Abstract
Pancreatic ductal adenocarcinoma (ie, pancreatic cancer) is among the most devastating of human malignancies. It is commonly diagnosed at advanced, already metastatic, and, hence, incurable stages. Despite extensive research efforts in recent decades, pancreatic cancer remains resistant to almost all clinically available therapy regimens. Recent advances in our understanding of the underlying pathophysiology and molecular biology have opened up avenues for the development of novel diagnostic and therapeutic strategies, some of which have shown highly promising preclinical results and are currently being translated into clinical application. Here in we present a review of recent literature on the molecular genetics of pancreatic cancer and emphasize clinical implications for the development of novel diagnostic and therapeutic approaches.
Pancreatic ductal adenocarcinoma (PDAC), which constitutes over 90% of pancreatic cancers in humans, is a devastating and virtually unexceptionally lethal malignancy, afflicting an estimated 213,000 individuals worldwide every year. Accounting for more than 33,000 fatalities annually in the United States, it represents the fourth most common cause of cancer-related deaths.1 Despite considerable recent research efforts aimed at better understanding the etiology and underlying pathophysiology and at the development of novel diagnostic and therapeutic strategies, this gain in understanding has not yet led to improvement of the overall prognosis of patients suffering from PDAC.2,3 Herein we present a review of the literature on the molecular genetics of PDAC, with an emphasis on recent data implicating promising novel diagnostic and therapeutic approaches for preclinical evaluation and, possibly, subsequent clinical application for the benefit of patients suffering from this dire malady. In the following text the terms pancreatic cancer and PDAC are used synonymously.
Molecular Abnormalities of Pancreatic Cancer
Genomic (DNA) Alterations
Genomic DNA abnormalities frequently found in pancreatic cancers comprise chromosomal aberrations, copy number changes, activating mutations of oncogenes, as well as specific silencing mutations of tumor suppressor and caretaker genes, epigenetic silencing, telomeric alterations, and mutations in mitochondrial DNA (mtDNA).
Copy Number Aberrations
Due to chromosomal instability, a common feature of most solid tumors, almost every pancreatic cancer harbors several numerical or structural chromosomal alterations revealed by cytogenetic analysis.4,5 The most common numerical changes observed in pancreatic cancer are losses on chromosomes 6, 12, 13, and 18, as well as gains on chromosomes 7 and 20; chromosomal breaks and rearrangements most frequently occur in regions involving 1p, 1q, 3p, 6q, 7q, 11p, 17p, and 19q.
Another technique that is commonly applied to identify regions of genomic losses at a “higher resolution” by means of polymorphic microsatellite markers is called allelotyping. In a recent study Iacobuzio-Donahue et al analyzed ∼80 pancreatic cancer xenografts by means of 386 microsatellite markers.6 Allelic losses found most commonly involved chromosomal regions 9p, 17p, and 18q, covering the tumor suppressor genes CDKN2A, TP53, and DPC4/SMAD4/MADH4, respectively. Additional losses were frequently found in 3p, 4q, 5q, 6q, 8p, 12q, 14q, 21q, and 22q. Many of these regions have been linked to candidate tumor suppressor genes, eg, the stress-activated protein kinase MKK4 (17p), which has been suggested to be involved in metastatic spread,7 or receptors of the transforming growth factor (TGF)-β signaling pathway TGFBR1 (9q), TGFBR2 (3p),8 and ACVR1B (12q).9 Interestingly, allelotype analysis of microdissected pancreatic intraepithelial neoplasia (PanIN) samples revealed loss of heterozygosity in several of the chromosomal regions also found in pancreatic cancer, including 9p, 17p, and 18q.10,11
Comparative genomic hybridization (CGH) can be used to discover genomic deletions as well as amplifications, providing the potential to uncover both potential tumor suppressor genes and oncogenes. For CGH, samples of non-neoplastic and tumor cell DNA are labeled with different dyes and hybridized against each other. Subsequently, the relative ratio of the two dyes indicates regions of cancer-associated gains or losses. Conventional CGH was originally performed using metaphase spreads, with the major drawbacks of relatively low resolution and frequent difficulties to map precisely the regions of genomic amplifications or losses.12 More recently, several array-based CGH techniques have been developed, using microarrays that are spotted with bacterial artificial chromosomes (BAC arrays),13 cDNAs (cDNA microarrays),14 or stretches of oligonucleotides (representational oligonucleotide microarray analysis),15 providing a significantly higher resolution than conventional CGH (up to 30 kb) and often allowing for precise mapping of deleted or amplified regions and genes included therein. In the setting of pancreatic cancer, array CGH revealed several genomic amplifications, including C-MYC (8q), EGFR (7p), KRAS22 (12p), AKT2 (19q), and AIB1 (20q), as well as deletions, including DPC4/SMAD4/MADH4 (18q), CDKN2A (9p), FHIT (3p), and MKK4 (17p).16,17 Using the example of thymidylate synthase, which has previously been linked to responsiveness to 5-fluorouracil treatment, a recent report by Brody et al suggests that results from studies examining copy number aberrations should be interpreted with particular caution, since copy numbers determined in tumor cells are not necessarily identical throughout the whole tumor and might vary over time, in response to chemotherapeutic agents or in metastatic foci as compared to primary tumors.18
Nuclear DNA Mutations: Oncogenes
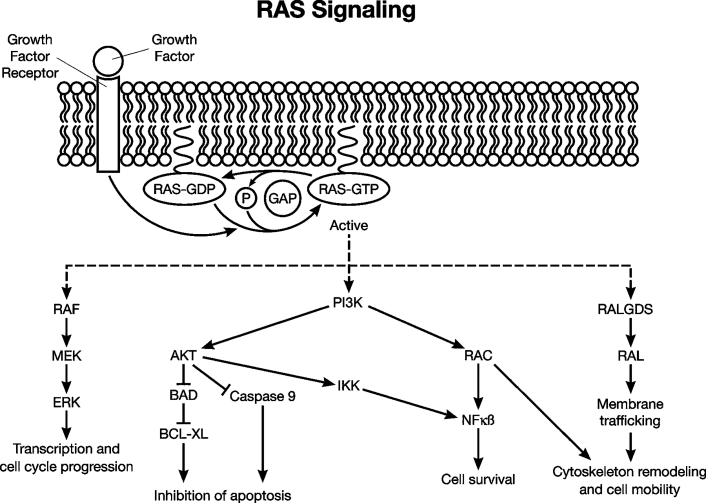
Activating point mutations within the KRAS oncogene (12p) are present in 80 to 90% of pancreatic cancers, most commonly affecting codon 12 but also 13 or 61.19 The activating mutations abolish the intrinsic GTPase activity of KRAS, resulting in constitutive activation of intracellular signal transduction (Figure 1). Of note, activating KRAS mutations are not only the most frequently found genetic abnormalities in pancreatic cancer but also seem to be among the earliest changes observed in nonmalignant precursor-lesions, already being present in about 30% of PanIN-1 lesions.21,22 Rare pancreatic cancers with wild-type KRAS usually harbor mutations of BRAF.23 Since both KRAS and BRAF function in activating the same Ras/Raf/MAP kinase signaling pathway, it explains why mutations of these two genes occur in a virtually exclusive pattern and further underscores the extraordinary importance of this signaling pathway in the genesis of pancreatic cancer. Other oncogenes involved in pancreatic cancer include CMYC, AKT2, and EGFR. CMYC amplifications and concomitant overexpression of CMYC can be detected in 50 to 60% of pancreatic cancers,17,24 providing a potential target for the development of future therapeutic options due to the recent development of compounds specifically inhibiting Myc signaling.25
Figure 1.
