate dehydrogenase: 30 years from discovery to identification. FEBS Lett. 1985 Oct 14;190(2):189–198. doi: 10.1016/0014-5793(85)81282-5. [DOI] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P. J., Morgan T. V., Devlin F., Penner-Hahn J. E., Hodgson K. O., Scott R. A., Stout C. D., Burgess B. K. [4Fe-4S]-cluster-depleted Azotobacter vinelandii ferredoxin I: a new 3Fe iron-sulfur protein. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5661–5665. doi: 10.1073/pnas.82.17.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Haniu M., Yasunobu K. T. The complete amino acid sequence of the Spirulina platensis ferredoxin. Biochem Biophys Res Commun. 1976 Apr 5;69(3):759–765. doi: 10.1016/0006-291x(76)90940-2. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tsunoda J. N., Yasunobu K. T., Whiteley H. R. Non-heme iron proteins. IX. The amino acid sequence of ferredoxin from Micrococcus aerogenes. J Biol Chem. 1968 Dec 10;243(23):6262–6272. [PubMed] [Google Scholar]
- Unden G., Kröger A. An essential sulfhydryl group at the substrate site of the fumarate reductase of Vibrio succinogenes. FEBS Lett. 1980 Aug 11;117(1):323–326. doi: 10.1016/0014-5793(80)80972-0. [DOI] [PubMed] [Google Scholar]
- Vik S. B., Hatefi Y. Possible occurrence and role of an essential histidyl residue in succinate dehydrogenase. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6749–6753. doi: 10.1073/pnas.78.11.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman P. D. Unity and diversity in some bacterial citric acid-cycle enzymes. Adv Microb Physiol. 1981;22:185–244. doi: 10.1016/s0065-2911(08)60328-8. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Wilde R. J., Jeyaseelan K., Guest J. R. Cloning of the aconitase gene (acn) of Escherichia coli K12. J Gen Microbiol. 1986 Jun;132(6):1763–1766. doi: 10.1099/00221287-132-6-1763. [DOI] [PubMed] [Google Scholar]
- Wood D., Darlison M. G., Wilde R. J., Guest J. R. Nucleotide sequence encoding the flavoprotein and hydrophobic subunits of the succinate dehydrogenase of Escherichia coli. Biochem J. 1984 Sep 1;222(2):519–534. doi: 10.1042/bj2220519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods S. A., Miles J. S., Roberts R. E., Guest J. R. Structural and functional relationships between fumarase and aspartase. Nucleotide sequences of the fumarase (fumC) and aspartase (aspA) genes of Escherichia coli K12. Biochem J. 1986 Jul 15;237(2):547–557. doi: 10.1042/bj2370547. [DOI] [PMC free article] [PubMed] [Google Scholar]