Abstract
Ticks evolved various mechanisms to modulate their host’s hemostatic and immune defenses. Differences in the anti-hemostatic repertoires suggest that hard and soft ticks evolved anti-hemostatic mechanisms independently, but raise questions on the conservation of salivary gland proteins in the ancestral tick lineage. To address this issue the sialome (salivary gland secretory proteome) from the soft tick, Argas monolakensis was determined by proteomic analysis and cDNA library construction of salivary glands from fed and unfed adult female ticks. The sialome is composed of ~130 secretory proteins, of which the most abundant protein folds are the lipocalin, BTSP, BPTI and metalloprotease families which also comprise the most abundant proteins found in the salivary glands. Comparative analysis indicates that the major protein families are conserved in hard and soft ticks. Phylogenetic analysis shows, however, that most gene duplications are lineage specific, indicating that the protein families analyzed possibly evolved most of their functions after divergence of the two major tick families. In conclusion, the ancestral tick may have possessed a simple (few members for each family), but diverse (many different protein families) salivary gland protein domain repertoire.
Keywords: Argas, blood-feeding, evolution, proteome, sialome
1. Introduction
Blood-feeding behavior evolved independently in insects in flies (Diptera), true bugs (Hemiptera), lice (Phthiraptera), fleas (Siphonaptera) and moths (Lepidoptera) (Ribeiro, 1995). In arachnids, hematophagous behavior evolved independently in ticks (Ixodida) and mesostigmatid mites (Mans and Neitz, 2004a; Radovsky, 1969). In each case the blood-feeding arthropod had to evolve mechanisms to modulate host defenses such as the hemostatic and immune systems. The mechanisms evolved by hematophagous arthropods mostly involve the secretion of salivary gland derived proteins during feeding and it is these proteins that target important host processes (Ribeiro, 1995). Adaptation to a blood-feeding environment by arthropods can thus be redefined as a study of the structural and functional evolution of the sialome (salivary gland proteomes of hematophagous organisms). From this perspective, the fact of independent adaptation to blood-feeding is clear when sialomes from different species are compared (Valenzuela et al., 2002a; Ribeiro and Francischetti, 2003; Champagne, 2004). In most cases the mechanisms to counteract the host’s immune and hemostatic defenses differ between various groups of hematophagous organisms. For the two major tick families, the soft (Argasidae) and hard (Ixodidae) ticks, this also holds and suggested that soft and hard ticks adapted to a blood-feeding environment independently (Mans et al., 2002a; Mans and Neitz, 2004a). Even so, phylogenetic analysis shows that the hard and soft tick families group as a monophyletic clade, indicating that they shared a common ancestor to the exclusion of other mites (Black and Piesman, 1994; Barker and Murrell, 2004). It was, however, also indicated that the ancestral tick lineage must have had some form of host-association, such as feeding on lymphatic fluid of the living host or as scavengers (Mans and Neitz, 2004a). This implies that the ancestral tick lineage must have features conserved in both families.
While the anti-hemostatic factors of soft and hard ticks differ in their mechanisms of action and protein families that they belong to, the two tick families share common protein folds in their salivary glands, such as the basic pancreatic trypsin inhibitor (BPTI) and lipocalin protein families (Mans and Neitz, 2004a). The BPTI/Kunitz family is composed of proteins that exhibit the basic pancreatic trypsin inhibitor fold (Laskowski and Kato, 1980). BPTI-like proteins act as thrombin, fXa and platelet aggregation inhibitors in soft ticks (Waxman et al., 1990; Karczewski et al., 1994; Van de Locht et al., 1998; Joubert et al., 1998; Mans et al., 2002b; Mans et al., 2007). In hard ticks, BPTI-like proteins inhibit the fVIIa/TF complex (Francischetti et al., 2002; Francischetti et al., 2004). The other major protein family so far described for ticks is the lipocalin family. In soft ticks, lipocalins function as anti-complement factors (Nunn et al., 2005), inhibitors of platelet aggregation (Waxman and Connolly 1993; Keller et al., 1993) and toxins (Mans et al., 2002c) and have been shown to be abundantly expressed in salivary glands (Mans et al., 2001; Mans et al., 2003; Oleaga et al., 2007). In hard ticks lipocalins that scavenge histamine and serotonin have also been described (Paesen et al., 1999; Paesen et al., 2000; Sangamnatdej et al., 2002). Recently high throughput sequencing of salivary gland transcripts from Ixodes scapularis and I. pacificus has shown that hard ticks possess more than 25 protein families in their sialomes (Valenzuela et al., 2002b; Francischetti et al., 2005; Ribeiro et al., 2006). The question raised is how many of these protein families are also represented in soft ticks? Data on this could indicate the number of conserved protein families found in the salivary glands of the ancestral tick lineage and whether any specific orthologs existed between hard and soft ticks before their divergence.
To address this, the salivary gland transcriptome and proteome of the soft tick Argas monolakensis were characterized. A. monolakensis is limited to islands located on Mono Lake, California where it feeds annually on the breeding Californian gull population (Larus californicus) (Schwan et al., 1992). It is problematic for ornithologists working on the islands and is a potential vector of Mono Lake virus (Rheoviridae: Orbivirus) (Schwan and Winkler, 1984; Schwan et al., 1988). We show that the major protein families are conserved between hard and soft ticks. Even so, the numerous gene duplication events observed within individual protein families appear to be limited to lineage specific expansions, indicating that most of the sialome diversity observed for the different tick families evolved after their divergence.
2. Materials and Methods
2.1. cDNA library construction and analysis
Argas monolakensis ticks were collected as described (Mans et al., 2007), salivary glands dissected and frozen at −70°C until use. Glands from unfed and fed (fed to repletion on chickens 4 days prior to dissection) adult females were thawed in RNAlater and cDNA libraries constructed as previously described (Valenzuela et al., 2002b). One thousand and seven hundred and twenty eight plaques were picked and sequenced for the unfed and fed library, respectively. Sequences were cleaned-up and clustered into contigs (synonymous with consensus sequences) using in-house bioinformatics programs as previously described (Ribeiro et al., 2006). Contigs obtained for individual clusters were analyzed using BLASTX to identify conserved proteins in the non-redundant database (Altschul et al., 1990). Potential protein coding sequences were manually identified from consensus sequences by the presence of an open reading frame, stop codon, poly-adenylation signal, poly-A tail and starting methionine. Protein domain data obtained from the BLASTX analysis were used to determine the coverage obtained for truncated ESTs that code for cytoplasmic proteins. Full-length secretory proteins were identified by the presence of a signal peptide using the SignalP3.0 server (Bendtsen et al., 2004). The presence of multiple cysteines that would suggest the presence of disulphide bonds were taken as evidence of secretion for truncated proteins. The assembly of the consensus sequences was manually inspected and full-length and truncated reading frames were confirmed by manual inspection.
2.2. Protein family analysis
BLASTP analysis was initially used to assign translated consensus sequences to protein families (Altschul et al., 1990). Subsequently, PSI-BLAST analysis (Altschul et al., 1997) was used to confirm these assignments, as this approach allows the retrieval of all family members, including those that are distantly related. This method allowed assignment of some contigs to protein families that could not be assigned using BLASTP analysis.
2.3. Comparative domain analysis of hard and soft tick sialomes
Most of the tick sequences derived from salivary gland transcriptomes have been deposited as ESTs with no corresponding data in the protein databank. As such, the data exist in a form that makes it difficult to perform a comparative analysis of salivary gland sialomes that will allow the assignment of relative domain numbers to different tick species. In order to derive a more representative data analysis of protein domains present in tick salivary glands, the EST datasets for different tick species were clustered as described above to obtain a non-redundant dataset of contigs for each. TBLASTN analysis using the predicted secretory proteins from A. monolakensis was then performed against these individual databases. Results were manually inspected and hits with e-values less than 10−5 were taken as positive hits to get a conservative estimate of protein domain numbers. Translated protein sequences were derived from the contigs for each species and used to perform a TBLASTN analysis against its own species specific database in order to find homologs.
2.4. Preparation of salivary gland extract
Salivary gland extract (SGE) was prepared as described previously (Mans et al., 2007). Briefly, salivary glands were dissected from female ticks in 20 mM Tris-HCl, 0.15 M NaCl, pH 7.4 by tearing the dorsal cuticle and removing the glands with forceps. Glands were washed in dissection buffer before being frozen at −70°C. Glands were reconstituted in 20 mM Tris-HCl, pH 7.4 and disrupted by sonication, before the cell debri were pelleted by centrifugation at 16000g for 20 minutes on a bench top centrifuge. Supernatant was removed and constituted SGE.
2.5. Proteomic analysis of salivary gland extract from A. monolakensis by one-dimensional electrophoresis and Edman sequencing
SGE (10 gland equivalents) was heat treated at 80°C and centrifuged at 16000g for 10 minutes to pellet denatured proteins. The pellet (P) was dissolved in SDS-PAGE sample buffer and processed for one-dimensional electrophoresis by heating at 70°C for 10 minutes. Of this sample the equivalent of 5 glands was loaded per well. The soluble protein (S) was processed for SDS-PAGE in a similar manner and 5 gland equivalents were loaded as well. Electrophoresis was performed using standard conditions and protein blotted onto PVDF membranes before staining with Coomassie Brilliant Blue. Bands were cut out and analyzed by Edman sequencing.