The RAS signaling pathway. RAS proteins are transiently activated by a wide range of extraneous signals, such as binding of growth factor ligands to the cognate growth factor receptor. In its active state, RAS is bound to GTP, and inactivation occurs through guanosine triphosphate-activating proteins (GAPs), which promote GTP hydrolysis. RAS proteins engage a number of downstream effector pathways, including RAF-mitogen activated protein kinase (RAF-MAPK), phosphoinsositide-3-kinase (PI3K), and RalGDS pathways. The proximal proteins in these signaling cascades, in turn, activate downstream intermediaries (eg, MEK/ERK, AKT, RAL) that mediate diverse cellular functions such as proliferation, cell survival, and cell motility. Oncogenic mutations of the KRAS2 gene, which compromises the intrinsic GTPase activity of the encoded protein, render the pathway constitutively active in pancreatic cancer. (Reproduced with permission from the International Society of Gastrointestinal Oncology).20
Although amplifications of AKT2 are found in less than 5% of cases,26 the Akt signaling pathway is activated in 30 to 40% of pancreatic cancers,27 suggesting additional underlying mechanisms of pathway activation, eg, loss of PTEN. Specific inhibitors of Akt signaling have been described,28 which might give rise to novel therapeutic strategies in the near future. Constitutive activation of the EGFR pathway through amplification of the EGFR gene or other mechanisms has been described in pancreatic cancer.16 Recent approaches were aimed to target this signaling pathway using monoclonal antibodies or small-molecule tyrosine kinase inhibitors.29
Tumor Suppressor Genes
The cyclin-dependent kinase CDKN2A/p16 on chromosome 9p inhibits cell cycle progression through the G1-S checkpoint and constitutes the most frequently inactivated gene in pancreatic cancers.19,30 More than 90% of pancreatic cancers show loss of CDKN2A/p16 function, which can occur due to homozygous deletions (∼40%), mutations with loss of the second allele (∼40%), or epigenetic silencing by promoter hypermethylation (10–15%). Interestingly, loss of nuclear p16 protein expression is already observed in 30% of PanIN-1, in 55% of PanIN-2, and in 71% of PanIN-3 lesions.31 Approximately 30% of homozygous CDKN2A/p16 mutations also include the MTAP gene, which has recently been proposed as potential therapeutic target.32
Deleted in pancreatic carcinoma 4 (DPC4/SMAD4/MADH4) on chromosome 18q21 is inactivated in about 55% of pancreatic cancers.33 This is due to mutations in one allele and loss of the second allele in about 25% of cases and due to homozygous mutations in 30% of cases. Of note, DPC4/SMAD4/MADH4 mutations are very rarely observed in other malignancies.34 Loss of DPC4/SMAD4/MADH4 function results in reduced growth inhibition and increased proliferation through interference with intracellular signaling cascades downstream of cell surface receptors of the TGF-β family (Figure 2). Abrogation of DPC4 function appears to be a rather late event in the pathogenesis of pancreatic carcinomas, as it is not observed in the majority of PanIN-3 lesions.35
Figure 2.
The TGF-β signaling pathway. The TGF-β signaling pathway is activated by binding of the ligand TGF-β to TGF-β type II receptor at the cell surface, which facilitates the recruitment and phosphorylation of type I TGF-β receptor. The latter activates the SMAD transcription factors SMAD 2 or 3 by phosphorylation, which in turn releases them from the cytoplasmic anchoring protein SMAD anchor for receptor activation (SARA). The phosphorylated SMAD 2/3 proteins complex with SMAD4 and translocate to the nucleus where they modulate the expression of multiple SMAD target genes. Note that TGF-β signaling can also occur via SMAD-independent mechanisms (not shown). (Reproduced with permission from the International Society of Gastrointestinal Oncology).20
Third, TP53 on chromosome 17p is inactivated in 50 to 75% of pancreatic cancers, almost exclusively due to intragenic mutations of one allele in combination with loss of the second allele. Wild-type TP53 induces cell cycle arrest and apoptosis in response to DNA damage, and therefore loss of TP53 function allows for accumulation of additional genetic aberrations. Nuclear accumulation of mutated TP53 is only observed in advanced PanIN-3 lesions, and loss of TP53 function is, like DPC4, considered to be a late event in the pathogenic cascade of pancreatic cancer development.35
While loss of these three (CDKN2A, DPC4 and TP53) genes is found in the majority of pancreatic cancers, other tumor suppressor genes are inactivated in smaller subsets (<10%) of PDAC, eg, LKB1/STK11 on chromosome 19p,36 TGF-βR1 on 9q, TGF-βR2 on 3p, RB1 on 13q,30 and MKK4 on 17p.37 Interestingly, MKK4 seems to be inactivated specifically in metastatic lesions, suggesting that wild-type MKK4 might have the ability to inhibit development of metastases through a yet unknown mechanism.
Caretaker Genes
As opposed to alterations of oncogenes and tumor suppressor genes, which both drive the neoplastic process by increasing tumor cell numbers through increased tumor cell growth or inhibition of cell death and cell cycle arrest, a third class of genes is commonly involved in the development of malignant neoplasias, such that when mutated, they act in a fundamentally different way. Such genes are commonly referred to as caretakers or stability genes.38 Caretaker genes minimize genetic alterations during DNA replication so that loss of their function can lead to accumulation of additional mutations in different other genes.39
The DNA damage repair genes hMLH1 and hMSH2 are inactivated in a small subset of familial pancreatic cancers, but rarely in sporadic cases,40 mostly medullary carcinomas of the pancreas.41 These tumors constitute a separate entity that is important to recognize, since on the one hand they tend to carry a more favorable prognosis than ductal adenocarcinomas and, on the other hand, they may indicate hereditary nonpolyposis colorectal cancer syndrome, in which case patients might benefit from genetic counseling. Medullary carcinomas of the pancreas show a typical medullary histology, which is characterized by poor differentiation, pushing borders, and syncytial growth pattern.41,42
Moreover, a subgroup of pancreatic cancers carries mutations in genes of the Fanconi's anemia DNA repair pathway. Mutations affecting the BRCA2 gene have been found in approximately 17% of familial pancreatic cancers,43 and FANCC and FANCG mutations have been described as rare findings in sporadic pancreatic cancers,44 all of which are thought to be part of the Fanconi's anemia DNA repair pathway. (See Kennedy and D'Andrea 45 and Taniguchi and D'Andrea46 for a recent review and for a schematic overview of the Fanconi's anemia pathway.)
Mutations in Mitochondrial DNA
As they harbor the enzymatic machinery of the respiratory cycle, mitochondria generally contain higher concentrations of free radicals, rendering mtDNA more prone to accumulation of genetic alterations. Moreover, mitochondrial DNA damage repair is overall less effective than in the nucleus, and therefore mutations of mtDNA are quite commonly observed in a variety of malignant neoplasias, including pancreatic cancers.47,48 Although mutations in mtDNA most likely do not contribute to PDAC pathogenesis but rather represent a bystander effect, they might nevertheless be an interesting diagnostic target. MtDNA is six- to eightfold more abundant in the cell than nuclear DNA, and thus it might be feasible to use the high frequency of mtDNA mutations for the development of screening tests for early detection of pancreatic cancer. A recent study from our own group has demonstrated in principle, that mtDNA mutations can be detected by means of a high-throughput mtDNA microarray of pancreatic juice aspiration samples from patients with pancreatic cancer.49 It is entirely possible that in the near future less expensive and more sensitive methods will become available, possibly allowing for screening using sample materials that are easier and less invasively to obtain, eg, peripheral blood samples, so that ideally even patients at average or only slightly increased risk could be tested regularly as a tool of secondary prophylaxis.
