Abstract
We describe the identification of Rex, a novel redox-sensing repressor that appears to be widespread among Gram-positive bacteria. In Streptomyces coelicolor Rex binds to operator (ROP) sites located upstream of several respiratory genes, including the cydABCD and rex-hemACD operons. The DNA-binding activity of Rex appears to be controlled by the redox poise of the NADH/NAD+ pool. Using electromobility shift and surface plasmon resonance assays we show that NADH, but not NAD+, inhibits the DNA-binding activity of Rex. However, NAD+ competes with NADH for Rex binding, allowing Rex to sense redox poise over a range of NAD(H) concentrations. Rex is predicted to include a pyridine nucleotide-binding domain (Rossmann fold), and residues that might play key structural and nucleotide binding roles are highly conserved. In support of this, the central glycine in the signature motif (GlyXGlyXXGly) is shown to be essential for redox sensing. Rex homologues exist in most Gram-positive bacteria, including human pathogens such as Staphylococcus aureus, Listeria monocytogenes and Streptococcus pneumoniae.
Keywords: pyridine nucleotide/redox/repressor/Streptomyces/transcription
Introduction
Nicotinamide adenine dinucleotide (NAD+) is a major acceptor of electrons in the oxidation of fuel molecules. During aerobic respiration, the resulting NADH is reoxidized by oxygen via an electron transport chain, and the released free energy is captured in the form of an electrochemical proton gradient, which is subsequently used to drive ATP synthesis. Since the NADH/NAD+ pool is relatively small compared with its turnover, the continued availability of NAD+ for substrate oxidation relies on concomitant NADH reoxidation, and hence on a continuous supply of oxygen. Bacteria cannot rely on a continuous oxygen supply, and use several adaptive strategies to allow them to cope with oxygen limitation. These include: the expression of alternative terminal cytochrome oxidases with a higher affinity for oxygen; slowing the rate of the TCA cycle, the major drain on NAD+; and ultimately, in the absence of oxygen, the exploitation of alternative electron acceptors such as nitrate and fumarate. The regulatory mechanisms involved are best understood for the facultative anaerobe Escherichia coli, where two major systems exist: the ArcAB two component system that functions mainly to repress aerobic respiratory genes in response to microaerobic conditions; and FNR that functions mainly as an activator of anaerobic pathways (Patschkowski et al., 2000). The ArcAB system consists of a membrane-anchored sensor kinase protein, ArcB, that autophosphorylates under reducing conditions, then transfers its phosphate to ArcA, allowing ArcA to repress several genes that encode TCA cycle enzymes (e.g. icd and mdh; Park et al., 1995; Chao et al., 1997), NADH dehydrogenase I (nuoA-N; Bongaerts et al., 1995) and the aerobic cytochrome bo terminal oxidase (cyoABCDE; Iuchi et al., 1990). Phosphorylated ArcA also activates a few genes, including the cydAB operon that encodes cytochrome bd terminal oxidase, an alternative oxidase with a high affinity for oxygen (Iuchi et al., 1990; Cotter et al., 1997). FNR, in its active form, will bind DNA and either activate transcription (e.g. of the nitrate and fumarate reductase genes) or repress transcription (e.g. of the nuoA-N operon; Meng et al., 1997; reviewed in Guest et al., 1996).
The regulatory systems involved in sensing and responding to oxygen limitation in Gram-positive bacteria are less well understood, although there have been recent developments in understanding how Bacillus subtilis senses oxygen limitation. This follows the discovery that B.subtilis is not strictly aerobic, as was previously considered, but can in fact grow anaerobically in the presence of alternative electron acceptors such as nitrate (Cruz Ramos et al., 1995; Hoffmann et al., 1995; Nakano and Zuber, 1998). Anaerobic nitrate respiration in B.subtilis is dependent on at least two regulatory systems with overlapping function: an FNR homologue (Cruz Ramos et al., 1995) and the ResDE two-component system (Nakano et al., 1996, 2000).
A molecular understanding of the sensing mechanisms involved in these different pathways is starting to emerge. Best understood is E.coli FNR, which appears to sense oxygen directly (Becker et al., 1996; Jordan et al., 1997). The active holo-form of FNR contains a 4Fe4S cluster that is essential for dimerization and DNA binding, and is rapidly disassembled in the presence of oxygen to a 2Fe2S form that is monomeric and unable to bind DNA (Lazazzera et al., 1996). However, oxygen does not directly modulate the autophosphorylation of ArcB. Rather, it appears that oxidized quinones can inhibit ArcB autophosphorylation and that the reduction of the quinone pool, caused by oxygen deprivation, relieves this inhibition, thereby leading to ArcA phosphorylation (Georgellis et al., 2001).
This study is concerned with the mechanism for sensing and responding to oxygen deprivation in the Gram-positive antibiotic-producing Streptomyces. These soil-dwelling bacteria are considered to be strictly aerobic, although the discovery of anaerobic dissimilatory genes (e.g. narGHJI) in the Streptomyces coelicolor genome sequence calls this view into question (Bentley et al., 2002; G.Sawers, personal communication). It is clear, however, that these organisms must flourish despite wide fluctuations in oxygen tension in their soil environment, for example, due to rapid changes in water content. From an industrial standpoint, oxygen supply during fermentation for antibiotic production is often a crucial factor in determining product yield (e.g. Rollins et al., 1990). However, nothing is known about how Streptomyces senses and responds to changes in oxygen tension and no regulatory genes have previously been identified. In this study we identify a key regulator in S.coelicolor that appears to directly sense changes in the cellular NADH/NAD+ redox poise that result from changes in oxygen status, and in turn modulates transcription of several components of the respiratory chain. We also present evidence that this novel regulatory system is present in most Gram-positive bacteria.
Results
The cydABCD operon is induced by oxygen limitation and respiratory inhibitors
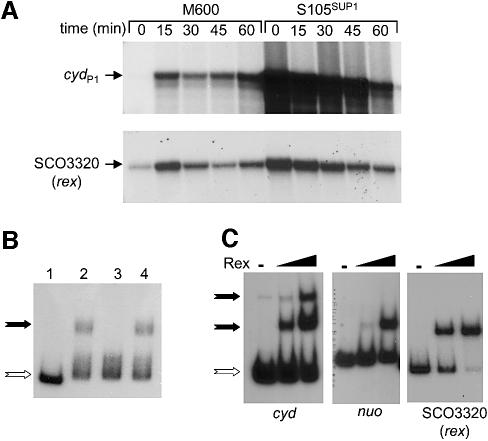
As a first step towards understanding the molecular mechanisms involved in the response of S.coelicolor to oxygen deprivation, we sought a promoter that was induced by oxygen limitation. The S.coelicolor genome contains a cydABCD operon that is predicted to encode both subunits of a cytochrome bd terminal oxidase complex (cydA and cydB) together with an ABC transporter (cydCD; Bentley et al., 2002). Since the cyd genes from several Gram-negative and -positive bacteria are induced by oxygen limitation, we tested whether this might be the case in S.coelicolor. S1 nuclease mapping revealed two promoters, cydP1 and cydP2, that initiate transcription 113 and ∼245 bp upstream from cydA, respectively. Whereas cydP2 was constitutively expressed, cydP1 was strongly induced within 15 min of oxygen limitation, falling slightly below its maximal level over the next 45 min (Figure 1).