2.6. Proteomic analysis of salivary gland extract from A. monolakensis by two-dimensional electrophoresis and peptide mass fingerprinting
Two-dimensional gel electrophoresis was performed using the ZOOM IPGRunner System (Invitrogen). Briefly, approximately 50 μg of salivary gland extract (5 salivary gland equivalents ) were solubilized with 155 μl rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 20 mM DTT, 0.5% carrier ampholytes, pH3-10). The samples were absorbed overnight at room temperature on rehydration ZOOM strips (7 cm; pH3-10NL) before focusing under manufacturer’s recommended conditions. The focused IPG strips were reduced/alkylated/equilibrated with reducing and then alkylation reagents dissolved in the sample buffer. The strips were then applied onto NuPAGE 4–12% Bis-Tris ZOOM gels (Invitrogen). The gels were run under MOPS buffer and stained with SeeBlue staining solution (Bio-Rad). A total of 78 spots were selected for tryptic digestion, based on their staining intensity. Spots picked were processed for tryptic digestion by standard mass spectrometry protocols that included reduction and alkylation before tryptic-digestion. Tryptic digests were analyzed by coupling the Nanomate (Advion BioSciences) – an automated chip-based nano-electrospray interface source – to a quadrupole time-of-flight mass spectrometer, QStarXL MS/MS System (Applied Biosystems/Sciex). Computer-controlled, data-dependent automated switching to MS/MS provided peptide sequence information. AnalystQS software (Applied Biosystems/Sciex) was used for data acquisition. Data processing and databank searching were performed with Mascot software (Matrix Science). The non-redundant protein database from the NCBI, National Library of Medicine, NIH, was used for the search analysis, as was a protein database generated during the course of this work.
2.7. Proteomic analysis of salivary gland extract from A. monolakensis by liquid chromatography and peptide mass fingerprinting
Analysis of the A. monolakensis proteome by liquid chromatography followed the same approach previously used to identify the anti-clotting and anti-platelet agents from salivary gland extract (Mans et al., 2007). Briefly, SGE (100 gland equivalents) was applied to an anion exchange column and fractionated. The flow-through was applied to a cation exchange column and fractionated under the same conditions. Fractions that represented major peaks were pooled and applied to reversed phase chromatography. Individual peaks (116 fractions) were collected and analyzed by peptide mass fingerprinting as described above.
2.8. Phylogenetic analysis of protein families
Protein families were aligned using ClustalX and manually checked and adjusted (Jeanmougin et al., 1998). Neighbor-joining analysis was performed using the Mega package (Kumar et al., 1994). The Poisson model for amino acid substitution was used and 10000 bootstraps were performed with complete deletion of gapped positions.
3. Results
3.1. cDNA library properties
A total of 3087 EST sequences were obtained and their analysis and Genbank accession numbers can be found in the supplementary materials. Clusterization of ESTs yielded 1472 contigs. Unknown sequences represented 38.1% of all sequences and 67% of all clusters, with an average of 1.2 sequences per cluster, indicating that most of them were singletons. House-keeping sequences represented 23.9% of all sequences, 21% of all clusters with an average of 2.4 sequences per cluster, indicating only a slight increase above the singleton background. In contrast, sequences coding for potential secreted proteins represented 37.8% of all sequences, 11% of all clusters and on average 7.2 sequences per cluster. The unknown sequences are mostly singletons that do not yield an open reading frame and were considered non-sense sequences with no informative value for cDNA library analysis. If these sequences are removed from the library analysis, house-keeping sequences represent 38% of all sequences and 64% of all clusters, while potential secretory sequences represent only 33% of all clusters, but 61% of all sequences. This indicates that potential secreted proteins, although being the lowest in numbers of clusterized contigs, are the most highly represented sequences in the library. A final number of 127 potential secretory components were deposited in the sequence databank (Table 1).
Table 1.
Secretory proteins identified in the sialome of Argas monolakensis.
| Protein family | GI number | Best hit in NRDB | GI number of best hit | E-value | Family assignment | Number of Transcripts | Proteome (E, LC, 2D) / % | Status | Mr (Da) | pI |
|---|---|---|---|---|---|---|---|---|---|---|
| TSP1 type 1 superfamily | ||||||||||
| Single BTSP domains** | ||||||||||
| AM-7 ** | 114152932 | AM-15 | 114152942 | 6e-16 | BLASTP | 81 | E, LC / 5.7% | Full-length | 9290.9 | 4.32 |
| AM-15 ** | 114152942 | AM-7 | 114152932 | 1e-13 | BLASTP | 9 | LC / 0.19% | Full-length | 7321.0 | 4.35 |
| AM-17 *** | 114152946 | AM-121 | 114153038 | 0.34 | BLASTP # | 21 | E, LC, 2D / 0.13% | Full-length | 16046.3 | 9.28 |
| AM-21 ** | 114152952 | AM-15 | 114152942 | 6e-10 | BLASTP | 22 | E, LC, 2D / 0.13% | Full-length | 10440.4 | 5.12 |
| AM-44 ** | 114152986 | AM-46 | 114152988 | 5e-31 | Cysteines | 14 | E, LC / 0.22% | Full-length | 8428.9 | 8.84 |
| AM-46 ** | 114152988 | AM-44 | 114152986 | 4e-24 | Cysteines | 3 | 2D | Full-length | 8554.0 | 9.30 |
| AM-60 ** | 114153006 | - | - | - | Cysteines | 3 | - | Full-length | 8250.1 | 8.24 |
| AM-71** | 114153018 | AM-185 | 114153060 | 0.001 | Cysteines | 4 | - | Full-length | 10473.8 | 4.80 |
| AM-78** | 114153024 | AM-233 | 114153114 | 0.087 | Cysteines | 12 | E, 2D | Full-length | 10653.1 | 9.19 |
| AM-121 ** | 114153038 | Isca | 67083315 | 0.001 | BLASTP | 1 | - | Full-length | 14078.7 | 4.14 |
| AM-185** | 114153060 | AM-233 | 114153114 | 0.001 | Cysteines | 2 | - | Full-length | 8814.9 | 5.19 |
| AM-199 *** | 114153078 | - | - | - | PSI-BLAST | 6 | - | Full-length | 15389.5 | 10.3 |
| AM-233** | 114153114 | AM-71 | 114153018 | 6e-04 | Cysteines | 4 | - | Full-length | 8545.5 | 4.93 |
| AM-256 *** | 114153132 | AM-121 | 114153038 | 7e-04 | BLASTP | 3 | - | Full-length | 14287.4 | 4.43 |
| AM-286 *** | 114153158 | AM-121 | 114153038 | 2e-04 | BLASTP | 2 | - | Truncated | - | - |
| 7DBF family – double BTSP domains | ||||||||||
| AM-59 | 114153004 | AM-670 | 114153250 | 7e-21 | BLASTP | 6 | 1D | Truncated | - | - |
| AM-63 | 114153008 | AM-634 | 114153244 | 2e-35 | BLASTP | 14 | 1D | Full-length | 19838.4 | 8.70 |
| AM-279 | 114153150 | AM-63 | 114153004 | 5e-14 | BLASTP | 3 | - | Truncated | - | - |
| AM-310 | 114153172 | AM-670 | 114153250 | 3e-22 | BLASTP | 2 | - | Full-length | 15636.4 | 6.45 |
| AM-459 | 114153212 | AM-670 | 114153250 | 3e-22 | BLASTP | 1 | - | Full-length | 16957 | 8.32 |
| AM-462 | 114153214 | AM-459 | 114153212 | 4e-12 | BLASTP | 1 | - | Truncated | - | - |
| AM-634 | 114153244 | AM-63 | 114153008 | 1e-35 | BLASTP | 1 | - | Full-length | 17234.5 | 8.36 |
| AM-670 | 114153250 | AM-310 | 114153172 | 2e-24 | BLASTP | 1 | - | Full-length | 16150.5 | 8.50 |
| BPTI/Kunitz family | ||||||||||
| AM-6A | 114152928 | AM-6B | 114152930 | 5e-51 | PHYRE | 65 | E, LC, 2D, 1D /1.6% | Full-length | 9961.2 | 8.99 |
| AM-6B | 114152930 | AM-6A | 114152928 | 5e-51 | PHYRE | E, LC, 2D, 1D | Full-length | 9930.1 | 8.82 | |
| AM-20 | 114152950 | TFPI-like | 3205213 | 3e-20 | BLASTP | 22 | E, LC, 2D / 0.53% | Full-length | 18487.4 | 5.29 |
| AM-28 | 114152962 | Ornithodorin | 3024318 | 2e-07 | BLASTP | 12 | LC / 0.73% | Full-length | 13678.4 | 4.21 |
| AM-30a | 114152966 | Monogrin 1B | 114152968 | 5e-36 | BLASTP | 15 | E, LC, 2D / 1.2% | Full-length | 9702.9 | 8.28 |
| AM-30b | 114152968 | Monogrin 1A | 114152966 | 5e-36 | BLASTP | E, LC, 2D / 1.7% | Full-length | 9817.1 | 8.96 | |
| AM-42 | 114152984 | Chelonianin | 124076 | 0.002 | BLASTP | 5 | - | Full-length | 17166.4 | 9.55 |
| AM-56 | 114153000 | AMBP | 2497695 | 0.007 | BLASTP | 18 | LC / 0.06% | Full-length | 7665.6 | 9.04 |