Telomere Length Abnormalities
Physiologically, telomeres, which consist of repeats of the sequence motif TTAGGG at the ends of each chromosome, confer chromosomal integrity during cell replication and prevent chromosome ends from fusing together.50 Marked telomere shortening can already be found in more than 90% of even the lowest grade PanIN lesions, so that occurrence of telomere abnormalities are among the earliest known events in the cascade of pancreatic cancer development.51 In the setting of cancer progression, telomeres play a role similar to that of caretaker genes in maintaining genomic integrity. It is assumed that loss of telomere function might permit subsequent accumulation of additional genomic changes at the chromosomal level that confer progression toward a fully malignant phenotype.52,53
Epigenetic Abnormalities
In addition to intragenic mutations and allelic loss, silencing of tumor suppressor genes through epigenetic mechanisms is a frequent finding in many cancers.54 Transcriptional abrogation by epigenetic silencing is mediated most commonly by hypermethylation of CpG islands in promoter regions of tumor suppressor genes. In pancreatic cancer, epigenetic silencing often affects genes that function as tumor suppressors or are involved in key homeostatic pathways, including P16/CDKN2A, E-cadherin, retinoic acid-β, osteonectin, suppressor of cytokine signaling-1, and tumor suppressor in lung cancer 1.55,56 More recently, promoter hypermethylation of human Hedgehog interacting protein was found in the majority of examined pancreatic cancer cell lines and primary tumor samples,57 potentially contributing to increased Hedgehog signaling observed in pancreatic cancers. Aberrant DNA methylation occurs already in PanIN-2 and PanIN-3 lesions,58 and thus diagnostic exploitation of this phenomenon, ie, detection of aberrantly methylated DNA in clinical samples as a strategy for early detection of pancreatic cancer, is currently an area of active research efforts. For example a recent study was successful in detecting methylation of proenkephalin sequences in about 60% of pancreatic juice samples obtained from pancreatic cancer patients but not from controls.59
Interestingly, promoter hypermethylation, though most common, is not the only epigenetic modification involved in carcinogenesis. More recently it was found that the reverse means of aberrant gene regulation (hypomethylation) is also exploited by pancreatic cancer cells, ie, some genes, including maspin, S100P, mesothelin, prostate stem cell antigen, and claudin-4, can be overexpressed due to promoter hypomethylation.60
Transcriptomic Changes
Development and better availability of new global gene expression analysis tools such as serial analysis of gene expression and oligonucleotide and cDNA microarrays in recent years have catalyzed an enormous increase in the generation of data characterizing abnormalities at the transcriptional (RNA) level in a variety of human cancers. In particular, several studies were performed to identify genes that are differentially expressed in pancreatic cancers as compared to normal, non-neoplastic pancreatic tissues.61,62,63,64,65,66,67,68
Interestingly, six genes—keratin 19, retinoic acid-induced 3, secretory leukocyte protease inhibitor, stratifin, tetraspan 1, and transglutaminase 2—were found to be overexpressed by all three techniques (serial analysis of gene expression and oligonucleotide and cDNA microarrays). It has yet to be elucidated whether up-regulation of these genes is only a bystander phenomenon or is mechanistically involved in malignant transformation and might pose a potential target for the development of novel therapeutic strategies. Moreover, differential gene expression might be used to establish new diagnostic tools such as tumor imaging or early detection of pancreatic cancer.
The tight junction protein claudin 4 was found to be overexpressed in PanIN lesions and in fully invasive pancreatic cancer tissue by microarray analysis, and overexpression was confirmed by immunohistochemistry.69 Radiolabeled anti-claudin 4 antibodies have recently been tested successfully in a preclinical setting as imaging tools as well as for therapeutic purposes in murine xenograft models of human pancreatic cancer.70,71
Mesothelin was originally identified by serial analysis of gene expression as being overexpressed in pancreatic cancer, and validation by immunohistochemistry revealed it is almost exclusively expressed in neoplastic cells but not in neighboring nonmalignant tissue.72 As a result of this initial finding, both an experimental tumor vaccine against mesothelin and a conjugated immunotoxin directed against mesothelin are currently undergoing initial evaluation in clinical trials.73
Prostate stem cell antigen, originally thought to be restricted to prostatic basal cells and prostate carcinomas, was found by serial analysis of gene expression to be expressed in about 60% of pancreatic cancers, but not in normal, non-neoplastic pancreas tissues.61 Following up on these findings, prostate stem cell antigen has thereafter been successfully tested as a potential target for immunotherapy74 as well as for diagnostic imaging in murine xenograft models of human pancreatic cancer.71
Yes-associated protein, the mammalian homologue of Yorkie, the main effector of the Hippo pathway, has recently found to be overexpressed at the RNA and protein level in pancreatic cancer cells, suggesting a possible role of this pathway in pancreatic carcinogenesis,75 likely through interaction with TGF-β signaling.76 Survivin, a major suppressor of apoptosis, is expressed at the RNA and protein level in low- to high-grade PanIN lesions and fully invasive pancreatic ductal adenocarcinomas in increasing levels, but not in neighboring non-neoplastic tissue, and it is thought to be involved in carcinogenesis as well as drug resistance.77,78 It might therefore be a promising target both as diagnostic marker for early detection as well as for therapeutic intervention.
Moreover, recently aberrant reactivation of the Hedgehog and Notch signaling pathways, and concomitant overexpression of their respective target genes, has been described in the majority of pancreatic cancers.79,80,81,82 Inhibition of Notch-1 lead to growth inhibition and increased apoptosis in pancreatic cancer cell lines in vitro,83,84 and ligand overexpression has been linked to neovascularization in vivo.85 To our knowledge, studies examining in vivo effects of Notch inhibition, eg, using xenograft model systems of pancreatic cancer, are still lacking at the time of this manuscript's preparation.
Hedgehog inhibition with the small molecule smoothened inhibitor cyclopamine has been found to increase cytotoxic effects of paclitaxel treatment and radiation on pancreatic cancer cells in vitro86 and to inhibit growth of pancreatic cancer xenografts and metastases in vivo.79,80,87 A simplified, schematic overview of the Hedgehog signaling pathway in mammals is given in Figure 3.
Figure 3.

The Hedgehog signaling pathway. The Hedgehog (Hh) signaling pathway involves a series of inhibitory interactions between secreted ligands, cell surface receptors, and cytoplasmic proteins, culminating in the activation of transcription and up-regulation of Hh target genes by the DNA-binding transcription factor GLI. The Hh signaling pathway is activated by binding of Hh ligands—Sonic, Desert, or Indian hedgehog (SHH, DHH, IHH)—to the 12-transmembrane receptor Patched (PTCH). This interaction releases the inhibitory effects of PTC on the 7-transmembrane smoothened (SMO) receptor, and the conformational change with activation that SMO undergoes, in turn, facilitates its inhibition of the cytoplasmic protein Suppressor of Fused (SUFU). SUFU inhibition allows GLI to escape cytoplasmic sequestration, on which it undergoes nuclear translocation and activates the transcription of Hh target genes.