Fig. 1. Transcriptional analysis of the S.coelicolor cyd operon using S1 nuclease mapping. Strains were grown to mid-late exponential phase prior to oxygen limitation, or the addition of respiratory inhibitors, ZnCl2 (0.5 mM) or KCN (5 mM), as indicated. RNA was isolated at the indicated time points. Arrows indicate the protected DNA fragments that correspond to the 5′ end of transcripts that initiate at cydP1 and cydP2. Whereas cydP2 remains relatively constant, cydP1 is induced by oxygen limitation, zinc treatment and cyanide treatment. The position of marker fragments from a HpaII digestion of pBR322 are indicated for oxygen limitation.
Although cytochrome bd terminal oxidase is not essential in S.coelicolor, a cydA mutant displayed a zinc-sensitive phenotype (our unpublished data). Escherichia coli cyd mutants show a similar phenotype, which is thought to result from the sensitivity of the major cytochrome bo terminal oxidase to zinc (Poole et al., 1989). Presumably, the E.coli cytochrome bd terminal oxidase plays a crucial role under conditions where the major aerobic system is inhibited. We considered that if the S.coelicolor cyd operon also plays an important role in the presence of zinc, then it might be induced under these conditions. To test this, RNA was isolated from aerobically growing S.coelicolor cultures following the addition of 0.5 mM ZnCl2. The cydP2 promoter was again constitutively expressed, whereas cydP1 was induced by zinc (Figure 1). As in E.coli, it is conceivable that zinc inhibits a zinc-sensitive terminal oxidase, thereby changing the redox state of the respiratory chain, or a linked redox couple, which may in turn act as a signal in the control of cydP1. In support of this, cyanide, a potent inhibitor of respiration in S.coelicolor (data not shown), also induced cydP1 (Figure 1).
Mutagenesis of the cydP1 promoter reveals a repressor binding site, ROP
The cydP1 promoter region was cloned into the multicopy promoter-probe vector pIJ487, which contains a promoter-less kanamycin resistance gene, neo. Streptomyces coelicolor containing pIJ487-cydP1 was resistant to 2–3 µg/ml kanamycin, which is barely above the level seen with pIJ487 alone. As a step towards identifying regulatory elements, a contiguous set of mutations was constructed, each replacing 6 bp of native cydP1 DNA with the BamHI restriction site. The mutants, M1–M13, covering the cydP1 promoter region from –65 to +15, were then assessed for promoter activity in pIJ487 (Figure 2A). Strikingly, two adjacent mutations, M12 and M13, both downstream from the transcription initiation site, conferred at least 3-fold increased levels of kanamycin resistance, thereby identifying a possible operator site for a cydP1 repressor (Figure 2A).
Fig. 2. Mutational analysis of the cydP1 promoter and the identification of a repressor binding site (ROP). (A) The non-template strand of the cydP1 promoter is indicated, together with the location of the cydP1 transcript 5′ end. DNA replaced by the BamHI restriction site in each of the mutations, M1–M13, is underlined. The activity of each promoter, as judged by the level of kanamycin resistance conferred on S.coelicolor when fused to neo in pIJ487, is indicated: –, no detectable activity; +, 2–3 µg/ml; +++, 10–12 µg/ml. The wild-type promoter conferred resistance to 2–3 µg/ml kanamycin. An inverted repeat (ROP), believed to be a repressor binding site is marked by inverted arrows. A putative overlapping half-site is also marked by an arrow. The non-template sequence corresponding to the region of the template strand protected from DNase I by the ROP-binding protein Rex (see Figure 4) is shaded in grey. (B) EMSA using a 168 bp cydP1 fragment as probe and crude extract prepared from S.coelicolor M600. Assays were conducted in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 1000-fold molar excess of non-homologous herring sperm DNA. The open arrow indicates unbound probe and the closed arrows indicate protein–DNA complexes. (C) Alignment of ROP-related sites located upstream from the cydABCD (cyd), SCO3320-hemACD (SCO3320), and nuoA-N (nuo) operons. The distance from the ROP site to the translation start codon of the first gene of each operon is indicated.
To investigate this further, we performed electromobility shift assays (EMSAs). Two protein–cydP1 complexes were detected using crude cell lysate from aerobically growing cultures (Figure 2B). Both complexes were resistant to 1000-fold molar excess of non-homologous DNA but were out-competed with unlabelled cydP1, confirming that the interaction was specific (data not shown). However, neither complex was observed using the cydP1-M12 promoter fragment, suggesting that the mutation had disrupted a binding site for a putative repressor(s) (Figure 2B). Analysis of the cydP1 sequence revealed a perfect 8 bp inverted repeat sequence that was centred 13 bp downstream from the transcription start site and which overlapped the site of the M12 and M13 mutations (Figure 2A). This sequence was considered the likely binding site for the putative repressor. We designated the unknown repressor Rex (redox regulator; see below) and the repressor binding site ROP (Rex operator). Interestingly, an 8 bp half-site was identified upstream and partially overlapping the ROP site, which was also disrupted in cydP1-M12 (see below).
To identify genes that might be co-regulated with the cyd operon, we searched for ROP-like sequences in the S.coelicolor genome. Two particularly good matches were found, one located 80 bp upstream from the first gene of the SCO3320-hemACD operon, and one located 107 bp upstream from the first gene of the nuoA-N operon, which is predicted to encode all 14 subunits of the 530 kDa membrane bound proton translocating NADH dehydrogenase NDH-1 (Figure 2C).
Identification of Rex, the ROP-binding repressor
The SCO3320-hemACD operon encodes three heme biosynthetic enzymes: glutamyl tRNA reductase (hemA); porphobilinogen deaminase (hemC); and the bifunctional uroporphyrin-IIIC-methyltransferase/uroporphyrinogen- III synthase (hemD). The 258 amino acid (26.8 kDa) product of the first gene of the operon (SCO3320) is homologous to an AT-rich DNA-binding protein from Thermus aquaticus of no known function (45% identity; Du and Pène, 1999). S1 nuclease mapping identified a single promoter, initiating transcription 62 bp upstream from the GTG start codon of SCO3320 and that was, as suspected, induced both by oxygen deprivation and zinc treatment (Figure 3A; data not shown).