| AM-138 | 114153042 | Amyloid A4 | 109484147 | 8e-12 | BLASTP | 1 | 2D | Full-length | 10349.9 | 9.75 |
| AM-176 | 114153048 | Collagen-like | 119890741 | 0.12 | BLASTP | 13 | LC, 2D / 0.92% | Truncated | - | - |
| Pro-Gly-rich peptides | ||||||||||
| AM-174 | 114153046 | Isca | 56159959 | 5e-10 | BLASTP | 1 | - | Full-length | 12199.5 | 6.38 |
| AM-217 | 114153098 | AM-174 | 114153046 | 4e-04 | BLASTP | 4 | - | Full-length | 10116.1 | 9.69 |
| AM-793 | 114153264 | - | - | - | - | 1 | - | Full-length | 6351.4 | 5.92 |
| AM-811 | 114153268 | - | - | - | - | 1 | - | Full-length | 13668.3 | 10.5 |
| AM-1065 | 114153294 | Isca | 67083773 | 0.03 | BLASTP | 1 | - | Full-length | 14797.5 | 6.15 |
| 5.3 kDa | ||||||||||
| AM-65 | 114153012 | Isca | 67083579 | 0.58 | TBLASTX | 2 | - | Full-length | 5314.0 | 8.92 |
| Metalloproteases | ||||||||||
| AM-32 | 114152972 | AM-49 | 114152992 | 3e-08 | BLASTP | 19 | - | Truncated | - | - |
| AM-49 | 114152992 | Bmic | 71726986 | 2e-26 | BLASTP | 7 | 2D | Truncated | - | - |
| AM-80 | 114153028 | Isca | 60729624 | 6e-34 | BLASTP | 6 | 1D | Truncated | - | - |
| AM-211 | 114153092 | AM-317 | 114153178 | 9e-04 | BLASTP | 5 | - | Truncated | - | - |
| AM-214 | 114153096 | AM-504 | 114153226 | 8e-16 | BLASTP | 4 | - | Truncated | - | - |
| AM-224 | 114153104 | AM-225 | 114153106 | 1e-33 | BLASTP | 3 | - | Truncated | - | - |
| AM-225 | 114153106 | AM-225 | 114153106 | 1e-33 | BLASTP | 1 | 2D | Truncated | - | - |
| AM-234 | 114153116 | AM-1119 | 114153298 | 0.069 | BLASTP | 4 | LC, 1D / 0.47% | Truncated | - | - |
| AM-267 | 114153144 | AM-299 | 114153162 | 1e-12 | BLASTP | 3 | - | Truncated | - | - |
| AM-277 | 114153148 | AM-478 | 114153220 | 1e-19 | BLASTP | 3 | - | Truncated | - | - |
| AM-299 | 114153162 | Isca | 22164296 | 2e-35 | BLASTP | 2 | - | Truncated | - | - |
| AM-317 | 114153178 | Ipac | 51011584 | 2e-09 | BLASTP | 2 | - | Truncated | - | - |
| AM-478 | 114153220 | AM-277 | 114153148 | 5e-19 | BLASTP | 1 | - | Full-length | 25669.2 | 9.12 |
| AM-504 | 114153226 | AM-214 | 114153096 | 8e-16 | BLASTP | 1 | - | Truncated | - | - |
| AM-642 | 114153246 | Isca | 60729624 | 0.16 | BLASTP | 1 | - | Truncated | - | - |
| AM-804 | 114153266 | AM-478 | 114153220 | 6e-08 | BLASTP | 1 | - | Truncated | - | - |
| AM-1119 | 114153298 | Isca | 60729624 | 5e-43 | BLASTP | 1 | 1D | Truncated | - | - |
| ADAM domain | ||||||||||
| AM-229 | 114153112 | ADAM | 118101316 | 0.033 | BLASTP | 3 | - | Full-length | 10604.1 | 9.17 |
| Lipocalins | ||||||||||
| AM-10 | 114152936 | Arg r1 | 58371884 | 7e-34 | BLASTP | 151 | LC, 2D / 23.8% | Full-length | 15725.0 | 5.46 |
| AM-14 | 114152940 | AM-218 | 114153100 | 3e-37 | PSI-BLAST | 4 | E, 2D | Full-length | 31682.1 | 5.33 |
| AM-26A | 114152958 | AM-26B | 114152960 | 9e-83 | BLASTP | 27 | E, LC, 2D / 3.8% | Full-length | 18526.4 | 7.76 |
| AM-26B | 114152960 | AM-26A | 114152958 | 9e-83 | BLASTP | E, LC, 2D | Full-length | 18375.1 | 5.57 | |
| AM-33 | 114152974 | TSGP4 | 25991438 | 13e-15 | BLASTP | 35 | E, LC, 2D / 3.3% | Full-length | 18427.1 | 9.03 |
| AM-38 | 114152976 | AM-182 | 114153056 | 1e-48 | BLASTP | 34 | E, LC, 2D / 8.8% | Full-length | 16559.1 | 4.58 |
| AM-41 | 114152982 | AM-118 | 114153036 | 8e-46 | BLASTP | 9 | - | Truncated | - | - |
| AM-47 | 114152990 | Bmic | 45360102 | 1e-26 | BLASTP | 12 | - | Full-length | 21574.6 | 8.88 |
| AM-50 | 114152994 | AM-51 | 114152996 | 4e-101 | BLASTP | 27 | E | Full-length | 23682.9 | 4.86 |
| AM-51 | 114152996 | AM-50 | 114152994 | 2e-105 | BLASTP | 2 | - | Full-length | 23673.9 | 4.61 |
| AM-53 | 114152998 | AM-26A | 114152958 | 6e-38 | BLASTP | 11 | LC / 0.23% | Full-length | 18812.6 | 5.05 |
| AM-118 | 114153036 | AM-41 | 114152982 | 8e-46 | BLASTP | 1 | 2D | Truncated | - | - |
| AM-180 | 114153054 | Arg r1 | 58371884 | 1e-10 | BLASTP | 1 | LC / 0.6% | Full-length | 20147.4 | 9.22 |
| AM-182 | 114153056 | AM-38 | 114152976 | 5e-52 | BLASTP | 9 | - | Full-length | 17094.0 | 7.00 |
| AM-194 | 114153072 | AM-47 | 114152990 | 4e-12 | BLASTP | 6 | - | Truncated | - | - |
| AM-209 | 114153090 | AM-47 | 114152990 | 0.024 | BLASTP | 5 | LC, 2D,1D | Truncated | - | - |
| AM-218 | 114153100 | AM-14 | 114152940 | 9e-42 | PSI-BLAST | 4 | 2D | Truncated | - | - |
| AM-249 | 114153124 | AM-47 | 114152990 | 4e-10 | BLASTP | 3 | - | Truncated | - | - |
| AM-304 | 114153166 | Isca | 114153166 | 5e-06 | BLASTP | 2 | - | Truncated | - | - |
| AM-378 | 114153198 | AM-304 | 114153166 | 0.01 | BLASTP | 2 | - | Truncated | - | - |
| AM-925 | 114153282 | Bmic | 45360102 | 1e-11 | BLASTP | 1 | 2D | Truncated | - | - |
| AM-1124 | 114153300 | AM-38 | 114152976 | 4e-42 | BLASTP | 1 | - | Full-length | 18543.9 | 8.99 |
| Novel 8 kDa Cys-rich domain – related to ixodegrins | ||||||||||
| AM-40 | 114152980 | AM-98 | 114153030 | 3e-17 | Cysteine | 2 | - | Full-length | 5759.2 | 7.81 |
| AM-98 | 114153030 | AM-40 | 114152980 | 3e-17 | Cysteine | 1 | - | Full-length | 5537.1 | 8.70 |
| AM-183 | 114153058 | - | - | - | Cysteine | 9 | - | Full-length | 4556.9 | 4.75 |
| AM-191A | 114153068 | AM-191B | 114153070 | 5e-19 | Cysteine | 6 | - | Full-length | 5568.0 | 8.33 |
| AM-191B | 114153070 | AM-191A | 114153068 | 4e-17 | Cysteine | 2 | - | Full-length | 5568.0 | 8.33 |
| Anti-microbial peptides | ||||||||||
| Defensins | ||||||||||
| AM-135 | 114153040 | AM-1246 | 114153306 | 6e-10 | BLASTP | 1 | - | Full-length | 5487.1 | 7.81 |
| AM-383 | 114153200 | Hlon | 62122377 | 1e-16 | BLASTP | 2 | - | Full-length | 6003.6 | 8.33 |
| AM-1173 | 114153302 | Omou | 22324328 | 2e-13 | BLASTP | 1 | - | Full-length | 5984.7 | 8.62 |
| AM-1246 | 114153306 | AM-135 | 114153040 | 6e-10 | BLASTP | 1 | - | Full-length | 5019.6 | 8.70 |
| Anti-microbial-Histidine-rich | ||||||||||
| AM-189 | 114153066 | Microplusin | 28864185 | 2e-013 | BLASTP | 7 | - | Full-length | 11062.2 | 5.82 |
| C3-like complement protein | ||||||||||
| AM-582 | 114153236 | Crot | 33325642 | 1e-04 | BLASTP | 1 | - | Truncated | - | - |
| Serpin | ||||||||||
| AM-385 | 114153202 | Iric | 115334937 | 3e-36 | BLASTP | 2 | - | Truncated | - | - |
| Chitinase | ||||||||||
| AM-24 | 114152956 | Agam | 118778781 | 4e-15 | BLASTP | 2 | - | Truncated | - | - |
| Chitinase binding domain – Mucin | ||||||||||
| AM-334 | 114153184 | Tcas | 91086069 | 6e-05 | BLASTP | 2 | - | Full-length | 22157.3 | 4.05 |
| AM-575 | 114153234 | Omou | 41352541 | 3e-017 | BLASTP | 1 | - | Full-length | 19486.7 | 5.07 |
| Antigen 5/SCP domain | ||||||||||
| AM-12 | 114152938 | AM-177 | 114153050 | 4e-49 | BLASTP | 26 | LC, 1D / 2.3% | Truncated | - | - |
| AM-177 | 114153050 | AM-12 | 114152938 | 2e-49 | BLASTP | 10 | LC, 2D / 0.95% | Truncated | - | - |
| Phospholipase A2 | ||||||||||
| AM-263 | 114153140 | AM-857 | 114153278 | 2e-23 | BLASTP | 3 | - | Truncated | - | - |
| AM-857 | 114153278 | AM-263 | 114153140 | 9e-24 | BLASTP | 1 | 2D | Truncated | - | - |
| Lysophospholipase | ||||||||||
| AM-515 | 114153228 | Amel | 48101936 | 3e-67 | BLASTP | 1 | - | Truncated | - | - |
| Procathepsin B | ||||||||||
| AM-632 | 114153242 | Longipain | 121309133 | 2e-23 | BLASTP | 1 | - | Truncated | - | - |
| Aspartic protease | ||||||||||
| AM-1215 | 114153304 | Hlon | 83319201 | 1e-25 | BLASTP | 1 | - | Truncated | - | - |
| 5′-Nucleotidase | ||||||||||
| AM-918 | 114153280 | Dpsu | 125981113 | 0.001 | BLASTP | 1 | - | Truncated | - | - |
| Cimex apyrase | ||||||||||
| AM-906 | Xlae | 57282934 | 2e-07 | BLASTP | 1 | - | Truncated | - | - | |
| 7CF | ||||||||||
| AM-186 | 114153062 | AM-1013 | 114153292 | 6e-38 | BLASTP | 8 | LC, 2D / 0.06% | Full-length | 12785.0 | 4.85 |
| AM-1013 | 114153292 | 1 | - | Full-length | 12803.5 | 8.49 | ||||
| Novel 15kDa | Related to | Der f2 allergens | ||||||||
| AM-197A | 114153074 | AM-197B | 114153076 | 5e-62 | BLASTP | 6 | LC, 2D / 0.66% | Full-length | 15116.9 | 5.68 |
| AM-197B | 114153076 | AM-197A | 114153074 | 9e-72 | BLASTP | 6 | LC / 1.16% | Full-length | 15115.8 | 5.17 |
| AM-201 | 114153082 | AM-197B | 114153076 | 0.18 | BLASTP | 5 | LC / 0.298% | Full-length | 14022.2 | 8.51 |