It has been suggested that reactivation of embryonic signaling pathways might be involved in maintaining a subset of cancer cells with stem cell-like properties, ie, putative cancer stem cells. A recent study has described a subpopulation of CD24+, CD44+, and ESA+ cells with cancer stem cell-like properties, including self-renewal and strikingly enhanced tumorigenic potential in soft agar as well as in athymic mice. Interestingly, this subpopulation was also characterized by marked overexpression of Sonic Hedgehog ligand (SHH) by more than 50-fold as compared to normal tissue, whereas overexpression in “bulk” tumor cells was found to be only about fivefold.88 Another study by our own group identified a subpopulation of aldehyde dehydrogenase-“bright” cells with enhanced tumorigenic potential that showed increased expression of the Hedgehog target gene Gli1 and were diminished by Hedgehog inhibition with cyclopamine.87 Presently, development of new, clinically applicable Hedgehog inhibitors is being pursued by several pharmaceutical companies and will hopefully soon be available for trials in a clinical setting.
Proteomic Abnormalities
The word proteome denotes the ensemble of proteins synthesized by a cell or, mutatis mutandis, a certain type of cells, eg, pancreatic cancer cells, making up the bulk neoplastic tissue. Studying gene expression differences at the protein level can in a way be considered as the most straightforward approach, as it bypasses the need to identify possibly different underlying mechanisms, eg, occurrence of intragenic mutations, hetero- or homozygous deletions, or epigenetic alterations, as discussed above.
The past decade has seen great progress in the development of high-throughput techniques addressing protein expression changes, including chip-based arrays that allow determination of a plethora of proteins within a given liquid sample in parallel, or tissue microarrays, enabling examination of expression of selected proteins in a great number of tissue specimens with antibody-based protocols like immunohistochemistry or immunofluorescence in one assay.
It has become evident from proteomic studies that during the multi-stepwise development of pancreatic cancer, changes in the protein expression pattern do not occur in a random manner but can be grouped into early, intermediate, and late changes, which mirror the stepwise accumulation of genomic alterations as discussed previously.35 (See Feldmann et al89 and Singh and Maitra90 for more detailed reviews of molecular abnormalities observed in precursor lesions of PDAC.) This observation might have direct clinical implications, eg, in the quest to identify novel tumor markers for early detection. While a protein like prostate stem cell antigen, which is already secreted by the earliest PanIN-1 lesions, might be an extremely sensitive marker for early detection, it might nevertheless be of limited clinical value due to the common occurrence of low-grade PanINs in pancreata of older individuals. Detection of mesothelin, which is expressed only in late PanIN-3 lesions and fully invasive cancer tissues, from pancreatic juice could on the other hand have much more severe prognostic implications.
Examination of protein expression patterns from pancreatic juice samples using surface-enhanced laser desorption and ionization mass spectrometry-based protein chips has led to the discovery of hepatocarcinoma-intestine-pancreas/ pancreatitis-associated-protein-1 (HIP/PAP-1) overexpression in pancreatic cancers. It has been shown that individuals with high HIP/PAP-1 concentrations >20 μg/ml had a more than 20-fold increased risk of developing pancreatic cancer.91 Surface-enhanced laser desorption and ionization mass spectrometry has recently successfully been tested to predict pancreatic cancer from patient serum samples with 78% sensitivity and a specificity of 97%.92
Another experimental approach to determine altered protein expression from liquid samples exploits liquid chromatography tandem mass spectrometry, which enables identification of proteins based on their individual charge/mass ratios. This technique has recently been used to identify 170 genes expressed in pancreatic juice of patients with pancreatic cancer, including several previously known tumor markers like CEA, MUC1, or HIP/PAP-1.93
Mouse Models of Pancreatic Cancer
Xenograft Models
Research aimed at better pathophysiological understanding and development of novel therapeutic strategies against pancreatic cancer has long been hampered by the lack of suitable small animal models that satisfactorily resemble clinical features of human disease. Traditionally, initial screening of potential new drugs for in vivo efficacy is most commonly performed on subcutaneous (athymic nude or SCID) mouse xenograft models, derived from either human pancreatic cancer cell lines or primary cancer tissue specimens, which accurately mirror genotypic features of the parental tumors and susceptibility or resistance to anticancer agents even after serial transplantation.94 While these in vivo models are relatively easy to generate and allow for easy screening of putative new drugs on several xenograft tumor lines, and thus are undoubtedly of immense value for initial pharmacological studies, they also suffer from some fundamental drawbacks.95,96 First, as these xenogenic transplantation models lack T cell or both B and T cell responses, effects of the tumor environment including immunological effects are poorly mirrored. Second, as these models are heterotopic, they do not reflect the physiological microenvironment in the pancreas, including anatomical conditions and effects of direct tumor invasion into neighboring structures, cytokine and chemokine secretion patterns, vascularization and tumor-induced neoangiogenesis. Third, and related to the first two points, subcutaneous xenografts of pancreatic cancers hardly ever metastasize, so that mechanisms involved in metastatic spread cannot be studied using these models, although this is one of the most prominent features of pancreatic cancer in humans that de facto determines the overall survival.
Therefore, recent years have seen increasing use of orthotopic xenograft models, which are more tedious to generate but are able to overcome many of these shortcomings. They are particularly useful to study drug effects on tumor microenvironment including neoangiogenesis and metastatic spread.87,97 Tumor cells are either injected directly into the mouse pancreas, often in the form of concentrated tumor cell suspensions containing Matrigel, or alternatively are surgically implanted into the murine pancreas as chunks of primary tumor tissues or xenografts. While the first variant is faster and allows generation of xenografts from cell lines without the need to first grow subcutaneous xenografts, the second technique is more reliable in preventing intraperitoneal leakage of tumor cells after injection and also allows for the use of primary tissue samples, thus bypassing the need for in vitro established cell lines.
