Figure 3.

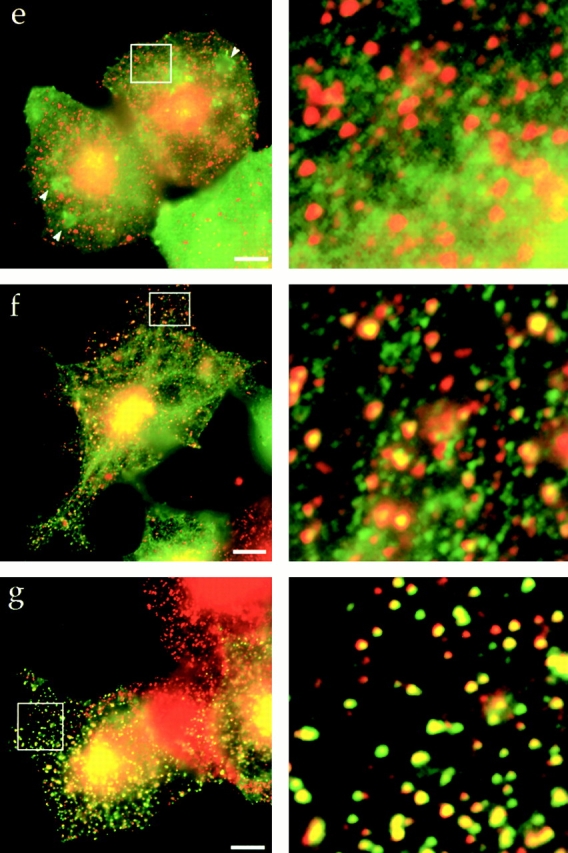
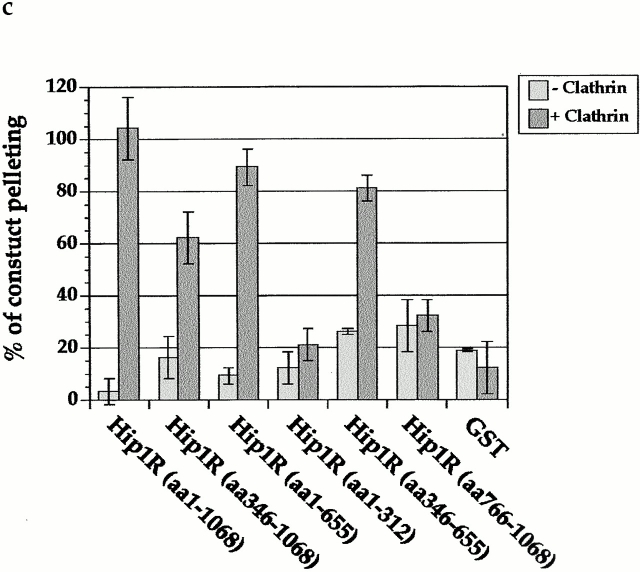
The central coiled-coil region of Hip1R binds to clathrin in vitro and in vivo. (a) Schematic diagrams of Hip1R domains expressed. (b) Coomassie-stained gel of the cage-pelleting assays. (T) shows the total amount of protein added in the assay, (−) represents pelleting without clathrin cages, and (+) represents pelleting with clathrin cages. For each assay, 600 nM of clathrin cages was used with ∼200 nM of HipR constructs. Because GST–Hip1R (amino acids 1–312) was partially degraded, a higher concentration of this construct was added to give ∼200 nM of nondegraded GST–Hip1R (amino acids 1–312). GST alone and His6LacI were used at ∼400 nM. (c) Graphical representation of results from cage-pelleting assays. The graph shows the percentage of Hip1R fragments pelleting with or without clathrin cages using data from three independent experiments. Error bars denote ±SE. (d) Endogenous clathrin specifically coimmunoprecipitated with Hip1R (amino acids 325–655)-6myc from Cos-7 cell extracts (lane 3). Clathrin was not detected using beads alone (lanes 2 and 6) or when Hip1R (amino acids 1–324)-6myc was immunoprecipitated (lane 7). The clathrin–Hip1R interaction was diminished when the beads were washed with buffer containing 0.5 M Tris-HCl (lane 4). Lanes 1 and 5 show extracts and lanes 2–4 and lanes 6 and 7 show pellets. (e–g and enlargements) Indirect immunofluorescence of endogenous clathrin HC (red) and myc-tagged Hip1R constructs (green) in Cos-7 cells. (e) Hip1R (amino acids 1–324)-6myc. (f) Hip1R (amino acids 325–655)-6myc. (g) Hip1R (amino acids 1–655)-6myc. Bar, 10 μm.