| AM-236 | 114153118 | AM-197A | 114153074 | 3e-08 | BLASTP | 4 | - | Full-length | 13409.9 | 8.17 |
| Novel singletons | ||||||||||
| AM-5 | 114152926 | - | - | - | - | 3 | - | Full-length | 6436.4 | 12.3 |
| AM-16 | 114152944 | - | - | - | 22 | - | Full-length | 5567.4 | 4.59 | |
| AM-19 | 114152948 | - | - | - | - | 5 | - | Full-length | 5393.1 | 6.39 |
| AM-22 | 114152954 | AM-113 | 114153034 | 2e-13 | 47 | - | Full-length | 4694.1 | 3.66 | |
| AM-69 | 114153014 | AM-70 | 114153016 | 4e-06 | 7 | - | Full-length | 4411.1 | 5.21 | |
| AM-70 | 114153016 | AM-69 | 114153014 | 4e-06 | 4 | - | Full-length | 4318.1 | 9.03 | |
| AM-112 | 114153032 | AM-22 | 114152954 | 6e-13 | 1 | - | Full-length | 4694.1 | 3.66 | |
| AM-113 | 114153034 | AM-22 | 114152954 | 2e-06 | 1 | - | Full-length | 5427.7 | 3.25 | |
| AM-170 | 114153044 | - | - | - | - | 14 | - | Truncated | - | - |
| AM-204 | 114153084 | - | - | - | - | 5 | - | Full-length | 7587.5 | 5.76 |
| AM-213 | 114153094 | - | - | - | - | 4 | - | Full-length | 10442.4 | 6.45 |
| AM-240 | 114153120 | - | - | - | - | 3 | - | Truncated | - | - |
| AM-285 | 114153156 | - | - | - | - | 3 | - | Full-length | 7861.6 | 9.87 |
| AM-314 | 114153176 | - | - | - | - | 2 | - | Full-length | 13083.2 | 8.62 |
| AM-320 | 114153180 | - | - | - | - | 2 | - | Truncated | - | - |
| AM-321 | 114153182 | - | - | - | - | 2 | - | Full-length | 8787.0 | 8.82 |
| AM-428 | 114153206 | - | - | - | - | 1 | - | Truncated | - | - |
| AM-852 | 114153274 | - | - | - | 1 | Full-length | 6544.6 | 9.38 | ||
Potential secretory proteins were identified based on the presence of a secretory peptide, or when truncated based on known secretory properties of other family members, or when novel based on the presence of cysteines and high transcript numbers. Indicated are identified proteins, their genbank accession numbers, the best hits in the non-redundant databank, number of transcripts in the cDNA library and presence in the proteome based on Edman degradation (E), peptide mass fingerprinting after liquid-chromatography (LC) and peptide mass fingerprinting after one-dimensional (1D) or two-dimensional electrophoresis (2D) of salivary gland extracts. Molecular masses and iso-electric points of mature proteins are also indicated. Additional data for the analysis of the cDNA libraries and proteome analysis can be found in the supplementary materials.
Proteins that did not retrieve additional best hits in the NRDB were assigned to protein families by PSI-BLAST analysis. In the case of the BTSP family, all members in the database were retrieved using AM-7 as seed protein.
BTSP proteins that possess the cysteine-rich N-terminal thrombospondin repeat motif domain, but lack the C-terminal low-complexity lysine- or glutamic acid-rich tail.
BTSP that possess the cysteine-rich N-terminal thrombospondin repeat motif domain as well as the C-terminal low-complexity lysine- or glutamic acid-rich tail. # BLASTP analysis was performed with the conserved N-terminal thrombospondin repeat motif.
3.2. Analysis of the proteome of A. monolakensis by Edman degradation
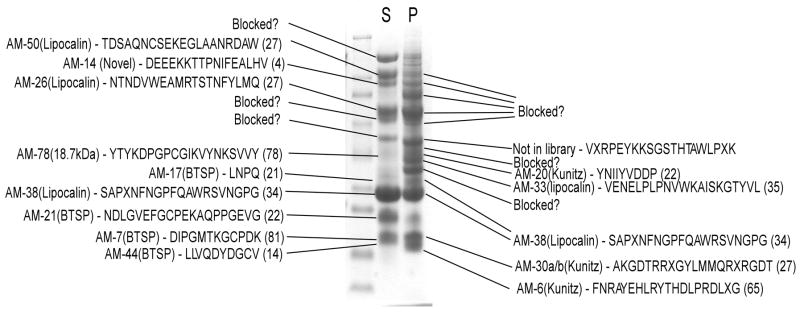
The salivary proteome of A. monolakensis as analyzed by one-dimensional SDS-PAGE is complex and consists of several highly abundant bands (Fig. 1). Temperature denaturation and precipitation allowed for differential fractionation of proteins for easier interpretation of Edman sequencing data. Fourteen sequences were obtained, of which thirteen corresponded to sequences found in the cDNA database (Table 1). In all cases, the N-terminal sequences obtained corresponded to the predicted N-terminal sequences using SignalP.
Fig. 1.
Proteome analysis of SGE by Edman degradation. Salivary gland extract from A. monolakensis was heat-inactivated at 80°C, precipitated (P) and soluble (S) proteins were separated by centrifugation. The soluble and insoluble fractions were then fractionated by one-dimensional SDS-PAGE, blotted onto PVDF membranes and bands were cut-out for Edman sequencing. Edman sequences are indicated, as well as their corresponding sequences found in the cDNA library. The protein families and the number of transcripts are indicated in brackets.
3.3. Analysis of the proteome of A. monolakensis by two-dimensional electrophoresis
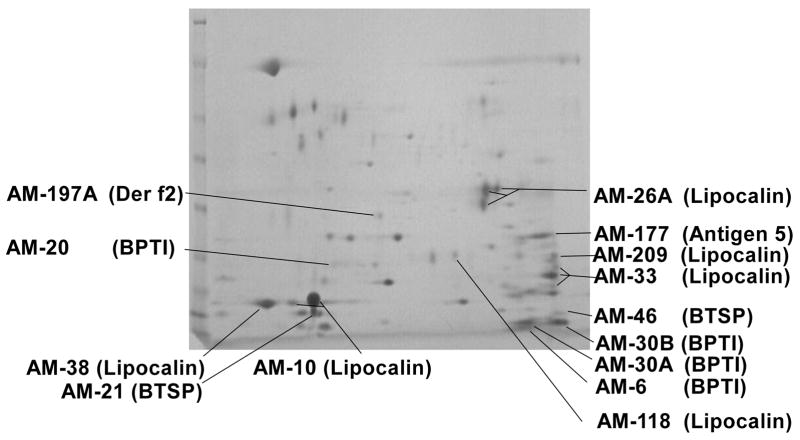
In most cases the Edman sequences correspond to the most abundant sequences found in the cDNA library. Even so, the lipocalin (AM-10) with the most transcripts (150) are not found with Edman sequencing. This is most probably due to the fact that many cellular proteins are N-terminally blocked. To address this issue, the proteome were also investigated by peptide mass fingerprinting. For this, the SGE was fractionated using two-dimensional electrophoresis (Fig. 2). This resulted in the resolution of 78 spots that were picked and analyzed. Of these, 18 spots matched cDNA transcripts with significant values (supplementary material) and 14 proteins were identified. All identified spots were secretory and 7 corresponded with Edman products previously identified (Fig. 2). As such, 7 additional proteins were identified that corresponded to sequences found in the cDNA database. Of these the most prominent spot corresponded with contig AM-10, the contig with the most abundant transcripts in the cDNA library.
Fig. 2.
Two-dimensional electrophoresis of salivary gland extract from A. monolakensis. Approximately 50 μg total SGE was resolved on the gel. All prominent proteins spots (78 spots) were excised for peptide mass fingerprinting. Spots that could be matched to cDNA library transcripts are indicated. Protein families are shown in brackets.
3.4. Proteome analysis by liquid chromatography followed by peptide mass fingerprinting
To corroborate the results obtained from Edman sequencing and 2D-analyses, SGE was fractionated by anion and cation exchange chromatography, followed by reversed phase chromatography (Fig. 3). This results in a two-dimensional separation of proteins based on their charge and hydrophobicity properties. Using in-house programs that allowed for the relative quantification of peaks obtained at 220 nm, relative concentrations of the peaks can also be estimated, thereby providing a measure of the relative yields for different identified proteins (Table 1). Salivary gland extract (100 glands, ~1 mg total protein) was first fractionated using anion exchange chromatography (Fig. 3, AEC). This resulted in the fractionation of 10 regions that were used for subsequent fractionation. The flow-through was subjected to cation exchange chromatography and yielded 8 regions that were used for further analysis (Fig. 3, CEC). The fractions from the ion exchange columns were applied to reversed phase chromatography and single peaks were collected for analysis by peptide mass fingerprinting (Fig. 3, RPC). This resulted in the analysis of 116 peaks, of which 52 could be identified with confidence and yielded 27 proteins. Eleven of these proteins were previously identified by Edman sequencing and 4 proteins were identified by 2D-SDS-PAGE that was not identified by Edman sequencing. Each method identified unique proteins not found with the other methods giving for Edman sequencing 4 unique proteins, for 2D-SDS-PAGE 2 unique proteins and for 2D-LC-MS 14 proteins. This yields a total of 35 proteins that were identified in the proteome of A. monolakensis and of these 29 were identified by liquid chromatography and quantified. Relative yields show that the 29 proteins identified correspond to ~ 60% of the total protein content of the salivary glands (supplementary table 6). The identified peaks cumulatively have 710 transcripts, which correspond to 20% of the secretory clusters and 60% of all secretory transcripts. Relative yields obtained after fractionation of salivary gland extract showed that the proteins identified by peptide mass fingerprinting represent 60% of the total protein extract. As such, the most highly abundant proteins in the proteome are also the most highly abundant transcripts.