Fig. 3. Evidence that SCO3320 encodes Rex, the ROP-binding repressor. (A) Transcriptional analysis of cydP1 and the SCO3320-hem operon promoter in M600 (wild type) and S105SUP1 (ΔSCO3320::apr) using S1 nuclease mapping (see Figure 1 for details). (B) EMSAs were performed using the SCO3320-hem operon promoter region as a probe. Lane 1 contains no added extract and 1anes 2–4 contain crude extracts prepared from M600, S105SUP1 or S105SUP1 (pSET152::SCO3320), respectively. (C) EMSAs using promoter fragments that include the ROP sites located upstream of the cyd, SCO3320-hem and nuo operons (1 nM each), mixed with purified SCO3320 (Rex) protein (25 or 100 nM). The open arrow indicates unbound probe and the closed arrows indicate protein–DNA complexes.
Attempts to titrate the Rex repressor by placing cydP1 on a multicopy number plasmid did not lead to increased activity of chromosomally located cydP1 (data not shown). This suggested either that Rex was present at a constitutively high level, or that the gene encoding Rex was negatively autoregulated, allowing the synthesis of sufficient Rex to occupy the increased copies of ROP. In view of the second possibility, we investigated whether SCO3320 encodes Rex. We constructed an S.coelicolor null mutant by replacing the SCO3320 reading frame with an apramycin resistance gene cassette to give strain S105. S105 grew poorly, was delayed in aerial mycelium development and produced faster-growing suppressor mutants at a high frequency. Preliminary analysis suggested that the phenotype was due to a polar effect of the mutation on the downstream heme biosynthetic genes. Nonetheless, the isolation of a stable S105 suppressor, named S105SUP1, allowed an initial investigation into the in vivo function of SCO3320. S1 mapping revealed that deletion of SCO3320 led to increased expression of both cydP1 and the SCO3320-hem operon promoter, even under aerobic conditions (Figure 3A). Indeed, the highest level of activity was seen before oxygen limitation was applied, the activity then decreasing over time in a SCO3320-independent manner. EMSAs using the SCO3320-hem operon promoter fragment as a probe revealed that the DNA-binding activity detected in M600 crude extracts was absent from S105SUP1 extracts, but was restored when SCO3320 was reintroduced on the integrative plasmid pSET152 (Figure 3B). This supports the idea that the product of SCO3320 was responsible for the ROP-binding activity detected in crude cell extracts.
To confirm that the SCO3320 protein interacts directly with ROP sites, we overproduced a His-tagged version. Following Ni2+ affinity purification, removal of the His tag and size exclusion chromatography, the SCO3320 protein was judged to be >95% pure based on Coomassie Blue staining after SDS–PAGE. EMSAs confirmed that the SCO3320 protein bound to the cydP1 and SCO3320-hem operon promoter fragments (Figure 3C). Furthermore, as seen with S.coelicolor crude cell extracts, two protein–DNA complexes were detected with the cydP1 fragment. Overall, these data indicate that the ROP-binding activity detected in crude extracts corresponds to the product of SCO3320, and the gene was therefore named rex.
As mentioned, a ROP-related site is situated upstream from the nuoA-N operon. EMSAs confirmed that Rex bound to a DNA fragment that contained this ROP-like site (Figure 3C). However, since the nuoA-N operon was not expressed under the growth conditions used, even under oxygen limitation, we were unable to assess the in vivo role of rex in its regulation. Therefore, although Rex might regulate nuoA-N, at least one other regulatory system is likely to be involved.
Binding of Rex to the cydP1 promoter region
Whereas Rex promotes the formation of a single protein–DNA complex with the rexP and nuoP promoter fragments, two complexes were detected with the cydP1 fragment, with the upper complex appearing in a concentration-dependent manner (Figure 3C). This might be due to a second Rex binding site in the cydP1 region. To investigate further, we mapped the Rex binding site(s) on cydP1 by DNase I footprinting. Rex protected cydP1 from –6 to +26 with respect to the transcription start point (Figures 2A and 4A). However, the ROP site was centred in the downstream half of the protected region, which suggested that Rex might also interact with the putative half-site that lies upstream from and partially overlapping the main ROP site (Figure 2A). To test this, EMSAs were performed using mutant promoter fragments (M11–M13) that cover this region (Figures 2A and 4B). As seen with crude cell extracts, no complexes were detected between Rex and cydP1-M12, in which both the half-site and the main ROP site were disrupted. The cydP1-M13 mutant, in which only the main ROP site was disrupted, also failed to bind Rex effectively. Strikingly, only the lower Rex–DNA complex was detected for cydP1-M11, in which only the upstream half-site was disrupted. Taken together, these data suggest that the lower protein–DNA complex was caused by the interaction of Rex with the complete ROP site and that the upper band was caused by Rex additionally binding to the upstream half-site.

Fig. 4. Binding of Rex to cydP1. (A) DNase I footprinting of the Rex–cydP1 complex. The 360 bp cydP1 fragment was 5′ end-labelled on the bottom (template) strand and mixed with increasing concentrations of Rex, prior to DNase I treatment, as follows: lane 3, no added Rex; lane 4, 1 nM; lane 5, 5 nM; lane 6, 10 nM; lane 7, 25 nM; lane 8, 50 nM. Lanes 1–2, A and G dideoxynucleotide sequencing reactions. The extent of the interaction between Rex and cydP1, with respect to the transcription initiation site, is indicated by a vertical grey bar (see Figure 2A for sequence). (B) EMSAs using cydP1 mutant promoter fragments (see Figure 2A) and purified Rex. Only the upstream ROP half-site is disrupted in cydP1-M11, both half-site and complete ROP site are disrupted in cydP1-M12, and only the complete ROP site is disrupted in cydP1-M13. Each assay contained 1 nM promoter DNA and 25 or 200 nM Rex. The open arrow indicates unbound probe and the closed arrows indicate Rex–DNA complexes.
Structural predictions and widespread occurrence of Rex in Gram-positive bacteria
Database searches revealed Rex-related proteins encoded by the genomes of most Gram-positive bacteria, including B.subtilis (41% identity), Staphylococcus aureus (37% identity), Listeria monocytogenes (41% identity), Streptococcus pneumoniae (35% identity) and Lactococcus lactis (36% identity) (Figure 5; data not shown). However, although Rex is found in the Streptomyces genus, it appears to be absent from other actinomycetes such as Mycobacterium tuberculosis, Corynebacterium diphtheriae and Bifidobacterium longum. Interestingly, Rex is not restricted to aerobic organisms, since a rex gene was identified in the genome of the strict anaerobe, Clostridium perfringens (38% identity). To date, no Rex homologue has been identified in Gram-negative bacteria, suggesting that Rex might be confined to Gram-positive organisms.