Transgenic Mouse Models
As opposed to subcutaneous or orthotopic xenografts, transgenic mouse models possess the potential to mimic human disease in a syngeneic system. While they can obviously not represent the whole spectrum of genetic aberrations observed in human pancreatic cancers, these models are of tremendous value for translational studies as they also include a syngeneic tumor environment as well as a fully intact immune system. The earliest models, described almost two decades ago, used acinar-specific elastase promoter to target oncogene expression to the murine pancreas, resulting in neoplasms of predominantly acinar histogenesis.98,99
The first mouse model most closely resembling histopathological features of human ductal adenocarcinoma was described in 2003.100 In this model, expression of oncogenic KrasG12D is suppressed by combination with Lox-STOP-Lox (LSL) constructs. Repression is released on Cre-mediated excision of the LSL cassettes and subsequent recombination. Targeting of transgene expression to the murine pancreas is achieved by expression of Cre recombinase under the control of pancreas-specific promoters Pdx1 or P48. It is commonly assumed that Pdx1-/P48-double positive cells give rise to virtually all mature cells in the pancreas.100,101,102 During embryonic development, expression of Pdx1/PF1 starts around E8.5, and P48/PTF1 expression begins slightly later. All of these mice develop ductal lesions resembling human PanIN lesions, which eventually progress into a fully invasive and metastatic adenocarcinoma phenotype in a small percentage (<10%) of animals. The long latency of 6 to 8 months and low frequency suggest the need of additional genetic alterations, likely including the INK4a-Rb or Arf-p53 pathways, whereas the majority of cells expressing oncogenic KrasG12D alone might undergo ras-induced senescence and thus fail to accumulate additional hits required to develop a fully malignant cancer phenotype.103 Similar to these observations, Grippo et al found multifocal acinar cell hyperplasia in Ela-KrasG12D mice 1 to 2 months of age. Interestingly, by the age of 6 to 18 months, some of these lesions underwent a process the authors referred to as acinar-to-ductal metaplasia and presented with a more duct-like phenotype, including expression of CK-19.104
In fact, newer transgenic models have recently been described, in that KrasG12D expression is directed to the pancreas by means of Cre recombinase under the control of a Pdx1 promoter in combination with inhibition of the INK4A/Arf or p53 pathways, respectively: LSL-KrasG12D; INK4a/Arflox/lox;Pdx1-Cre,105 LSL-Kras;p53lox/lox;Pdx1-Cre,106 and LSL-Kras;Trp53R172H;Pdx1-Cre.107 Mice with knockout of the INK4a/Arf locus, resulting in loss of both murine p16 and p19 function, develop poorly differentiated carcinomas very rapidly and start to die before 7 weeks of age,105 whereas inhibition of p53 function, either by genetic knockout106 or by introduction of a dominant negative p53 allele,107 preferentially leads to moderately to well-differentiated adenocarcinomas. Of note, abrogation of either INK4a/Arf or p53 signaling alone in the absence of oncogenic Kras does not lead to the development of pancreatic carcinomas or associated precursor lesions, underscoring the crucial importance of Kras signaling in initiating the cascade of events, eventually culminating in a fully malignant phenotype during pancreatic carcinogenesis.105
The described models, and especially the latter,107 to date also represent those recapitulating most closely the clinicopathological characteristics of human pancreatic adenocarcinomas and thus carry enormous potential for future translational studies in providing an excellent platform for preclinical evaluation of novel drugs and other therapeutic approaches. Several signaling pathways shown to be aberrantly activated in human pancreatic cancers are also found to be turned on in the discussed mouse models, including the Hedgehog and Notch signaling pathways. As these pathways can be targeted by using blocking antibodies or small-molecule inhibitors, for example, it is tempting to speculate whether therapeutic regimens exploiting blockade of these pathways might have an effect on survival in these preclinical models and might moreover eventually be translated into applications in a clinical setting.
Using a mouse model of TGF-α-induced pancreatic cancer previously described by Wagner and colleagues,108 a recent study from the same group found that pancreatic carcinomas occurring in C57BL/6-EL-TGF-alpha;Trp53−/− mice induced a distinct immune response including secretion of proinflammatory cytokines and occurrence of tumor-specific regulatory T lymphocytes in the host. Surprisingly, tumor-derived cell lines did not form xenograft tumors in immunocompetent mice of the same genetic background, a finding that is yet to be fully understood. From these results the authors conclude that spontaneous tumors arising in this mouse model are recognized by the host immune system and that therefore transgenic models might be more suitable than xenografts to evaluate certain immunotherapeutic regimens in a preclinical setting.109
An immunotherapeutic approach using tumor vaccination with tumor/dendritic cell fusion supplemented by injection of superantigen staphylococcal enterotoxin B leads to generation of tumor-specific cytotoxic T lymphocytes and increased survival in a previously described transgenic mouse model of acinar cell-type pancreatic cancer overexpressing MUC1,110,111 which is based on a model originally described by Tevethia et al, in which pancreatic acinar carcinomas are induced by pancreas-specific expression of a transgene containing the N-terminal amino acids 1–127 of large T-cell antigen under the control of THE elastase-1 promoter.112
In the last few years, several other transgenic mouse models targeting different pathways thought to be involved in pancreatic carcinogenesis, which allow deeper insight in underlying etiological and pathogenetic mechanisms, have been described. Three recent mouse models addressed the role of TGF-β signaling in pancreatic cancer (see Figure 2 for an overview of the TGF-β pathway) by pancreas-specific deletion of either SMAD4113,114 or TGF-β receptor type II (TGFBR2).115 Interestingly, as observed for interruption of INK4a/Arf and p53 signaling pathways, neither SMAD4 nor TGFBR2 deletion alone is sufficient to induce pancreatic neoplasia. However, when combined with oncogenic KrasG12D, development of fully malignant carcinomas is enhanced with shorter latencies than observed for KrasG12D alone. While mice lacking pancreatic TGFBR2 expression in the presence of KrasG12D all develop well-differentiated adenocarcinomas and die of their disease within 200 days, with a median survival of 59 days,115 loss of SMAD4 in KrasG12D-expressing pancreata leads to development of premalignant precursor lesions of intraductal papillary mucinous neoplasm or mucinous cystic neoplasm type, which can progress into fully malignant carcinomas. For the latter, median survival of 8 months114 and 7 to 12 weeks113 have been described. TGF-β signaling has previously been proposed to mediate epithelial-to-mesenchymal transition,116 and in line with this concept, INK4A/Arf-null;KrasG12D mice with wild-type SMAD4 usually present with poorly differentiated carcinomas, whereas mice that also lack SMAD4 expression, develop mostly well- to moderately differentiated pancreatic adenocarcinomas that express epithelial markers such as E-cadherin or cytokeratin-19.
Studies examining the role of Hedgehog signaling demonstrated that overexpression of the Hedgehog ligand Sonic Hedgehog (SHH) in the pancreas is sufficient to induce formation of PanIN lesions.79 Introduction of a dominant-active form of the activating Hedgehog transcription factor GLI2 (CLEG2) leads to formation of poorly differentiated pancreatic carcinomas that lack CK-19 expression in about 30% of PDX1-Cre;CLEG2 mice, which seem to develop without evidence of PanINs as precursor lesions.117 Of note, combined expression of CLEG2 and KrasG12D in the murine pancreas results in formation of PanIN lesions and pancreatic carcinomas in all studied mice, with shorter latency and dramatically decreased overall survival of only 3 to 8 weeks.
Overexpression of COX-2 in the pancreas under the control of a keratin 5 promoter leads to formation of IPMN- and PanIN-like lesions, enhanced ras signaling, inflammation, and fibrosis in a subset of mice.118 Clerc and colleagues described development of pancreatic carcinomas through acinar-to-ductal transition in three of 20 mice overexpressing CCK2/gastrin receptor in pancreatic acinar cells.119
Other Promising Translational Studies
Some novel therapeutic strategies that were developed based on recent advances in our understanding of molecular biology and pathophysiology of pancreatic cancer are actually on the verge of being tested in the clinics and might soon be beneficial, if not for all, for at least a subset of patients suffering from PDAC. Targeting aberrantly reactivated developmental signaling pathways, namely the Hedgehog,79,80 Notch,84,120 and Wnt121,122 signaling pathways, by means of blocking antibodies or small-molecule inhibitors are highly promising future treatment options in PDAC, as already mentioned above. These approaches are particularly exciting, as first evidence is emerging that at least some of these strategies might target distinct subpopulations of cancer cells with stem cell-like properties87,88 and could therefore carry the potential to overcome development of drug resistance and tumor recurrence commonly observed today with standard chemotherapy regimens in several malignancies. However, as evaluation of treatment strategies targeting these pathways has only just begun and, moreover, the concept of cancer stem cells is still highly and controversially discussed in the scientific community, it is not possible yet to give a final judgment as to the full therapeutic potential of these approaches.
Multiple immunotherapeutic trials have been performed in a clinical setting at several institutions, including our own, with variable success.123 Overall, immunotherapeutic regimens using monoclonal antibodies have so far been the most successful approaches. However, tumor vaccination protocols for pancreatic cancer are also increasingly translated into clinical therapies, some of which showed extremely encouraging initial results. A phase I study conducted at our own institution using a whole tumor cell vaccine coadministered with granulocyte macrophage–colony-stimulating factor was shown to be safe and potentially effective in patients with early stage disease.124 Follow-up phase II clinical trials are currently ongoing.