Fig. 3.
Two-dimensional liquid chromatography followed by peptide mass fingerprinting. Indicated is fractionation of SGE by anion exchange chromatography (AEC). The flow-through from the AEC was fractionated with cation exchange chromatography (CEC). Fractions (R1-R17) from AEC and CEC were fractionated using reversed phase chromatography (RPC). Peaks were collected and those identified by peptide mass fingerprinting are indicated.
3.5. Secretory components for the sialome of A. monolakensis
Of the 127 secretory proteins deposited and analyzed, 81 are full-length while 46 are N-terminally truncated (Table 1). The truncated proteins are useful, as they can still be assigned to protein families. This allows for the description of the sialome of A. monolakensis in terms of protein domain composition. Proteins were assigned to families using the BLAST suite of programs (Altschul et al., 1990; Altschul et al., 1997). In some cases, no homologies were found using BLASTP or PSI-BLAST analysis. However, inspection of conserved cysteine-patterns allowed the assignment of small cysteine-rich proteins to various families (Table 1). Approximately forty different protein families or domains can be identified at first analyses, of which thirteen have more than one member (Table 1). The most abundant protein family is the lipocalins (27% of all domains, 34% of all secretory transcripts, 40.5% of total protein). This correlates with previous proteomic analysis that showed that lipocalins are the most abundant proteins found in the salivary glands of the soft tick O. savignyi (Mans et al., 2001; Mans et al., 2003; Oleaga et al., 2007). Other abundant domains include the basic tail secretory protein (BTSP) family (28% of all domains, 21% of all secretory transcripts, 6.4% of total protein), kunitz-BPTI family (12% of all domains and 15% of all secretory transcripts, 6.7% of total protein) and metalloproteases (21% of all domains and 6% of all secretory transcripts). The lipocalin, kunitz and metalloprotease families have been previously identified as domains found in soft and hard ticks (Mans and Neitz, 2004a).
3.6. Functional proteins from the sialome of A. monolakensis
Functions for possible tick salivary gland proteins and families have been proposed in a variety of sialome papers and will not be stated here (Valenzuela et al., 2002b; Francischetti et al., 2005; Ribeiro et al., 2006). Biochemical analysis has indicated that A. monolakensis possess anti-thrombin (monobin) and anti-platelet (monogrins and apyrase) activities that are orthologous to that found in Ornithodoros soft ticks (Mans et al., 2007). Monobin and the monogrins belong to the Kunitz/BPTI-family as was previously found for soft tick inhibitors (Joubert et al., 1998; Mans et al., 2002b; van de Locht et al., 1996; Waxman et al., 1990). These are so far, the only confirmed functions for the salivary gland proteins from A. monolakensis. Other functions observed in SGE include fibrinogenase and collagenase activity (results not shown) that are most probably associated with metalloproteases (Francischetti et al., 2003). Other functions readily transferable via homology include anti-microbial activities of defensins and microplusin-like proteins previously found in ticks (Fogaça et al., 2004; Lai et al., 2004; Todd et al., 2007; Zhou et al., 2007).
3.7. Comparative analysis of sialome protein families in different tick species
The salivary transcriptomes from various hard tick species have been described. These include Amblyomma variegatum (Nene et al., 2002), Dermacentor andersoni (Alarcon-Chaidez et al., 2007), Haemaphysalis longicornis (Nakajima et al., 2005), I. pacificus (Francischetti et al., 2005), I. scapularis (Valenzuela et al., 2002b; Ribeiro et al., 2006), and Rhipicephalus appendiculatus (Nene et al., 2004). With the data obtained in the present study the question can be asked, as to how many salivary gland derived protein domains are conserved between hard and soft tick species? This will serve as an indication of the protein domain repertoire that was present in the salivary glands of the ancestral tick lineage. In order to address this, secretory proteins identified in this study were used in PSI-BLAST analysis to retrieve hard tick proteins that has also been identified in salivary glands from the non-redundant database. This search mostly identified proteins from Ixodes and Haemaphysalis species, as protein sequence data for these ticks are available in the non-redundant protein database. In cases where EST sequences were deposited instead of protein sequences, clusterized databases were constructed for each species after which TBLASTN analysis was performed to detect homologs. This analysis indicates that the major protein domains are conserved between the soft and hard tick families, indicating that most of the protein folds found in ticks were already present in the salivary glands of the ancestral tick lineage (Table 2; supplementary material). It is of interest, that in some cases where no positive hits were found in hard tick salivary gland databases, homologs were found in the transcriptome data of B. microplus which includes pooled transcripts from a variety of organs. This would indicate that although these specific proteins were not expressed in hard tick salivary glands they were probably present in the genome of the ancestral tick lineage. In these cases, expression might have been lost in hard tick salivary glands or have been co-opted in soft tick salivary glands at a later time. The overall trends observed for soft tick salivary glands are also apparent for the hard tick protein families, in that the major families seem to be the BTSP, kunitz, lipocalin and metalloprotease families.
Table 2.
Secretory protein families in the salivary glands of hard and soft ticks.
| Protein family | Amon | Ipac | Isca | Acaj | Aame | Avar | Dand | Rapp | Bmic |
|---|---|---|---|---|---|---|---|---|---|
| BTSP | 15 | 20 | 66 | 2 | 7 | 1 | 5 | 3 | 3 |
| 7DBF | 8 | - | - | - | - | - | - | - | - |
| BPTI | 8 | 8 | 50 | 3 | 23 | 7 | 17 | 11 | 39 |
| Pro/Gly | 5 | - | 1 | 9 | 2 | 2 | - | 1 | 27 |
| 5.3 kDa | 1 | - | 2 | - | - | - | - | - | - |
| Metalloprotease | 17 | 1 | 12 | 2 | 2 | 6 | 1 | 42 | 23 |
| ADAM | 1 | 1 | 3 | - | - | - | - | - | - |
| Lipocalin | 21 | 8 | 35 | - | 10 | - | 5 | 5 | 17 |
| 8 kDa Cys-rich | 5 | - | - | - | - | - | - | - | - |
| Defensin | 4 | - | 4 | 2 | - | - | 2 | - | 3 |
| Microplusin | 1 | - | 2 | 2 | 1 | - | - | 1 | 8 |
| C3-like complement | 1 | - | - | - | - | - | - | - | 2 |
| Serpin | 1 | - | 2 | 2 | 1 | 2 | - | 8 | 18 |
| Chitinase | 1 | - | - | - | 1 | - | - | 1 | 10 |
| CBD | 2 | - | 1 | - | - | 5 | - | 1 | 7 |
| AG5/SCP | 2 | - | 1 | - | - | - | - | 1 | 9 |
| Phospholipases | 1 | - | - | - | - | - | - | 1 | 5 |
| Procathepsin B | 1 | - | - | - | - | - | - | - | - |
| Aspartic protease | 1 | - | - | - | 1 | - | - | 1 | 10 |
| 5′-Nucleotidase | 1 | - | - | - | - | - | - | 1 | 5 |
| Cimex apyrase | 1 | - | - | - | - | - | - | - | 1 |
| 7CF | 2 | - | - | - | 1 | - | - | - | - |
| 15 kDa | 4 | - | - | - | - | - | - | - | - |
| AM-240 | 1 | - | - | - | - | - | - | - | 1 |
| AM-321 | 1 | - | - | - | - | - | - | 2 | 1 |
Secretory components identified in the present study (Table 1) were used to perform BLASTP analysis against protein databases that were constructed previously for I. pacificus and I. scapularis. EST data that were previously deposited for hard tick species were clusterized as described to yield unique EST contigs. TBLASTN analysis was performed against these new databases. BLAST results were inspected manually and unique positive hits (E<0.004) were tallied and grouped into specific families. Species for which sialome data have been deposited in the databank include I. pacificus (Ipac), I. scapularis (Isca), A. cajanensis (Acaj), A. americanum (Aame), A. variegatum (Avar), D. andersoni (Dand) and R. appendiculatus (Rapp). Data for R. (B.) microplus (Bmic) includes ESTs pooled from all internal organs.
3.8. Detection of homologies between protein families identifies a thrombospondin repeat superfamily
PSI-BLAST analysis of the various protein families allows for the elucidation of relationships between protein families that share the same fold, but are themselves too divergent for detection of homologous relationships (Altschul and Koonin, 1998). Using this approach, we found that the BTSP and the 7DBF family identified in this study are homologous (Table 1). Members of the 18.7 kDa previously identified in Ixodes ticks (Valenzuela et al., 2000b; Francischetti et al., 2005; Ribeiro et al., 2006), were also retrieved. PSI-BLAST analysis of this family also retrieved metalloproteases of the ADAM-TS family and the alignments produced during the BLAST analysis indicated similarity to a region not yet assigned to any domain structure. The cysteine arrangement is, however, similar to that of the thrombospondin type 1 repeat superfamily (Tang, 2001). The BTSPs possess a single TSP-1 repeat domain with a cysteine bond pattern similar to that found for TSP1 and TSP4 of F-spondin (Fig. 4A) (Pääkönen et al., 2006). The 7DBF family possesses two TSP-1 repeat domains, the first with a cysteine bond pattern that corresponds to that of TSP-1 repeats of thrombospondin (Tan et al., 2002), while the cysteine bond pattern of the second domain corresponds to that of F-spondin (Fig. 4B). Members also possess two extra cysteines at the N-terminus that presumably forms a disulphide bond. The 18.7 kDa family previously described for Ixodes species is composed of two TSP-1 repeats, both resembling the disulphide bond pattern of the F-spondin domain (Fig. 4C). Some members also possess two extra cysteines that are located between the two thrombospondin-like domains and presumably form a disulphide bond that will result in the formation of a hairpin-turn. This will bring the two thrombospondin domains in proximity, which could indicate that in these proteins inter-domain interaction occurs.