Fig. 5. Alignment of Rex-related proteins from Gram-positive bacteria using Clustal_W. The proteins are also aligned with rat biliverdin reductase (BVR) as predicted by sequence-structure comparisons using FUGUE (Shi et al., 2001). Identical residues are highlighted in black, and similar residues are outlined in grey. The secondary structure of the pyridine nucleotide-binding domain (Rossmann fold) of BVR, as determined from the crystal structure (Kikuchi et al., 2001), is indicated by black lines (helices) or arrows (strands). A secondary structure prediction of Rex, determined using PSIPRED (McGuffin et al., 2000), is indicated by grey lines (helices) or arrows (strands). A putative HTH motif in Rex is marked with dotted arrows. A conserved glycine-rich signature motif (Gly100XGly102XXGly105 in S.coelicolor) that is found in most NAD+-dependent dehydrogenases is indicated by asterisks. Also indicated is an aspartate (Asp126 in S.coelicolor) that might play a role in discriminating between NAD(H) and NADP(H). The amino acid sequence data was obtained from the SwissProt database. Sco, S.coelicolor (Q9WX14); Lmo, L.monocytogenes (Q929U6); Bsu, B.subtilis (O05521); Sau, S.aureus (Q99SK6). The non-conserved C-terminal 20 residues of S.coelicolor Rex are not included.
The DNA-binding domain of Rex is likely to reside in the first third of the protein (residues 1–90). A TetR-like helix–turn–helix (HTH) motif was identified by searching Pfam (residues 26–65; E-value 0.64; Bateman et al., 2000), and this was supported by secondary structure predictions using PSIPRED (Figure 5). Furthermore, the sequence structure homology recognition program FUGUE (Shi et al., 2001) predicted homology between Rex (residues 20–70) and the HTH DNA-binding domain of a transcriptional regulator from Thermotoga maritima (Protein Data Bank code 1J5Y; Z score 7.7). Taken together, these predictions suggest that a three-helix bundle forms the DNA-binding domain of Rex, with the second and third helices constituting a classic HTH motif (Figure 5).
Remarkably, the remainder of Rex is likely to constitute a ‘Rossmann fold’, which is common to many pyridine nucleotide-dependent dehydrogenases (e.g. d-lactate dehydrogenase; Rossmann et al., 1975). The S.coelicolor Rex sequence Gly100-Ile101-Gly102-Asn103-Leu104-Gly105 corresponds to the ‘fingerprint’ (GlyXGlyXXGly) of classic dinucleotide-binding proteins. Furthermore, FUGUE predicted homology between Rex (residues ∼90 to ∼190) and several dinucleotide-binding domains of known structure. One of the closest structural homologues was biliverdin reductase (BVR; Z score 11.6), which catalyses the last step in heme degradation and is aligned with Rex homologues in Figure 5. The Rossmann fold typically consists of two sets of β-α-β-α-β units, which together form a parallel β-sheet flanked by α-helices (Lesk, 1995). The first and third glycines in the GlyXGlyXXGly motif play important structural roles; the first allows a tight turn of the main chain from the β1 strand into the loop and the third allows the close packing of helix αA with β1 (Lesk, 1995; Figure 5). The role of the central glycine is to permit close contact between the main chain and the pyrophosphate of the nucleotide. Dehydrogenases that use NAD(H) rather than NADP(H) often contain an acidic residue (aspartate or glutamate) at the C-terminal edge of β2 that interacts with the 2′-OH group of the adenosine ribose of NAD(H) (Baker et al., 1992; Lesk, 1995). Asp126 is located in this position and is highly conserved in Rex homologues, supporting the idea that Rex binds NAD(H) via a Rossmann fold.
Rex DNA-binding activity is inhibited by NADH
To investigate whether Rex binds pyridine nucleotides and, if so, whether this interaction affects its DNA binding activity, we performed EMSAs using Rex and the rexP fragment in the presence of NAD+, NADH, NADP+ or NADPH. As shown in Figure 6A, both the redox and phosphorylation states of the pyridine nucleotides determined their influence on the DNA-binding activity of Rex. The oxidized nucleotides NAD+ and NADP+ did not decrease DNA-binding at concentrations up to 1 mM. On the other hand, both reduced nucleotides decreased Rex–DNA complex formation, although NADH was more effective than NADPH, in agreement with the sequence characterization of Rex described above. One millimolar NADPH caused ∼40% loss of the Rex–DNA complex, whereas 100 µM NADH was sufficient to completely abolish the complex. In order to test whether the conserved GlyXGlyXXGly motif is important in pyridine nucleotide binding, the central glycine was changed to alanine to generate RexG102A. In EMSAs RexG102A bound to the rexP fragment but, as predicted, was insensitive to the addition of either oxidized or reduced pyridine nucleotides at concentrations up to 1 mM (Figure 6B).
Fig. 6. The DNA-binding activity of Rex is inhibited by NADH. (A) EMSAs were performed using a rex (SCO3320) promoter fragment as probe (1 nM), purified Rex (35 nM) and 0.1 or 1 mM pyridine nucleotides, as indicated. The open arrow indicates unbound probe and the closed arrows indicate the Rex–DNA complex. –, no added Rex. (B) EMSAs were performed as in (A) but using RexG102A in which the central glycine in the GlyXGlyXXGly Rossmann fold fingerprint was changed to alanine. (C) EMSAs were performed as in (A) but with 50 nM Rex and a range of NADH concentrations, as indicated. (D) The data from (C) were quantified by phosphoimage analysis and plotted as % Rex–DNA complex versus NADH concentration. Values were normalized to 0 µM NADH (100% Rex–DNA complex). The concentration of NADH required to generate an ∼50% loss of the Rex–DNA complex (IC50) was determined from the best-fit curve.
More extensive EMSAs revealed that, at 50 nM Rex, the inhibitory concentration of NADH required to generate an ∼50% loss of the Rex–DNA complex (IC50) was ∼6 µM (Figure 6C and D). Therefore, Rex DNA-binding activity appears to be particularly sensitive to the concentration of NADH. Although further experiments were performed using only the NADH/NAD+ redox couple (the most likely physiological effector of Rex activity; see Discussion), the role of phosphorylated pyridine nucleotides in the control of Rex has not yet been dismissed.
