Abstract
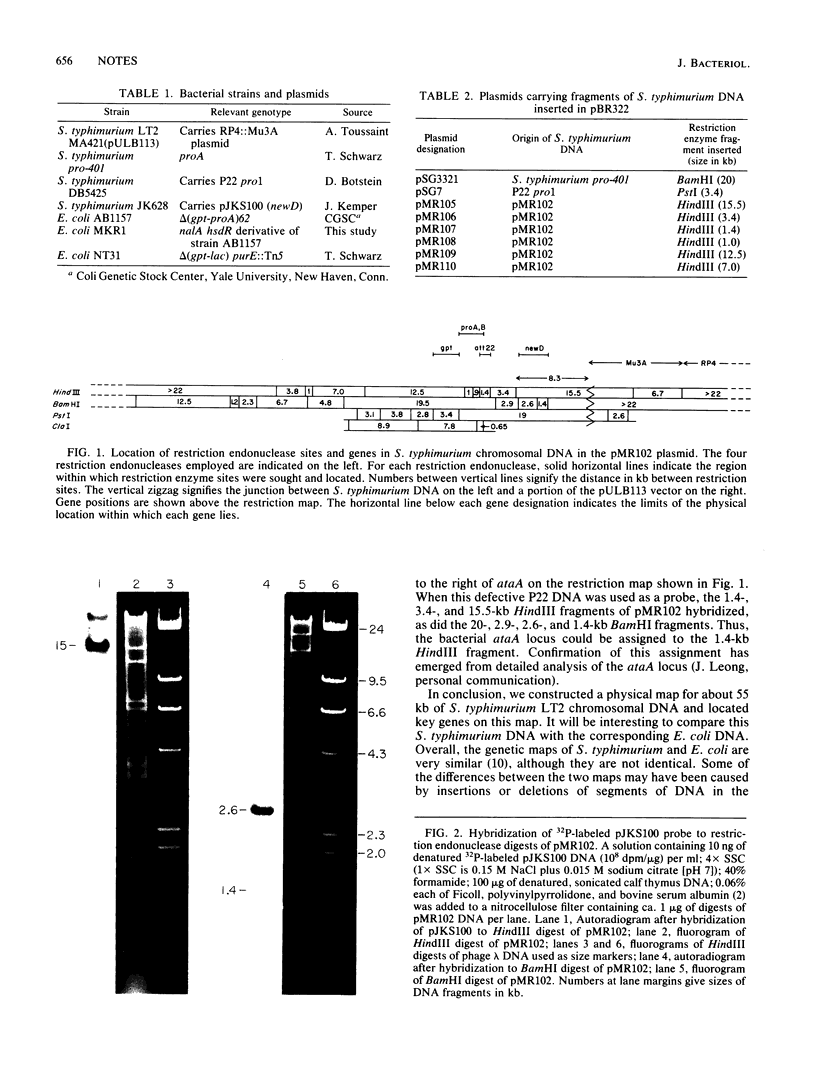
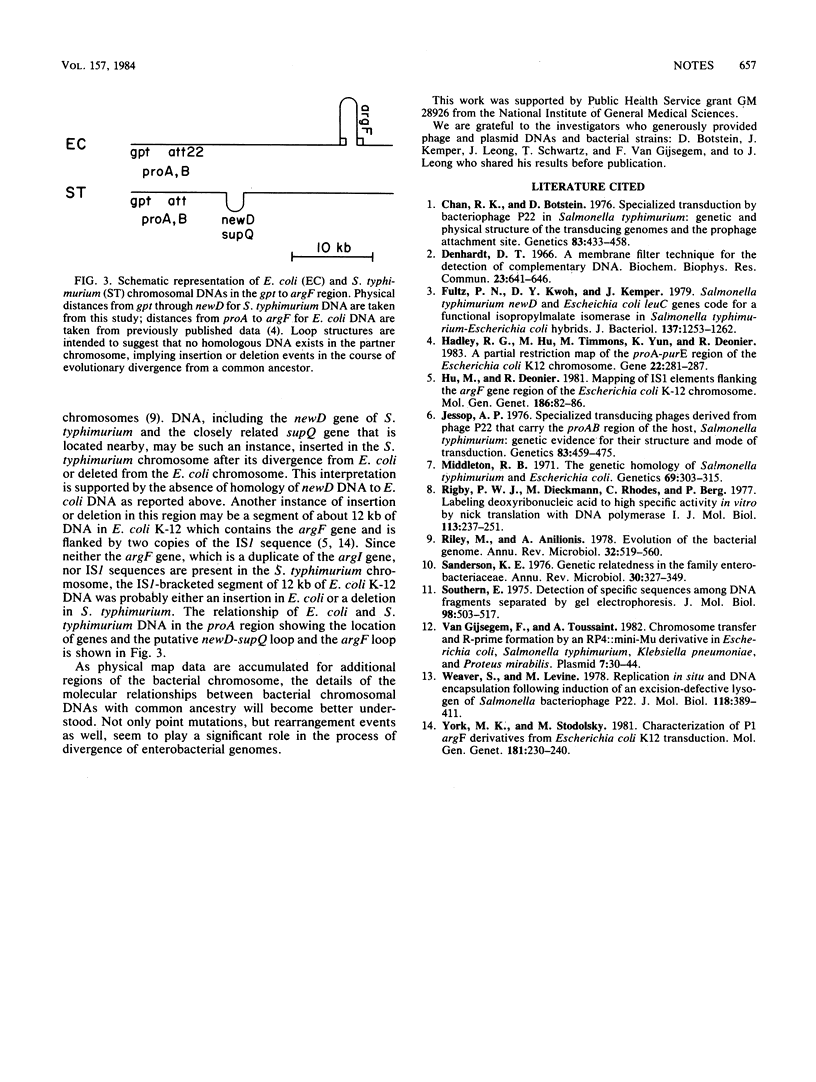
More than 55 kilobases of chromosomal DNA of Salmonella typhimurium LT2, including the gpt, proA, ataA, and newD genes, were cloned in plasmid vector pULB113. The locations of the genes and selected restriction endonuclease cleavage sites were established, and some of the restriction enzyme fragments were subcloned in plasmid vector pBR322.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chan R. K., Botstein D. Specialized transduction by bacteriophage P22 in Salmonella typhimurium: genetic and physical structure of the transducing genomes and the prophage attachment site. Genetics. 1976 Jul;83(3 PT2):433–458. [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Fultz P. N., Kwoh D. Y., Kemper J. Salmonella typhimurium newD and Escherichia coli leuC genes code for a functional isopropylmalate isomerase in Salmonella typhimurium-Escherichia coli hybrids. J Bacteriol. 1979 Mar;137(3):1253–1262. doi: 10.1128/jb.137.3.1253-1262.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley R. G., Hu M., Timmons M., Yun K., Deonier R. C. A partial restriction map of the proA-purE region of the Escherichia coli K12 chromosome. Gene. 1983 May-Jun;22(2-3):281–287. doi: 10.1016/0378-1119(83)90113-0. [DOI] [PubMed] [Google Scholar]
- Jessop A. P. Specialized transducing phages derived from phage P22 that carry the pro AB region of the host, Salmonella typhimurium: genetic evidence for their structure and mode of transduction. Genetics. 1976 Jul;83(3 PT2):459–475. [PMC free article] [PubMed] [Google Scholar]