Fig. 4.

The BTSP, 7DBF and 18.7 kDa families belong to the thrombospondin repeat superfamily. A) Single domain BTSP members were aligned with TSP1 and TSP4 domains of f-spondin as well as Salp14, a representative BTSP from the genus Ixodes. B) Alignment of the 7DBF family with human TSP2 (Tan et al., 2002) at its N-terminal domain and TSP4 from f-spondin at its C-terminal domain (Pääkönen et al., 2006). C) Alignment of 18.7 kDa with TSP1 and TSP4 domains of f-spondin (Pääkönen et al., 2006). TSP1 was aligned with the N-terminal domain and TSP4 with the C-terminal domain. Indicated are the conserved cysteine patterns and predicted disulphide bonds as obtained from the structures of human TSP2 (Tan et al., 2002) and TSP1 and TSP4 from f-spondin (Pääkönen et al., 2006). Accession numbers for sequences are included in sequence name.
3.9. The 8kDa cysteine-rich family is shared between hard and soft ticks
In some cases no positive hits were found between soft and hard ticks, for example, the 8kDa cysteine-rich family. However, small proteins might be divergent to an extent that even PSI-BLAST analysis might not detect homologies. In these cases, conserved features such as cysteine and disulphide bond patterns can be used to show that proteins possess the same fold. This study found at least 5 proteins that all share the same cysteine bond pattern but do not retrieve other proteins, i.e. the 8kDa cysteine-rich family. When compared to a group previously identified in Ixodes ticks (the ixodegrins), they clearly share the same cysteine pattern (Fig. 5). Some ixodegrins possess the RGD integrin recognition motif that correlates with that found for the snake derived disintegrin, dendroaspin and the hard tick platelet aggregation inhibitor variabilin (Francischetti et al., 2005). The Argas proteins, however, lack the RGD-motif. Proteins with the RGD-motif have been identified in the salivary glands of A. monolakensis and are orthologous to platelet aggregation inhibitors from the BPTI-like family found in the soft tick genus Ornithodoros (Mans et al., 2007).
Fig. 5.
Alignment of the 8kDa family from A. monolakensis and the ixodegrins from Ixodes. Indicated are the conserved cysteine pattern and RGD-motif. Accession numbers for sequences are included in sequence name.
3.10. The relative absence of orthologs between soft and hard ticks and lineage specific expansion of protein families
As has been indicated previously, most anti-hemostatic and immunomodulatory activities are not conserved between soft and hard ticks (Mans and Neitz, 2004a). Given the fact that soft and hard ticks did share a common ancestor at a given time to exclusion of other mites (Barker and Murrell, 2004), it stands to reason that they should share to some extent a number of orthologous proteins that was present before divergence of the major tick families. This is supported by the number of shared protein families found in soft and hard ticks (Table 2), but raises the question regarding the presence of orthologs shared between the two tick families. Phylogenetic analysis of the major protein families (lipocalins and BTSP) found in soft and hard ticks indicates that most of the gene duplications found in the protein families are lineage specific expansions (Figs. 6 and 7). This implies that most gene duplication events occurred after divergence of hard and soft ticks and that most orthologous relationships shared between the two families were probably restricted to a limited number of members in each protein family. The numbers on these might be as little as one or two orthologs present in the ancestral tick lineage for each family. This indicates that although the ancestral tick lineage possessed a protein repertoire that gave rise to the diversity observed in salivary transcriptomes of present day ticks, it was itself most probably a minimal sialome that had very few of the functional proteins found in modern day ticks. The high level of gene duplication (paralogous genes) observed within the major protein families, could also obscure the true orthologous relationships between hard and soft tick proteins. Functional analysis of protein families should therefore remedy this and allow for detection of orthologs that were present in the ancestral tick lineage.
Fig. 6.
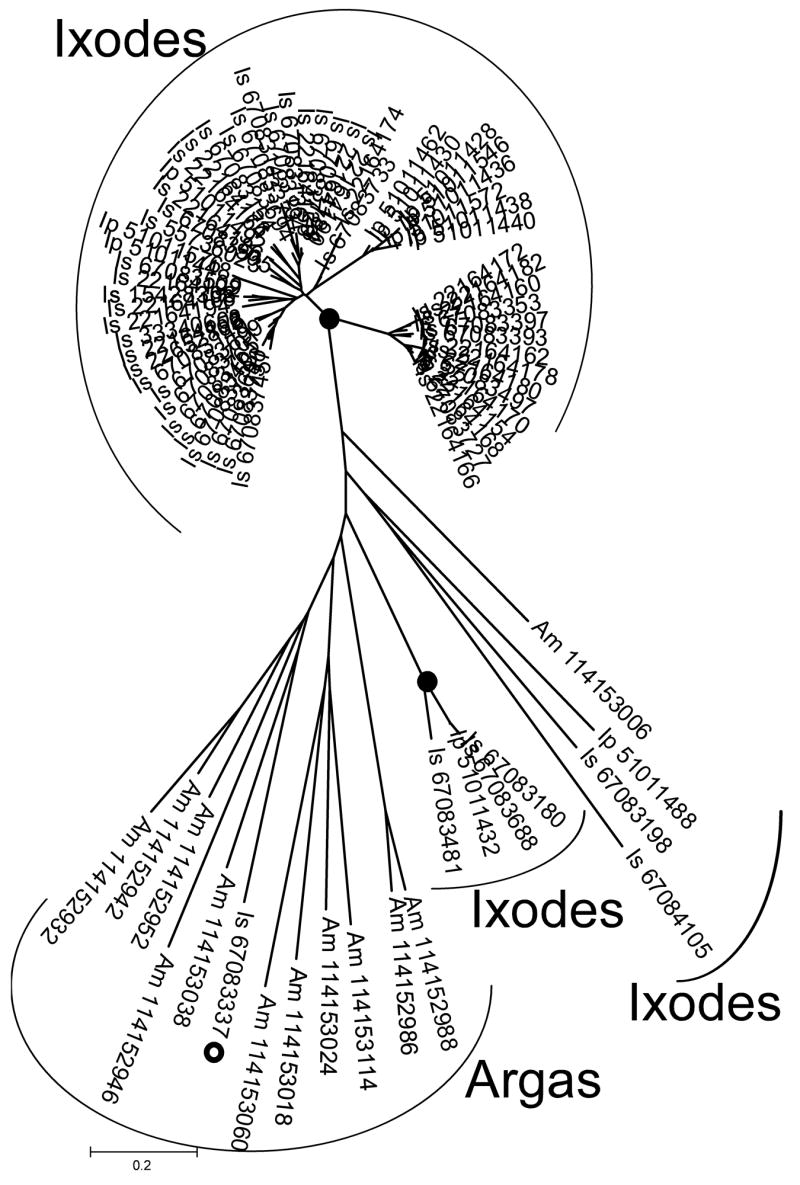
Phylogenetic analysis of the BTSP family. A neighbor-joining analysis shows nodes with more than 90% bootstrap support (10000 replicates) labeled with black dots. The gray dot indicates a single Ixodes sequence that grouped within the Argas clade with weak support.
Fig. 7.
Phylogenetic analysis of the tick lipocalin family. A neighbor-joining analysis shows nodes with more than 60% bootstrap support (10000 replicates) labeled with black dots.
4. Discussion
The field of vector-host interaction has gained tremendously by high-throughput analysis of salivary gland transcripts and proteomes, collectively called the sialome (Ribeiro and Francischetti, 2003). This approach allows for the description of secretory products involved in blood-feeding of hematophagous organisms. Heretofore, the identification of proteins active at the blood-feeding site was only accessible using biochemical purification techniques to isolate bio-active components from salivary glands or saliva. The present advances in high-throughput methodologies allows us to gain a glimpse of the sialome in its full complexity. Thus, while biochemistry will always be needed to confirm and validate functional predictions derived from sialomic databases, the analysis of sialomes in terms of their protein domain compositions allows for a comparative analysis of sialome complexity and diversity that gives us insights into the evolution of blood-feeding behavior in arthropods. In the present study we analyzed the sialome of a soft tick species (A. monolakensis) and compare it with hard tick sialomes previously described. We derive the general conclusion that hard and soft ticks shared a similar salivary gland protein repertoire in their last common ancestor.
Approximately 130 potential secretory transcripts were identified in the salivary gland transcriptome of A. monolakensis by cDNA library construction. This compares well with the proteomic analysis using 2D-electrophoresis and liquid chromatography that resolved 78 and 118 abundant proteins, respectively. It also indicates that this number is probably close to the real number of secretory components found in the salivary glands of this soft tick. In comparison, more than 500 proteins have been described for the salivary glands of I. scapularis (Ribeiro et al., 2006). In this latter study, the data were obtained from the analysis of ~8000 ESTs, while the current study only analyzed ~3000 ESTs. However, these estimates likely correlate with salivary gland complexity, with hard tick salivary glands being more complex than that of soft ticks, as evidenced by that higher number of secretory acini and cell types found in hard ticks (Coons and Alberti, 1999). Presumably, this is because hard ticks would need more components to control the host’s defense mechanisms and because they feed for longer periods of time and is exposed to the host’s immune system for extended periods of time.