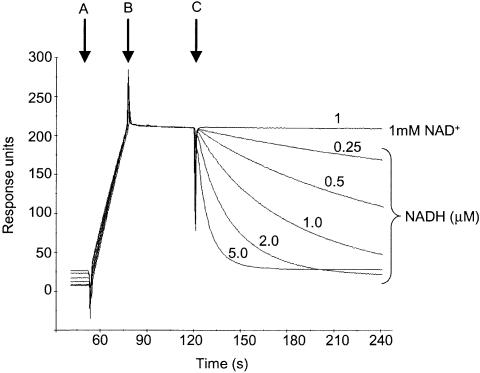
To determine whether NADH could actively dissociate a Rex–DNA complex, we developed a surface plasmon resonance (SPR)-based assay. A 37 bp biotinylated double stranded oligonucleotide, comprising the ROP site and surrounding DNA located upstream from the nuoA-N operon (ROPnuo), was attached to a streptavidin sensor chip. Rex was then bound to the sensor chip in the absence of pyridine nucleotides to give a response unit (RU) shift of ∼200 RU. Rex bound tightly to ROPnuo, allowing the effect of pyridine nucleotides on complex stability to be tested by subsequent injection (Figure 7). In support of the EMSA data, 1 mM NAD+ did not decrease the stability of the Rex-ROPnuo complex while 0.25–5 µM NADH increased the rate of dissociation in a dose-dependent manner. This confirms that NADH, but not NAD+, can actively dissociate Rex that is bound to its DNA target.
Fig. 7. NADH, but not NAD+, actively dissociates a Rex–ROP complex. SPR experiments were carried out at 25°C using HSB-T as running buffer and a flow rate of 20 µl/min. Sensorgrams were obtained by the subtraction of background values obtained from a DNA-free flow cell from the raw interaction data, then overlayed. The streptavidin sensor chip was charged with a 37 bp double stranded biotinylated oligonucleotide including the ROPnuo site. (A) Rex (SCO3320) was injected to give a response unit shift of ∼200 RU. (B) End of Rex injection. (C) NAD+ or NADH, at the concentrations indicated, were injected and the dissociation of the complex monitored over time.
Rex senses the NADH/NAD+ redox poise
There are two possible explanations for the failure of NAD+ to inhibit Rex DNA-binding activity. Either NAD+ does not bind to Rex, or an NAD+–Rex complex retains DNA-binding activity. If the second possibility was true, NAD+ might compete with NADH for Rex binding. To test this, we performed EMSAs using a range of NADH concentrations in the presence of NAD+. To reflect the cellular situation, where an increase in NADH would be balanced by a decrease in the NAD+, the total level of NAD(H) co-factors was held constant, at either 0.25 or 1 mM (Figure 8A and B). Rex remained sensitive to NADH even in the presence of NAD+, but NAD+ clearly influenced the inhibitory effect of NADH on Rex DNA-binding activity. The binding curves revealed that, under the conditions used, the IC50 for loss of Rex DNA-binding activity was ∼5 µM NADH at 0.25 mM NAD(H) and ∼20 µM at 1 mM NAD(H). In each case, this corresponded to ∼2% NADH as a proportion of the total NAD(H) co-factor pool. The discovery that a 4-fold increase in NAD+ led to a 4-fold increase in the IC50 strongly suggests that NAD+ competes with NADH for Rex binding, and also that NAD+-bound Rex retains DNA-binding activity. Furthermore, it suggests that the role of Rex is to sense the redox poise of the NAD(H) co-factor pool rather than the concentration of NADH per se.
Fig. 8. Rex DNA-binding activity is modulated by NADH/NAD+ redox poise. (A) EMSAs were performed using a rex (SCO3320) promoter fragment as probe (1 nM) and purified Rex (50 nM). Each assay contained NADH at the concentration indicated, balanced with the appropriate level of NAD+ to give final co-factor concentrations of either 0.25 or 1 mM. Open arrows indicate unbound probe and closed arrows indicate Rex–DNA complexes. (B) The data from (A) was quantified by phosphoimage analysis and plotted as percentage Rex–DNA complex versus NADH concentration. Values were normalized to 0 µM NADH (100% Rex–DNA complex). For each experiment, the concentration of NADH required to generate an ∼50% loss of the Rex–DNA complex (IC50) was determined from the best-fit curve.
Discussion
Here we have described the discovery of Rex, a novel redox-sensitive repressor in S.coelicolor that appears to modulate transcription in response to changes in cellular NADH/NAD+ redox poise. Our evidence suggests that Rex binds NADH and NAD+ via a Rossmann fold, but that these co-factors differ considerably in their effect on the interaction between Rex and its binding site, an 8 bp inverted repeat designated ROP. Whereas <5 µM NADH could inhibit DNA-binding in EMSAs and could actively dissociate Rex from a ROP site, 1 mM NAD+ had no apparent inhibitory effect. However, in the cellular environment, in which NAD+ and NADH are both present, the high sensitivity of Rex to NADH may be in part offset by NAD+, which is able to compete with NADH for Rex binding. This competition may be of physiological importance. In other organisms it has been found that the concentration of the total NAD(H) co-factor pool can vary depending on the growth conditions (e.g. van Keulen et al., 2000). If Rex was responsive solely to NADH levels, then changes in the total level of NAD(H) co-factors that occur independently of changes in redox poise would influence Rex activity. However, we found that 2% NADH, as a percentage of the total NAD(H) pool, led to 50% loss of DNA-binding activity, whether the total NAD(H) co-factor level was 0.25 or 1 mM. In other words, Rex appears to be more attuned to the redox poise of the NAD(H) pool than to the concentration of NADH per se, allowing Rex to function as a redox sensor over a range of total co-factor concentrations. Presumably, the poise at which the redox switch is set will depend on the relative binding affinities of the oxidized and reduced co-factors for Rex. Although equilibrium dissociation constants have not been investigated here, our data suggest that Rex has a much higher affinity for NADH than for NAD+.
The promoters cydP1 and rexP were repressed by Rex during aerobic growth suggesting that, under these growth conditions, NADH is likely to make up less than 2% of the total cellular NAD(H) pool. Although the level and poise of the NAD(H) pool has not been studied in Streptomyces, studies in other bacteria have found that the NADH/NAD+ ratio is maintained at a low level in rapidly growing aerobic cultures. For example, NADH comprised ∼3% of the total NAD(H) pool in steady state E.coli cultures grown at 10% dissolved oxygen in a chemostat (de Graef et al., 1999). This study (along with several others; e.g. Wimpenny and Firth, 1972) also showed that the NADH/NAD+ ratio increased significantly under oxygen-limited conditions, with 0.1% dissolved oxygen leading to ∼60% NADH. In a separate study, it was found that the respiratory inhibitor cyanide caused a 16-fold increase in NADH concentration in E.coli (Woodmansee and Imlay, 2002). In congruence with this, we found that cyanide induced the expression of cydP1. We believe that such studies, together with the in vitro data presented in this work, support the following model for Rex regulation. During aerobiosis the low cellular NADH/NAD+ ratio allows Rex to repress target genes by binding to their respective ROP sites. In response to oxygen limitation or any other condition that inhibits respiration, the subsequent increase in the NADH/NAD+ ratio, as a result of continued substrate oxidation, would cause Rex to lose affinity for DNA, leading to the expression of the Rex regulon. Since this regulon includes the cytochrome bd terminal oxidase operon, this might increase the ability to scavenge oxygen and thereby enable a return to redox homeostasis.