Another idea is based on the recent observation that a small subset of PDAC that carry mutations in the Fanconi anemia/BRCA2 pathway is highly sensitive to cross-linking agents such as mitomycin C or cisplatin in vitro and in vivo.44,125 Although affecting only 5 to 10% of human PDAC, these findings might soon be translated into effective chemotherapeutic treatment regimens for these cases. A phase II clinical trial is presently underway.
Overexpression of the chemokine receptor CXCR4 is thought to be involved in the development of metastases, possibly through attraction of CXCR4-positive tumor cells by SDF-1alpha/CXCL12.126 Moreover, tumor neoangiogenesis and growth of subcutaneous murine xenografts could be inhibited by administration of anti-CXCR4 blocking antibodies.127 Therefore, the SDF-1/CXCR4 signaling axis might be yet another promising therapeutic target for PDAC in the near future.
Using in vitro and in vivo model systems, treatment with tumor necrosis factor-related apoptosis-inducing ligand has been demonstrated to be a valid novel therapeutic strategy in several solid tumors, including pancreatic cancer, especially when combined with other substances sensitizing cancer cells to tumor necrosis factor-related apoptosis-inducing ligand by overcoming resistance to apoptosis.128,129 This strategy is also most likely soon to undergo initial evaluation in a clinical setting.
Clinical use of several commonly administered cytotoxic chemotherapeutics is often limited by systemic adverse effects. Utilization of nanotechnology carries the potential to minimize these problems, by encapsulating drugs in nanoparticles and via specific delivery to tumor cells by passive (through enhanced permeation and retention effect) or active targeting (nanoparticles coated with antibodies or receptor analogs directed against tumor-specific surface antigens), thereby drastically reducing the total amount of drug needed to achieve a therapeutic response.130 Moreover, encapsulation into nanoparticles can overcome poor water solubility, a common shortcoming of multiple substances being screened as potential drugs. An example is curcumin, a plant extract from Curcuma longa, which has long been known in traditional Indian medicine and has well-described anti-inflammatory and antineoplastic properties in vitro. While promising in vivo results have been achieved in animal models of PDAC in that comparably huge doses of curcumin could be administered,131 as well as in clinical trials in preventing progression of preneoplastic colon adenomas,132 broader application in different tumor types has been hampered by poor water solubility and almost zero resorption from the gastrointestinal tract and systemic bioavailability.133,134 Nanoencapsulation of curcumin has recently been shown to render the drug readily water soluble while retaining its biological properties in vitro.135,136 However, the in vivo efficacy of these new formulations has yet to be evaluated.
Conclusion
Pancreatic cancer continues to prove resistant against most clinically available treatment options to date, and it remains to be one of the worst killers among human malignancies. It seems unlikely that potent therapeutic options, which would enable us to definitely cure or, more desirably, prevent all pancreatic cancers, will be available in the near future. However, based on recent advances, as described in this article, there is increasing hope that tenacious research efforts and better understanding of underlying biological characteristics will finally yield enhanced survival and better quality of life for patients suffering from this disease. Multimodal strategies combining different therapeutic strategies are likely to have the greatest chance for improvement in the coming years.
Acknowledgements
We thank Jennifer Parsons-Brumbaugh and Joseph Dieter for preparation of the illustrations. We apologize to colleagues whose work could not be cited due to lack of space.
Footnotes
Supported by National Institutes of Health grants R01CA113669 and R21DK072532, The Sol Goldman Pancreatic Cancer Research Center, and an American Association for Cancer Research-PanCAN award (to A.M.). G.F. was supported by a fellowship grant within the Postdoc-Programme of the German Academic Exchange Service.
References
- 1.American Cancer Society . Cancer Facts & Figures 2007. American Cancer Society; Atlanta: 2007. [Google Scholar]
- 2.Carpelan-Holmstrom M, Nordling S, Pukkala E, Sankila R, Luttges J, Kloppel G, Haglund C. Does anyone survive pancreatic ductal adenocarcinoma? A nationwide study re-evaluating the data of the Finnish Cancer Registry. Gut. 2005;54:385–387. doi: 10.1136/gut.2004.047191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 4.Sirivatanauksorn V, Sirivatanauksorn Y, Gorman PA, Davidson JM, Sheer D, Moore PS, Scarpa A, Edwards PA, Lemoine NR. Non-random chromosomal rearrangements in pancreatic cancer cell lines identified by spectral karyotyping. Int J Cancer. 2001;91:350–358. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1049>3.3.co;2-3. [DOI] [PubMed] [Google Scholar]
- 5.Griffin CA, Hruban RH, Morsberger LA, Ellingham T, Long PP, Jaffee EM, Hauda KM, Bohlander SK, Yeo CJ. Consistent chromosome abnormalities in adenocarcinoma of the pancreas. Cancer Res. 1995;55:2394–2399. [PubMed] [Google Scholar]
- 6.Iacobuzio-Donahue CA, van der Heijden MS, Baumgartner MR, Troup WJ, Romm JM, Doheny K, Pugh E, Yeo CJ, Goggins MG, Hruban RH, Kern SE. Large-scale allelotype of pancreaticobiliary carcinoma provides quantitative estimates of genome-wide allelic loss. Cancer Res. 2004;64:871–875. doi: 10.1158/0008-5472.can-03-2756. [DOI] [PubMed] [Google Scholar]
- 7.Xin W, Yun KJ, Ricci F, Zahurak M, Qiu W, Su GH, Yeo CJ, Hruban RH, Kern SE, Iacobuzio-Donahue CA. MAP2K4/MKK4 expression in pancreatic cancer: genetic validation of immunohistochemistry and relationship to disease course. Clin Cancer Res. 2004;10:8516–8520. doi: 10.1158/1078-0432.CCR-04-0885. [DOI] [PubMed] [Google Scholar]
- 8.Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58:5329–5332. [PubMed] [Google Scholar]
- 9.Su GH, Bansal R, Murphy KM, Montgomery E, Yeo CJ, Hruban RH, Kern SE. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc Natl Acad Sci USA. 2001;98:3254–3257. doi: 10.1073/pnas.051484398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luttges J, Galehdari H, Brocker V, Schwarte-Waldhoff I, Henne-Bruns D, Kloppel G, Schmiegel W, Hahn SA. Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. Am J Pathol. 2001;158:1677–1683. doi: 10.1016/S0002-9440(10)64123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamano M, Fujii H, Takagaki T, Kadowaki N, Watanabe H, Shirai T. Genetic progression and divergence in pancreatic carcinoma. Am J Pathol. 2000;156:2123–2133. doi: 10.1016/S0002-9440(10)65083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahlamaki EH, Hoglund M, Gorunova L, Karhu R, Dawiskiba S, Andren-Sandberg A, Kallioniemi OP, Johansson B. Comparative genomic hybridization reveals frequent gains of 20q, 8q, 11q, 12p, and 17q, and losses of 18q, 9p, and 15q in pancreatic cancer. Genes Chromosomes Cancer. 1997;20:383–391. doi: 10.1002/(sici)1098-2264(199712)20:4<383::aid-gcc10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 13.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 14.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 15.Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, West JA, Rostan S, Nguyen KC, Powers S, Ye KQ, Olshen A, Venkatraman E, Norton L, Wigler M. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holzmann K, Kohlhammer H, Schwaenen C, Wessendorf S, Kestler HA, Schwoerer A, Rau B, Radlwimmer B, Dohner H, Lichter P, Gress T, Bentz M. Genomic DNA-chip hybridization reveals a higher incidence of genomic amplifications in pancreatic cancer than conventional comparative genomic hybridization and leads to the identification of novel candidate genes. Cancer Res. 2004;64:4428–4433. doi: 10.1158/0008-5472.CAN-04-0431. [DOI] [PubMed] [Google Scholar]