The transcript numbers for highly abundant contigs also correlate well with the highly abundant proteins identified during the proteomic analysis. This correlation is consistent with the emerging paradigm in vector salivary gland biology, that proteins secreted during feeding are generally the most abundant salivary gland proteins with the correlated highest numbers of mRNA salivary gland transcripts. Even so, it is of interest that soft tick glands show such a good correlation between transcript and protein abundance. Soft tick salivary glands are normally filled with large secretory granules where proteins are stored until secretion during feeding (Roshdy, 1972, Roshdy and Coons, 1975, Coons and Roshdy, 1981, El Shoura, 1985; El Shoura 1987, Mans et al., 2004). Ticks, such as A. monolakensis, may only feed once a year and in the periods in between, granules will be stationary. Salivary glands can presumably only accommodate a certain number of granules and as such, proteins can be stored over prolonged periods of time and will accumulate, so that a general correlation between protein concentration and transcript number would not necessarily follow. Comparison of the cDNA libraries obtained for fed and unfed ticks do not differ markedly in terms of transcript numbers for various contigs (supplementary material). This would indicate that transcription occurs at the same rate for secretory transcripts regardless of the feeding status of the tick. Thus, if there is regulation of protein levels or salivary granule number, it would occur at a post-transcriptional level.
Analysis of the protein families identified in the Argas sialome indicate that they share to a large extent a similar salivary gland proteome with hard ticks. This would imply that most of the shared protein domains were also present in the ancestral tick lineage to the two families. Even so, few clear-cut orthologs were identified for highly abundant protein folds found in the soft and hard tick families. This observation could be extended to the lesser abundant protein families (results not shown). In the case of very short sequences (BPTI-kunitz and defensin families), sequences may evolve fast so that phylogenetic information is lost (Mans et al., 2002a). Host immune pressure and divergence times extending back to 400 MYA may also account for the high divergence of sequence (Barker and Murrell, 2004). Numerous biochemical studies have also shown that specific functions involved in the regulation of the host’s immune and hemostatic systems are not conserved between hard and soft ticks (Mans and Neitz, 2004a; Mans et al., 2007). The possibility thus exists that once functions are found for many of the divergent proteins in the sialomes; they will differ between the tick families. Those protein families for which orthologous proteins exist, are most probably ones conserved throughout invertebrates and would include the metalloproteases and anti-microbials. In short, these proteins would have had generalized functions before adaptation to a blood-feeding environment.
The predicted presence of certain shared protein folds in the ancestral tick lineage from which salivary gland function evolved raises the question as to where these folds derived from. For most of the major salivary gland protein families, they clearly derived from much more ancient members of the same folds that are present in arthropods. The ancestor to the salivary BPTI proteins probably derived from a hemolymph BPTI-like protein, such as those common in the hemolymph of arthropods (Mans and Neitz, 2004a). These proteins would generally be involved in the regulation of serine proteases involved in various processes and their presence in the salivary glands might have assisted in the inhibition of hemolymph proteases at a stage when the ancestral tick lineage still scavenged dead arthropods. Lipocalins most probably derived from a Lazarillo-like ancestor that was involved in the development of the neural system (Mans and Neitz, 2004b). The metalloproteases are ancient enzymes that are conserved throughout the animal kingdom and are involved in all processes of extra-cellular matrix remodeling, a role it most probably also plays in arachnids. In the ancestral tick lineage these proteases must have played a role in digestion and liquefaction of a scavenged meal. It would seem a simple jump to re-adapt and retain them for blood-feeding use, especially when previous targets were collagen or fibrinogen-like. The presence of other “house-keeping, but adaptive” domains are also noted, for example the anti-microbials that would have a more ancient protective role, but would be co-opted for blood-feeding. Certain folds seem to be novel to tick salivary glands, most notably the BTSP-fold because of the high number of members found in both hard and soft tick salivary glands. The thrombospondin-repeat occurs in a number of mammalian proteins (Tucker, 2004). Thus far the only proteins outside of the BTSP family with TSP repeat that has been found in tick salivary glands are the ADAM-TS metalloproteases. This suggests that the BTSP fold and all its derivatives, i.e. 7DBF, 18kDa, 2CF, were most probably derived from a gene duplication of this domain from an existing metalloprotease. Whether this occurred more than once is difficult to ascertain, but would not be impossible, as the disulphide patterns from the 18.7kDa and 7DBF families are different and probably derived from existing TSP-1 families. Those domains which are currently orphan domains, i.e. proteins that cannot be assigned to any known protein family or domain, are most probably highly divergent members of known domains. It is foreseen that as more data become available, most of these orphan domains will eventually be assigned to well known families and that their origins will be easier to trace.
Certain recurring trends appear for many proteins found in tick salivary glands. They either belong to large families of which most genes seem to be lineage specific expansions, or their origins can be traced back to proteins that were already present in the salivary glands of the ancestral tick lineage. From this viewpoint, we propose that the ancestral tick lineage had a restricted set of salivary gland derived protein families which was not necessarily adapted to function within a blood-feeding environment. Subsequently the main tick families diverged and adapted to a blood-feeding environment. During this period, novel proteins involved in the modulation of the host’s hemostatic and immune systems evolved by gene duplication of full-length domains, as well as sub-domains. Proteins with functions that could affect the host’s defenses were also recruited during this period.
5. Conclusions
The present study shows that the protein domain repertoires of hard and soft ticks are similar. This suggests that most protein domains were already expressed in the salivary glands of the ancestral tick. Even so, most gene duplication events occurred after divergence of the major tick families. This suggests an adaptive response of the different families, most probably in response to a blood-feeding environment, after their divergence.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the government of the United States of America. We thank Paul Kelly and Kristie Nelson for logistic support in collecting the ticks on islands in Mono Lake.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcon-Chaidez FJ, Sun J, Wikel SK. Transcriptome analysis of the salivary glands of Dermacentor andersoni Stiles (Acari: Ixodidae) Insect Biochem Mol Biol. 2007;37:48–71. doi: 10.1016/j.ibmb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST - a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- Barker SC, Murrell A. Systematics and evolution of ticks with a list of valid genus and species names. Parasitology Suppl. 2004;129:15–36. doi: 10.1017/s0031182004005207. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Black WC, Piesman J. Phylogeny of hard- and soft-tick taxa (Acari:Ixodida) based on mitochondrial16S rDNA sequences. Proc Natl Acad Sci. 1994;91:10034–10038. doi: 10.1073/pnas.91.21.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne DE. Antihemostatic strategies of blood-feeding arthropods. Curr Drug Targets Cardiovasc Haematol Disord. 2004;4:375–396. doi: 10.2174/1568006043335862. [DOI] [PubMed] [Google Scholar]
- Coons LB, Roshdy MA. Ultrastructure of granule secretion in salivary glands of Argas (Persicargas) arboreus during feeding. Z Parasitenkd. 1981;65:225–234. [Google Scholar]
- Coons LB, Alberti G. The Acari-Ticks. In: Harrison FW, Foelix R, editors. Microscopic Anatomy of Invertebrates, Chelicerate Arthropoda. 8B. Wiley-Liss; New York: 1999. pp. 267–514. [Google Scholar]
- El-Shoura SM. Ultrastructure of salivary glands of Ornithodoros (Ornithodoros) moubata (Ixodoidea: Argasidae) J Morph. 1985;186:45–52. doi: 10.1002/jmor.1051860105. [DOI] [PubMed] [Google Scholar]
- El Shoura SM. Ultrastructural changes in the salivary alveoli of Argas (Persicargas) persicus (Ixodoidea: Argasidae) during and after feeding. Exp Appl Acarol. 1987;3:347–360. doi: 10.1007/BF01193170. [DOI] [PubMed] [Google Scholar]
- Fogaça AC, Lorenzini DM, Kaku LM, Esteves E, Bulet P, Daffre S. Cysteine-rich antimicrobial peptides of the cattle tick Boophilus microplus: isolation, structural characterization and tissue expression profile. Dev Comp Immunol. 2004;8:191–200. doi: 10.1016/j.dci.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Andersen JF, Mather TN, Ribeiro JM. Ixolaris, a novel recombinant tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick, Ixodes scapularis: identification of factor X and factor Xa as scaffolds for the inhibition of factor VIIa/tissue factor complex. Blood. 2002;99:3602–3612. doi: 10.1182/blood-2001-12-0237. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Mather TN, Ribeiro JM. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochem Biophys Res Commun. 2003;305:869–875. doi: 10.1016/s0006-291x(03)00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Mather TN, Ribeiro JM. Penthalaris, a novel recombinant five-Kunitz tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick vector of Lyme disease, Ixodes scapularis. Thromb Haemost. 2004;91:886–898. doi: 10.1160/TH03-11-0715. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, My Pham V, Mans BJ, Andersen JF, Mather TN, Lane RS, Ribeiro JM. The transcriptome of the salivary glands of the female western black-legged tick Ixodes pacificus (Acari: Ixodidae) Insect Biochem Mol Biol. 2005;35:1142–1161. doi: 10.1016/j.ibmb.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Joubert AM, Louw AI, Joubert F, Neitz AW. Cloning, nucleotide sequence and expression of the gene encoding factor Xa inhibitor from the salivary glands of the tick, Ornithodoros savignyi. Exp Appl Acarol. 1998;22:603–619. doi: 10.1023/a:1006198713791. [DOI] [PubMed] [Google Scholar]