Rex is the first bacterial regulator described that senses the NADH/NAD+ redox state as a signal in the modulation of respiratory gene expression. This signal, rather than oxygen itself, would allow Rex to sense oxygen-independent changes in the respiratory rate, for example, caused by natural respiratory inhibitors (e.g. the antibiotic antimycin D), and would allow Rex to play a general role as a sensor of cellular redox balance. The extent to which Rex influences global transcriptional patterns in response to redox imbalance remains to be seen. An interesting comparison can be made between Rex and the ArcAB system of E.coli. The ArcAB system, and most likely Rex, both play key regulatory roles at the interface between aerobic and microaerobic growth. For example, Rex and ArcAB both modulate expression of a cytochrome bd terminal oxidase complex, leading to its induction under microaerobic conditions. Both regulatory systems sense the poise of redox couples that are directly affected by the electron flow rate through the respiratory chain. However, the two systems differ in the nature of these redox couples. Whereas Rex senses the redox poise of the cytoplasmic NAD(H) pool, ArcB senses the redox poise of the membrane-bound quinone pool (Georgellis et al., 2001).
NAD(H) co-factors occupy a central position in metabolism, linking substrate oxidation to energy conservation, and they allosterically control many key metabolic steps, including several TCA cycle and glycolytic enzymes. However, the NAD(H) redox state has only recently emerged as a modulator of gene expression. There have been two recent reports of eukaryotic regulators that are controlled by the redox state of the pyridine nucleotide pool. In mammals, the DNA-binding activity of the Clock–BMAL1 and NPAS2–BMAL1 heterodimeric complexes, which are involved in controlling circadian rhythms as a function of the light–dark cycle, appear to be regulated by the redox status of NAD(H) and NADP(H) (Rutter et al., 2001). Both NADH and NADPH stimulated DNA-binding activity, whereas the oxidized forms of the co-factors inhibited DNA-binding. It was proposed that this might allow the molecular clock to be entrained by changes in cellular redox potential that might arise from changes in feeding patterns. The activity of the transcriptional co-repressor CtBP, on the other hand, appears to be exquisitely sensitive to NADH (Zhang et al., 2002), although this has been disputed (Kumar et al., 2002). CtBP interacts with and inhibits the transactivation function of the adenovirus E1A protein, as well as several other transcription factors involved in growth and development. NADH was found to be two to three orders of magnitude more effective than NAD+ at promoting formation of the CtBP–E1A complex, with nanomolar levels generating a response (Zhang et al., 2002). CtBP contains a Rossmann fold for NAD(H) binding and, intriguingly, possesses intrinsic dehydrogenase activity, although the natural substrate is not known (Kumar et al., 2002). In bacteria, although no other NAD(H) sensors have been described, there has been a report of a LysR-type regulator in Xanthobacter flavus that appears to be modulated by the redox status of the NADP(H) pool. CbbR is a regulator that activates the expression of several Calvin cycle genes. The addition of NADPH but not NADH stimulated the DNA-binding activity of CbbR, although CbbR does not contain a recognizable pyridine nucleotide-binding motif (van Keulen et al., 1998).
Finally, we have identified genes that encode Rex homologues in the sequenced genomes of most Gram-positive bacteria, including several important human and animal pathogens such as S.aureus, L.monocytogenes and S.pneumoniae. However, Rex homologues appear to be absent from the mycobacteria, as well as from all Gram-negative organisms. Rex is predicted to include a Rossmann fold for pyridine nucleotide binding, and residues that might play key structural and nucleotide binding roles in this putative fold are highly conserved. We propose therefore that Rex homologues are likely to act as regulatory sensors of NADH/NAD+ redox poise in most Gram-positive bacteria.
Materials and methods
Bacterial strains and culture conditions
Experiments were performed using S.coelicolor A3(2) strain M600 grown on MS agar or in NMMP + glucose liquid media as described (Kieser et al., 2000). Overexpression of rex was carried out using E.coli BL21λDE3 (pLysS) (Studier and Moffat, 1986).
RNA isolation and S1 nuclease protection analysis
RNA was isolated from mid-late exponential liquid cultures (OD450 0.6–0.8) following oxygen limitation or treatment with zinc or cyanide. For oxygen-limited RNA, the culture was used to fill 15 ml tubes, sealed, and static incubation was continued at 30°C. At 15 min intervals, following brief centrifugation, RNA was extracted from mycelium. Streptomyces coelicolor grows as small mycelial clumps that, during static incubation, sink to the bottom of the sealed tube. Although it was not possible to measure the localized oxygen tension around the sunken clumps, it is reasonable to assume that continued respiration leads to oxygen limitation. For zinc- or cyanide-treated RNA, ZnCl2 (0.5 mM) or KCN (5 mM) were added to cultures prior to continued aerobic incubation, and at specified time points mycelium was collected by rapid filtration. RNA was extracted as detailed by Paget et al. (2001). Probes for S1 nuclease mapping were generated by PCR using 5′ end-labelled oligonucleotides internal to the gene of interest, unlabelled oligonucleotides positioned 300–500 nt upstream and M600 chromosomal DNA as template. Primers were labelled and S1 nuclease protection assays were set up using 30 µg total RNA as described previously (Kieser et al., 2000). Protected DNA bands were quantified with a Storm phosphoimager (Amersham Pharmacia).
Construction and analysis of cydP1::neo transcriptional fusions
The cydP1 promoter was amplified by PCR using the primers CDP1a (5′-CGGGAATTCCGGACTCAACGGTCTGTTG) and CDP1b (5′-CGGT CTAGAGCGTGTTTGTCACACCCATC), and cloned into pGEM T-easy (Promega), to give pSX131. The cydP1 fragment was then subcloned into the neo reporter plasmid pIJ487 (Kieser et al., 2000) to yield pSX133. Expression of neo in S.coelicolor was determined on minimal agar plates containing 0.2% casamino acids and various concentrations of kanamycin. The cydP1 mutants (M1–M13) were constructed by replacing contiguous sections of 6 bp native DNA with a BamHI restriction site (GGATCC) using inverse PCR mutagenesis.