- Middleton R. B. The genetic homology of Salmonella typhimurium and Escherichia coli. Genetics. 1971 Nov;69(3):303–315. doi: 10.1093/genetics/69.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Riley M., Anilionis A. Evolution of the bacterial genome. Annu Rev Microbiol. 1978;32:519–560. doi: 10.1146/annurev.mi.32.100178.002511. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E. Genetic relatedness in the family Enterobacteriaceae. Annu Rev Microbiol. 1976;30:327–349. doi: 10.1146/annurev.mi.30.100176.001551. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Van Gijsegem F., Toussaint A. Chromosome transfer and R-prime formation by an RP4::mini-Mu derivative in Escherichia coli, Salmonella typhimurium, Klebsiella pneumoniae, and Proteus mirabilis. Plasmid. 1982 Jan;7(1):30–44. doi: 10.1016/0147-619x(82)90024-5. [DOI] [PubMed] [Google Scholar]
- Weaver S., Levine M. Replication in situ and DNA encapsulation following induction of an excision-defective lysogen of Salmonella bacteriophage P22. J Mol Biol. 1978 Jan 25;118(3):389–411. doi: 10.1016/0022-2836(78)90235-8. [DOI] [PubMed] [Google Scholar]
- York M. K., Stodolsky M. Characterization of P1argF derivatives from Escherichia coli K12 transduction. I. IS1 elements flank the argF gene segment. Mol Gen Genet. 1981;181(2):230–240. doi: 10.1007/BF00268431. [DOI] [PubMed] [Google Scholar]