- 17.Aguirre AJ, Brennan C, Bailey G, Sinha R, Feng B, Leo C, Zhang Y, Zhang J, Gans JD, Bardeesy N, Cauwels C, Cordon-Cardo C, Redston MS, DePinho RA, Chin L. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci USA. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brody JR, Hucl T, Gallmeier E, Winter JM, Kern SE, Murphy KM. Genomic copy number changes affecting the thymidylate synthase (TYMS) gene in cancer: a model for patient classification to aid fluoropyrimidine therapy. Cancer Res. 2006;66:9369–9373. doi: 10.1158/0008-5472.CAN-06-2165. [DOI] [PubMed] [Google Scholar]
- 19.Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 20.Hruban RH, Maitra A, Schulick R, Laheru D, Herman J, Kern SE, Goggins M. Emerging molecular biology of pancreatic cancer. Gastrointestinal Cancer Res. 2007 (in press) [PMC free article] [PubMed] [Google Scholar]
- 21.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- 22.Hruban RH, Wilentz RE, Kern SE. Genetic progression in the pancreatic ducts. Am J Pathol. 2000;156:1821–1825. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calhoun ES, Jones JB, Ashfaq R, Adsay V, Baker SJ, Valentine V, Hempen PM, Hilgers W, Yeo CJ, Hruban RH, Kern SE. BRAF and FBXW7 (CDC4. FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol. 2003;163:1255–1260. doi: 10.1016/S0002-9440(10)63485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- 25.Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci USA. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruggeri BA, Huang L, Wood M, Cheng JQ, Testa JR. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol Carcinog. 1998;21:81–86. [PubMed] [Google Scholar]
- 27.Altomare DA, Tanno S, De Rienzo A, Klein-Szanto AJ, Tanno S, Skele KL, Hoffman JP, Testa JR. Frequent activation of AKT2 kinase in human pancreatic carcinomas. J Cell Biochem. 2003;88:470–476. doi: 10.1002/jcb.10287. [DOI] [PubMed] [Google Scholar]
- 28.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Kleeff J, Giese N, Buchler MW, Korc M, Friess H. Gefitinib (“Iressa”. ZD1839), a selective epidermal growth factor receptor tyrosine kinase inhibitor, inhibits pancreatic cancer cell growth, invasion, and colony formation. Int J Oncol. 2004;25:203–210. [PubMed] [Google Scholar]
- 30.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W, Baylin SB, Kern SE, Herman JG. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 31.Wilentz RE, Geradts J, Maynard R, Offerhaus GJ, Kang M, Goggins M, Yeo CJ, Kern SE, Hruban RH. Inactivation of the p16 (INK4A) tumor-suppressor gene in pancreatic duct lesions: loss of intranuclear expression. Cancer Res. 1998;58:4740–4744. [PubMed] [Google Scholar]
- 32.Chen ZH, Zhang H, Savarese TM. Gene deletion chemoselectivity: codeletion of the genes for p16(INK4), methylthioadenosine phosphorylase, and the alpha- and beta-interferons in human pancreatic cell carcinoma lines and its implications for chemotherapy. Cancer Res. 1996;56:1083–1090. [PubMed] [Google Scholar]
- 33.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 34.Schutte M, Hruban RH, Hedrick L, Cho KR, Nadasdy GM, Weinstein CL, Bova GS, Isaacs WB, Cairns P, Nawroz H, Sidransky D, Casero RA, Jr, Meltzer PS, Hahn SA, Kern SE. DPC4 gene in various tumor types. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 35.Maitra A, Adsay NV, Argani P, Iacobuzio-Donahue C, De Marzo A, Cameron JL, Yeo CJ, Hruban RH. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16:902–912. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 36.Sahin F, Maitra A, Argani P, Sato N, Maehara N, Montgomery E, Goggins M, Hruban RH, Su GH. Loss of Stk11/Lkb1 expression in pancreatic and biliary neoplasms. Mod Pathol. 2003;16:686–691. doi: 10.1097/01.MP.0000075645.97329.86. [DOI] [PubMed] [Google Scholar]
- 37.Su GH, Hilgers W, Shekher MC, Tang DJ, Yeo CJ, Hruban RH, Kern SE. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 1998;58:2339–2342. [PubMed] [Google Scholar]
- 38.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 39.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 40.Ghimenti C, Tannergard P, Wahlberg S, Liu T, Giulianotti PG, Mosca F, Fornaciari G, Bevilacqua G, Lindblom A, Caligo MA. Microsatellite instability and mismatch repair gene inactivation in sporadic pancreatic and colon tumours. Br J Cancer. 1999;80:11–16. doi: 10.1038/sj.bjc.6690314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilentz RE, Goggins M, Redston M, Marcus VA, Adsay NV, Sohn TA, Kadkol SS, Yeo CJ, Choti M, Zahurak M, Johnson K, Tascilar M, Offerhaus GJ, Hruban RH, Kern SE. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: a newly described and characterized entity. Am J Pathol. 2000;156:1641–1651. doi: 10.1016/S0002-9440(10)65035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goggins M, Offerhaus GJ, Hilgers W, Griffin CA, Shekher M, Tang D, Sohn TA, Yeo CJ, Kern SE, Hruban RH. Pancreatic adenocarcinomas with DNA replication errors (RER+) are associated with wild-type K-ras and characteristic histopathology. Poor differentiation, a syncytial growth pattern, and pushing borders suggest RER+ Am J Pathol. 1998;152:1501–1507. [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy KM, Brune KA, Griffin C, Sollenberger JE, Petersen GM, Bansal R, Hruban RH, Kern SE. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17% Cancer Res. 2002;62:3789–3793. [PubMed] [Google Scholar]
- 44.van der Heijden MS, Yeo CJ, Hruban RH, Kern SE. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–2588. [PubMed] [Google Scholar]
- 45.Kennedy RD, D'Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24:3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 46.Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 47.Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 48.Jones JB, Song JJ, Hempen PM, Parmigiani G, Hruban RH, Kern SE. Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass”-ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61:1299–1304. [PubMed] [Google Scholar]
- 49.Maitra A, Cohen Y, Gillespie SE, Mambo E, Fukushima N, Hoque MO, Shah N, Goggins M, Califano J, Sidransky D, Chakravarti A. The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res. 2004;14:812–819. doi: 10.1101/gr.2228504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gisselsson D. Chromosome instability in cancer: how, when, and why? Adv Cancer Res. 2003;87:1–29. doi: 10.1016/s0065-230x(03)87164-6. [DOI] [PubMed] [Google Scholar]
- 51.van Heek NT, Meeker AK, Kern SE, Yeo CJ, Lillemoe KD, Cameron JL, Offerhaus GJ, Hicks JL, Wilentz RE, Goggins MG, De Marzo AM, Hruban RH, Maitra A. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meeker AK. Telomeres and telomerase in prostatic intraepithelial neoplasia and prostate cancer biology. Urol Oncol. 2006;24:122–130. doi: 10.1016/j.urolonc.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Hackett JA, Greider CW. Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene. 2002;21:619–626. doi: 10.1038/sj.onc.1205061. [DOI] [PubMed] [Google Scholar]