Construction of a rex mutant
The SCO3320 (rex) open reading frame was replaced with an apramycin resistance cassette using the PCR-directed approach described by Gust et al. (2003). Briefly, a gene replacement cassette containing an apramycin resistance gene (apr) and an RK2 origin of transfer (oriT) was amplified from pIJ773 (Gust et al., 2003) by PCR using primers containing 39 nt 5′ homology extensions corresponding to the N- or C-terminal regions of rex, and 20 nt 3′ homology to the unique priming sites at each end of the cassette. The PCR product was recombined into the rex-containing cosmid SCE68 using E.coli BW25113/pIJ790 as host, thereby creating a replacement mutation in which only the first 50 and last 36 codons of rex were present. The mutant allele was recombined into the M600 genome as described previously (Gust et al., 2003). A ΔSCO3320::apr mutant was identified and named S105. However, since S105 was unstable, further studies used the stable suppressor strain S105SUP1. To complement S105SUP1, SCO3320 (rex), together with its promoter region, was amplified by PCR and cloned into the conjugative plasmid pSET152 (Bierman et al., 1992) to yield pSET152::SCO3320.
EMSAs and DNase I footprinting assays
Probes (∼200 bp) that included the relevant ROP site upstream of the cyd, SCO3320-hem (rex-hem) and nuo operons were generated using PCR. Following purification, probes were 5′ end-labelled using [γ-32P]ATP. EMSAs contained in a final volume of 10 µl: DNA probe (∼1 nM); binding buffer (20 mM Tris–HCl pH 8.0, 5% v/v glycerol, 1 mM MgCl2, 40 mM KCl); crude extract or pure Rex protein; 1 µg herring sperm DNA (Promega); and pyridine nucleotides, as specified. Following 15 min incubation at room temperature the binding reactions were separated on a 6% polyacrylamide gel. Bands were quantified using a Storm phosphoimager. Experiments were performed at least twice and representative data sets are presented. Crude extract was prepared from S.coelicolor M600 grown in NMMP + 0.5% glucose liquid medium to an OD450 of 0.5–0.6. Mycelium was lysed by sonication in lysis buffer (25 mM Tris–HCl, pH 8.0, 5 mM EDTA, 5% v/v glycerol, 1 mM DTT, 150 mM NaCl, 0.1 mM Prefabloc; Roche), then cleared by centrifugation at 4°C.
For DNase I footprinting the cydP1 template was prepared by PCR using a [γ-32P]ATP 5′end-labelled oligonucleotide (CD2, 5′-GTAGAC GGTCGTGATGCCGAAC) internal to cydA and an unlabelled oligonucleotide (CD1, 5′-CGAGGCGAACGACATCTTCCTG) positioned ∼360 nt upstream). Binding and DNase I reactions were carried out according to Takano et al. (2001), using 4.5 nM template and various concentrations of Rex. Dideoxynucleotide sequencing reactions (A and G) were performed using an fmol DNA sequencing kit (Promega) and 5′ end-labelled CD2 as primer.
Overexpression and purification of Rex
The rex (SCO3320) gene was amplified from M600 DNA by PCR, simultaneously engineering an NdeI start codon, an upstream EcoRI site and a downstream BamHI site. The EcoRI–BamHI digested PCR product was cloned into pBluescript II SK+ (Stratagene) to give pSX136, then subcloned into pET15b (Novagen) to give pSX135. Overexpression of rex was performed using E.coli BL21λDE3 (pLysS), and the overproduced His-tagged Rex was purified using nickel affinity chromatography. Following removal of the His tag using thrombin, Rex was further purified to >95% purity by gel filtration using a Superdex 200 HiLoad 16/60 column. The RexG102A mutant was constructed using inverse PCR mutagenesis, converting the GGC glycine codon to a GCC alanine codon.
SPR
Experiments were conducted in HBS-T buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3.4 mM EDTA, 0.005% v/v Tween-20) using a BIAcore 2000 instrument (BIAcore AB, Uppsala, Sweden) at a flow rate of 20 µl/min and 25°C. The 5′ biotinylated oligonucleotide (5′-AGATCG CGAACATGTGAAGCAGGTCACAAGCCCAACT) and the non-biotinylated oligonucleotide (5′-AGTTGGGCTTGTGACCTGCTTCACA TGTTCGCGATCT) were annealed at a ratio of 1:4, diluted to 1 ng/µl, then injected over a streptavidin sensor chip to give a final increase of ∼300 RU. Non-specifically bound DNA was removed using 1 M NaCl. At least two of the four flow cells were used as DNA-free controls. Rex was pre-bound to the DNA by injecting 0.3 µM aliquots to give a final increase of ∼200 RU. Pyridine nucleotides, made up in HBS-T, were then injected, and the change in RU monitored over time. Following each experiment, residual protein was removed from the DNA using 1 M NaCl.
Acknowledgments
Acknowledgements
We would like to thank Andrew Smith, Gary Sawers, Philip Doughty and Mark Buttner for helpful comments, and the University of Sussex for providing a Graduate Teaching Assistantship to D.B.
References
- Baker P.J., Britton,K.L., Rice,D.W., Rob,A. and Stillman,T.J. (1992) Structural consequences of sequence patterns in the fingerprint region of the nucleotide binding fold. Implications for nucleotide specificity. J. Mol. Biol., 228, 662–671. [DOI] [PubMed] [Google Scholar]
- Bateman A. et al. (2000) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S., Holighaus,G., Gabrielczyk,T. and Unden,G. (1996) O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J. Bacteriol., 178, 4515–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley S.D. et al. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature, 417, 141–147. [DOI] [PubMed] [Google Scholar]
- Bierman M., Logan,R., O’Brien,K., Seno,E.T., Rao,R.N. and Schoner,B.E. (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene, 116, 43–49. [DOI] [PubMed] [Google Scholar]
- Bongaerts J., Zoske,S., Weidner,U. and Unden,G. (1995) Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol. Microbiol., 16, 521–534. [DOI] [PubMed] [Google Scholar]
- Chao G., Shen,J., Tseng,C.P., Park,S.J. and Gunsalus,R.P. (1997) Aerobic regulation of isocitrate dehydrogenase gene (icd) expression in Escherichia coli by the arcA and fnr gene products. J. Bacteriol., 179, 4299–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.A., Melville,S.B., Albrecht,J.A. and Gunsalus,R.P. (1997) Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol. Microbiol., 25, 605–615. [DOI] [PubMed] [Google Scholar]