- Karczewski J, Endris R, Connolly TM. Disagregin is a fibrinogen receptor antagonist lacking the Arg-Gly-Asp sequence from the tick, Ornithodoros moubata. J Biol Chem. 1994;269:6702–6708. [PubMed] [Google Scholar]
- Keller PM, Waxman L, Arnold BA, Schultz LD, Condra C, Connolly TM. Cloning of the cDNA and expression of moubatin, an inhibitor of platelet aggregation. J Biol Chem. 1993;268:5450–5456. [PubMed] [Google Scholar]
- Klompen JSH. Comparative morphology of argasid larvae (Acari:Ixodida:Argasidae), with notes on phylogenetic relationships. Ann Entomol Soc Am. 1992;85:541–560. [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comp Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- Lai R, Takeuchi H, Lomas LO, Jonczy J, Rigden DJ, Rees HH, Turner PC. A new type of antimicrobial protein with multiple histidines from the hard tick, Amblyomma hebraeum. FASEB J. 2004;18:1447–1449. doi: 10.1096/fj.03-1154fje. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Jr, Kato I. Protein inhibitors of proteinases. Annu Rev Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Venter JD, Vrey PJ, Louw AI, Neitz AW. Identification of putative proteins involved in granule biogenesis of tick salivary glands. Electrophoresis. 2001;22:1739–1746. doi: 10.1002/1522-2683(200105)22:9<1739::AID-ELPS1739>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Louw AI, Neitz AW. Evolution of hematophagy in ticks: common origins for blood coagulation and platelet aggregation inhibitors from soft ticks of the genus Ornithodoros. Mol Biol Evol. 200a;19:1695–705. doi: 10.1093/oxfordjournals.molbev.a003992. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Louw AI, Neitz AW. Savignygrin, a platelet aggregation inhibitor from the soft tick Ornithodoros savignyi, presents the RGD integrin recognition motif on the Kunitz-BPTI fold. J Biol Chem. 2002b;277:21371–21378. doi: 10.1074/jbc.M112060200. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Steinmann CM, Venter JD, Louw AI, Neitz AW. Pathogenic mechanisms of sand tampan toxicoses induced by the tick, Ornithodoros savignyi. Toxicon. 2002c;40:1007–1016. doi: 10.1016/s0041-0101(02)00098-3. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Louw AI, Neitz AW. The major tick salivary gland proteins and toxins from the soft tick, Ornithodoros savignyi, are part of the tick Lipocalin family: implications for the origins of tick toxicoses. Mol Biol Evol. 2003;20:1158–1167. doi: 10.1093/molbev/msg126. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Venter JD, Coons LB, Louw AI, Neitz AW. A reassessment of argasid tick salivary gland ultrastructure from an immuno-cytochemical perspective. Exp Appl Acarol. 2004;33:119–129. doi: 10.1023/b:appa.0000030012.47964.b3. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Neitz AW. Adaptation of ticks to a blood-feeding environment: evolution from a functional perspective. Insect Biochem Mol Biol. 2004a;34:1–17. doi: 10.1016/j.ibmb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Neitz AW. Exon-intron structure of outlier tick lipocalins indicate a monophyletic origin within the larger lipocalin family. Insect Biochem Mol Biol. 2004b;34:585–594. doi: 10.1016/j.ibmb.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Andersen JF, Schwan TG, Ribeiro JM. Conservation of anti-hemostatic factors in the soft tick family (Ixodida: Argasidae): Implications for the evolution of blood-feeding in ticks. 2007 doi: 10.1016/j.ibmb.2007.09.002. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleaga A, Escudero-Población A, Camafeita E, Pérez-Sánchez R. A proteomic approach to the identification of salivary proteins from the argasid ticks Ornithodoros moubata and Ornithodoros erraticus. Insect Biochem Mol Biol. 2007 doi: 10.1016/j.ibmb.2007.07.003. In press. [DOI] [PubMed] [Google Scholar]
- Nakajima C, da Silva Vaz I, Jr, Imamura S, Konnai S, Ohashi K, Onuma M. Random sequencing of cDNA library derived from partially-fed adult female Haemaphysalis longicornis salivary gland. J Vet Med Sci. 2005;67:1127–1131. doi: 10.1292/jvms.67.1127. [DOI] [PubMed] [Google Scholar]
- Nene V, Lee D, Quackenbush J, Skilton R, Mwaura S, Gardner MJ, Bishop R. AvGI, an index of genes transcribed in the salivary glands of the ixodid tick Amblyomma variegatum. Int J Parasitol. 2002;32:1447–1456. doi: 10.1016/s0020-7519(02)00159-5. [DOI] [PubMed] [Google Scholar]
- Nene V, Lee D, Kang’a S, Skilton R, Shah T, de Villiers E, Mwaura S, Taylor D, Quackenbush J, Bishop R. Genes transcribed in the salivary glands of female Rhipicephalus appendiculatus ticks infected with Theileria parva. Insect Biochem Mol Biol. 2004;34:1117–1128. doi: 10.1016/j.ibmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Nunn MA, Sharma A, Paesen GC, Adamson S, Lissina O, Willis AC, Nuttall PA. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J Immunol. 2005;174:2084–2091. doi: 10.4049/jimmunol.174.4.2084. [DOI] [PubMed] [Google Scholar]
- Pääkönen K, Tossavainen H, Permi P, Rakkolainen H, Rauvala H, Raulo E, Kilpelainen I, Guntert P. Solution structures of the first and fourth TSR domains of F-spondin. Proteins. 2006;64:665–672. doi: 10.1002/prot.21030. [DOI] [PubMed] [Google Scholar]
- Paesen GC, Adams PL, Harlos K, Nuttall PA, Stuart DI. Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure. Mol Cell. 1993;3:661–671. doi: 10.1016/s1097-2765(00)80359-7. [DOI] [PubMed] [Google Scholar]
- Paesen GC, Adams PL, Nuttall PA, Stuart DL. Tick histamine-binding proteins: lipocalins with a second binding cavity. Biochim Biophys Acta. 2000;1482:92–101. doi: 10.1016/s0167-4838(00)00168-0. [DOI] [PubMed] [Google Scholar]
- Radovsky FJ. Adaptive radiation in the parasitic mesostigmata. Acarologia. 1969;11:450–483. [PubMed] [Google Scholar]
- Ribeiro JM. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Alarcon-Chaidez F, Francischetti IM, Mans BJ, Mather TN, Valenzuela JG, Wikel SK. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem Mol Biol. 2006;36:111–129. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Roshdy MA. The subgenus Persicargas (Ixodoidea, Argasidae, Argas). 15 Histology and histochemistry of the salivary glands of A (P.) persicus (Oken) J Med Entomol. 1972;9:143–148. doi: 10.1093/jmedent/9.2.143. [DOI] [PubMed] [Google Scholar]
- Roshdy MA, Coons LB. The subgenus Persicargas (Ixodoidea: Argasidae: Argas). 23 Fine structure of the salivary glands of unfed A (P.) arboreus Kaiser, Hoogstraal, and Kohls. J Parasitol. 1975;61:743–752. [PubMed] [Google Scholar]
- Sangamnatdej S, Paesen GC, Slovak M, Nuttall PA. A high affinity serotonin- and histamine-binding lipocalin from tick saliva. Insect Mol Biol. 2002;11:79–86. doi: 10.1046/j.0962-1075.2001.00311.x. [DOI] [PubMed] [Google Scholar]
- Schwan TG, Corwin MD, Brown SJ. Argas (Argas) monolakensis, new species (Acari: Ixodoidea: Argasidae), a parasite of California gulls on islands in Mono Lake, California: description, biology, and life cycle. J Med Entomol. 1992;29:78–97. doi: 10.1093/jmedent/29.1.78. [DOI] [PubMed] [Google Scholar]
- Schwan TG, Winkler DW. Ticks parasitizing humans and California gulls at Mono Lake, California. In: Griffiths DA, Bowman CE, editors. Acarology VI. Horwood; Chichester: 1984. pp. 1193–1199. [Google Scholar]
- Schwan TG, Oprandy JJ, Main AJ. Mono Lake virus infecting Argas ticks (Acari: Argasidae) associated with California gulls breeding on islands in Mono Lake, California. J Med Entomol. 1988;25:381–387. doi: 10.1093/jmedent/25.5.381. [DOI] [PubMed] [Google Scholar]
- Tan K, Duquette M, Liu JH, Dong Y, Zhang R, Joachimiak A, Lawler J, Wang JH. Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. J Cell Biol. 2002;159:373–382. doi: 10.1083/jcb.200206062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL. ADAMTS: a novel family of extracellular matrix proteases. Int J Biochem Cell Biol. 2001;33:33–44. doi: 10.1016/s1357-2725(00)00061-3. [DOI] [PubMed] [Google Scholar]
- Todd SM, Sonenshine DE, Hynes WL. Tissue and life-stage distribution of a defensin gene in the Lone Star tick, Amblyomma americanum. Med Vet Entomol. 2007;21:141–147. doi: 10.1111/j.1365-2915.2007.00682.x. [DOI] [PubMed] [Google Scholar]
- Tucker RP. The thrombospondin type 1 repeat superfamily. Int J Biochem Cell Biol. 2004;36:969–974. doi: 10.1016/j.biocel.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG. High-throughput approaches to study salivary proteins and genes from vectors of disease. Insect Biochem Mol Biol. 2002a;32:1199–1209. doi: 10.1016/s0965-1748(02)00083-8. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Mather TN, Ribeiro JM. Exploring the sialome of the tick Ixodes scapularis. J Exp Biol. 2002b;205:2843–2864. doi: 10.1242/jeb.205.18.2843. [DOI] [PubMed] [Google Scholar]
- van de Locht A, Stubbs MT, Bode W, Friedrich T, Bollschweiler C, Hoffken W, Huber R. The ornithodorin-thrombin crystal structure, a key to the TAP enigma? EMBO J. 1996;15:6011–6017. [PMC free article] [PubMed] [Google Scholar]
- Waxman L, Smith DE, Arcuri KE, Vlasuk GP. Tick anticoagulant peptide (TAP) is a novel inhibitor of blood coagulation factor Xa. Science. 1990;248:593–596. doi: 10.1126/science.2333510. [DOI] [PubMed] [Google Scholar]
- Waxman L, Connolly TM. Isolation of an inhibitor selective for collagen-stimulated platelet aggregation from the soft tick Ornithodoros moubata. J Biol Chem. 1993;268:5445–5449. [PubMed] [Google Scholar]
- Zhou J, Liao M, Ueda M, Gong H, Xuan X, Fujisaki K. Sequence characterization and expression patterns of two defensin-like antimicrobial peptides from the tick Haemaphysalis longicornis. Peptides. 2007;28:1304–1310. doi: 10.1016/j.peptides.2007.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.