- Cruz Ramos H., Boursier,L., Moszer,I., Kunst,F., Danchin,A. and Glaser,P. (1995) Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J., 14, 5984–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graef M.R., Alexeeva,S., Snoep,J.L. and Teixeira de Mattos,M.J. (1999) The steady-state internal redox state (NADH/NAD+) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol., 181, 2351–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X. and Pène,J.J. (1999) Identification, cloning and expression of p25, an AT-rich DNA-binding protein from the extreme thermophile, Thermus aquaticus YT-1. Nucleic Acids Res., 27, 1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgellis D., Kwon,O. and Lin,E.C. (2001) Quinones as the redox signal for the arc two-component system of bacteria. Science, 292, 2314–2316. [DOI] [PubMed] [Google Scholar]
- Guest J.R., Green,J., Irvine,S. and Spiro,S. (1996) The FNR modulon and FNR-regulated gene expression. In Lin,E.C.C. and Lynch,A.S. (eds), Regulation of Gene Expression in Escherichia coli. R.G. Landes Company, Austin, TX, pp. 317–342. [Google Scholar]
- Gust B., Challis,G.L., Fowler,K., Kieser,T. and Chater,K.F. (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl Acad. Sci. USA, 100, 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T., Troup,B., Szabo,A., Hungerer,C. and Jahn D. (1995) The anaerobic life of Bacillus subtilis: cloning of the genes encoding the respiratory nitrate reductase system. FEMS Microbiol. Lett., 131, 219–225. [DOI] [PubMed] [Google Scholar]
- Iuchi S., Chepuri,V., Fu,H.A., Gennis,R.B. and Lin,E.C. (1990) Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J. Bacteriol., 172, 6020–6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P.A., Thomson,A.J., Ralph,E.T., Guest,J.R. and Green,J. (1997) FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett., 416, 349–352. [DOI] [PubMed] [Google Scholar]
- Kieser T., Bibb,M.J., Buttner,M.J., Chater,K.F. and Hopwood,D.A. (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich, UK. [Google Scholar]
- Kikuchi A., Park,S.Y., Miyatake,H., Sun,D., Sato,M., Yoshida,T. and Shiro,Y. (2001) Crystal structure of rat biliverdin reductase. Nat. Struct. Biol., 8, 221–225. [DOI] [PubMed] [Google Scholar]
- Kumar V., Carlson,J.E., Ohgi,K.A., Edwards,T.A., Rose,D.W., Escalante,C.R., Rosenfeld,M.G. and Aggarwal,A.K. (2002) Transcription corepressor CtBP is an NAD+-regulated dehydrogenase. Mol. Cell., 10, 857–869. [DOI] [PubMed] [Google Scholar]
- Lazazzera B.A., Beinert,H., Khoroshilova,N., Kennedy,M.C. and Kiley,P.J. (1996) DNA-binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem., 271, 2762–2768. [DOI] [PubMed] [Google Scholar]
- Lesk A.M. (1995) NAD-binding domains of dehydrogenases. Curr. Opin. Struct. Biol., 5, 775–783. [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson,K. and Jones,D.T. (2000) The PSIPRED protein structure prediction server. Bioinformatics, 16, 404–405. [DOI] [PubMed] [Google Scholar]
- Meng W., Green,J. and Guest,J.R. (1997) FNR-dependent repression of ndh gene expression requires two upstream FNR-binding sites. Microbiology, 43, 1521–1532. [DOI] [PubMed] [Google Scholar]
- Nakano M.M. and Zuber,P. (1998) Anaerobic growth of a ‘strict aerobe’ (Bacillus subtilis). Annu. Rev. Microbiol., 52, 165–190. [DOI] [PubMed] [Google Scholar]
- Nakano M.M., Zuber,P., Glaser,P., Danchin,A. and Hulett,F.M. (1996) Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J. Bacteriol., 178, 3796–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M.M., Zhu,Y., Lacelle,M., Zhang,X. and Hulett,F.M. (2000) Interaction of ResD with regulatory regions of anaerobically induced genes in Bacillus subtilis. Mol. Microbiol., 37, 1198–1207. [DOI] [PubMed] [Google Scholar]
- Paget M.S.B., Bae,J.B., Hahn,M.Y., Li,W., Kleanthous,C., Roe,J.H. and Buttner,M.J. (2001) Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Mol. Microbiol., 39, 1036–1047. [DOI] [PubMed] [Google Scholar]
- Park S.J., Cotter,P.A. and Gunsalus,R.P. (1995) Regulation of malate dehydrogenase (mdh) gene expression in Escherichia coli in response to oxygen, carbon, and heme availability. J. Bacteriol., 177, 6652–6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patschkowski T., Bates,D.M. and Kiley,P.J. (2000) Mechanisms for sensing and responding to oxygen deprivation. In Storz,G. and Hengge-Aronis,R. (eds), Bacterial Stress Responses. ASM Press, Washington, DC, pp. 61–78. [Google Scholar]
- Poole R.K., Williams,H.D., Downie,J.A. and Gibson,F. (1989) Mutations affecting the cytochrome d-containing oxidase complex of Escherichia coli K12: identification and mapping of a fourth locus, cydD. J. Gen. Microbiol., 135, 1865–1874. [DOI] [PubMed] [Google Scholar]
- Rollins M.J., Jensen,S.E., Wolfe,S. and Westlake,D.W. (1990) Oxygen derepresses deacetoxycephalosporin C synthase and increases the conversion of penicillin N to cephamycin C in Streptomyces clavuligerus. Enzyme Microb. Technol., 12, 40–45. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., Liljas,A., Brändén,C.-I. and Banaszak,L.J. (1975) Evolutionary and structural relationships among dehydrogenases. In Boyer P.D. (ed.), The Enzymes. Vol 11: Oxidation Reduction Part A. Academic Press, New York, NY, pp. 61–102. [Google Scholar]
- Rutter J., Reick,M., Wu,L.C. and McKnight,S.L. (2001) Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science, 293, 510–514. [DOI] [PubMed] [Google Scholar]
- Shi J., Blundell,T.L. and Mizuguchi,K. (2001) FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol., 310, 243–257. [DOI] [PubMed] [Google Scholar]
- Studier F.W. and Moffatt,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- Takano E., Chakraburtty,R., Nihira,T., Yamada,Y. and Bibb,M.J. (2001) A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol., 41, 1015–1028. [DOI] [PubMed] [Google Scholar]
- van Keulen G., Girbal,L., van den Bergh,E.R., Dijkhuizen,L. and Meijer,W.G. (1998) The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus is an NADPH sensor. J. Bacteriol., 180, 1411–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Keulen G., Dijkhuizen,L. and Meijer,W.G. (2000) Effects of the Calvin cycle on nicotinamide adenine dinucleotide concentrations and redox balances of Xanthobacter flavus. J. Bacteriol., 182, 4637–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimpenny J.W. and Firth,A. (1972) Levels of nicotinamide adenine dinucleotide and reduced nicotinamide adenine dinucleotide in facultative bacteria and the effect of oxygen. J. Bacteriol., 111, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodmansee A.N. and Imlay,J.A. (2002) Reduced flavins promote oxidative DNA damage in non-respiring Escherichia coli by delivering electrons to intracellular free iron. J. Biol. Chem., 277, 34055–34066. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Piston,D.W. and Goodman,R.H. (2002) Regulation of corepressor function by nuclear NADH. Science, 295, 1895–1897. [DOI] [PubMed] [Google Scholar]