- 54.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 55.Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, Goggins M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–1839. [PubMed] [Google Scholar]
- 56.Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–3742. [PubMed] [Google Scholar]
- 57.Martin ST, Sato N, Dhara S, Chang R, Hustinx SR, Abe T, Maitra A, Goggins M. Aberrant methylation of the human Hedgehog interacting protein (HHIP) gene in pancreatic neoplasms. Cancer Biol Ther. 2005;4:728–733. doi: 10.4161/cbt.4.7.1802. [DOI] [PubMed] [Google Scholar]
- 58.Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, Wilentz RE, Yeo CJ, Hruban RH, Goggins M. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–1581. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fukushima N, Walter KM, Uek T, Sato N, Matsubayashi H, Cameron JL, Hruban RH, Canto M, Yeo CJ, Goggins M. Diagnosing pancreatic cancer using methylation specific PCR analysis of pancreatic juice. Cancer Biol Ther. 2003;2:78–83. doi: 10.4161/cbt.183. [DOI] [PubMed] [Google Scholar]
- 60.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 61.Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SR, Leach SD, Ryu B, Skinner HG, Goggins M, Jaffee EM, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Discovery of new markers of cancer through serial analysis of gene expression: prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001;61:4320–4324. [PubMed] [Google Scholar]
- 62.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–8622. [PubMed] [Google Scholar]
- 63.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryu B, Jones J, Blades NJ, Parmigiani G, Hollingsworth MA, Hruban RH, Kern SE. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002;62:819–826. [PubMed] [Google Scholar]
- 66.Ryu B, Jones J, Hollingsworth MA, Hruban RH, Kern SE. Invasion-specific genes in malignancy: serial analysis of gene expression comparisons of primary and passaged cancers. Cancer Res. 2001;61:1833–1838. [PubMed] [Google Scholar]
- 67.Geng M, Wallrapp C, Muller-Pillasch F, Frohme M, Hoheisel JD, Gress TM. Isolation of differentially expressed genes by combining representational difference analysis (RDA) and cDNA library arrays. Biotechniques. 1998;25:434–438. doi: 10.2144/98253st05. [DOI] [PubMed] [Google Scholar]
- 68.Crnogorac-Jurcevic T, Efthimiou E, Capelli P, Blaveri E, Baron A, Terris B, Jones M, Tyson K, Bassi C, Scarpa A, Lemoine NR. Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene. 2001;20:7437–7446. doi: 10.1038/sj.onc.1204935. [DOI] [PubMed] [Google Scholar]
- 69.Nichols LS, Ashfaq R, Iacobuzio-Donahue CA. Claudin 4 protein expression in primary and metastatic pancreatic cancer: support for use as a therapeutic target. Am J Clin Pathol. 2004;121:226–230. doi: 10.1309/K144-PHVD-DUPD-D401. [DOI] [PubMed] [Google Scholar]
- 70.Michl P, Buchholz M, Rolke M, Kunsch S, Lohr M, McClane B, Tsukita S, Leder G, Adler G, Gress TM. Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology. 2001;121:678–684. doi: 10.1053/gast.2001.27124. [DOI] [PubMed] [Google Scholar]
- 71.Foss CA, Fox J, Feldmann G, Maitra A, Iacobuzio-Donahue C, Kern SE, Hruban RH, Pomper MG. Radiolabeled anti-claudin 4 and anti-prostate stem cell antigen: initial imaging in experimental models of pancreatic cancer. Molecular Imaging. 2007:6. [PubMed] [Google Scholar]
- 72.Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, Wilentz RE, Murugesan SR, Leach SD, Jaffee E, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE) Clin Cancer Res. 2001;7:3862–3868. [PubMed] [Google Scholar]
- 73.Thomas AM, Santarsiero LM, Lutz ER, Armstrong TD, Chen YC, Huang LQ, Laheru DA, Goggins M, Hruban RH, Jaffee EM. Mesothelin-specific CD8(+) T cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. J Exp Med. 2004;200:297–306. doi: 10.1084/jem.20031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wente MN, Jain A, Kono E, Berberat PO, Giese T, Reber HA, Friess H, Buchler MW, Reiter RE, Hines OJ. Prostate stem cell antigen is a putative target for immunotherapy in pancreatic cancer. Pancreas. 2005;31:119–125. doi: 10.1097/01.mpa.0000173459.81193.4d. [DOI] [PubMed] [Google Scholar]
- 75.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo J, Kleeff J, Zhao Y, Li J, Giese T, Esposito I, Buchler MW, Korc M, Friess H. Yes-associated protein (YAP65) in relation to Smad7 expression in human pancreatic ductal adenocarcinoma. Int J Mol Med. 2006;17:761–767. [PubMed] [Google Scholar]
- 77.Bhanot U, Heydrich R, Moller P, Hasel C. Survivin expression in pancreatic intraepithelial neoplasia (PanIN): steady increase along the developmental stages of pancreatic ductal adenocarcinoma. Am J Surg Pathol. 2006;30:754–759. doi: 10.1097/00000478-200606000-00013. [DOI] [PubMed] [Google Scholar]
- 78.Satoh K, Kaneko K, Hirota M, Masamune A, Satoh A, Shimosegawa T. Expression of survivin is correlated with cancer cell apoptosis and is involved in the development of human pancreatic duct cell tumors. Cancer. 2001;92:271–278. doi: 10.1002/1097-0142(20010715)92:2<271::aid-cncr1319>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 79.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, Yajnik V, Antoniu B, McMahon M, Warshaw AL, Hebrok M. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, Beachy PA. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 81.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, Hruban RH, Ball DW, Schmid RM, Leach SD. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 82.Kimura K, Satoh K, Kanno A, Hamada S, Hirota M, Endoh M, Masamune A, Shimosegawa T. Activation of Notch signaling in tumorigenesis of experimental pancreatic cancer induced by dimethylbenzanthracene in mice. Cancer Sci. 2007;98:155–162. doi: 10.1111/j.1349-7006.2006.00369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Notch-1 down-regulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer. 2006;106:2503–2513. doi: 10.1002/cncr.21904. [DOI] [PubMed] [Google Scholar]
- 84.Wang Z, Zhang Y, Li Y, Banerjee S, Liao J, Sarkar FH. Down-regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol Cancer Ther. 2006;5:483–493. doi: 10.1158/1535-7163.MCT-05-0299. [DOI] [PubMed] [Google Scholar]
- 85.Buchler P, Gazdhar A, Schubert M, Giese N, Reber HA, Hines OJ, Giese T, Ceyhan GO, Muller M, Buchler MW, Friess H. The Notch signaling pathway is related to neurovascular progression of pancreatic cancer. Ann Surg. 2005;242:791–800. doi: 10.1097/01.sla.0000189115.94847.f1. discussion 800–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shafaee Z, Schmidt H, Du W, Posner M, Weichselbaum R. Cyclopamine increases the cytotoxic effects of paclitaxel and radiation but not cisplatin and gemcitabine in Hedgehog expressing pancreatic cancer cells. Cancer Chemother Pharmacol. 2006;58:765–770. doi: 10.1007/s00280-006-0227-4. [DOI] [PubMed] [Google Scholar]
- 87.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, Karikari C, Alvarez H, Iacobuzio-Donahue C, Jimeno A, Gabrielson KL, Matsui W, Maitra A. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 89.Feldmann G, Beaty R, Hruban RH, Maitra A. Molecular genetics of pancreatic intraepithelial neoplasia. J Hepatobiliary Pancreat Surg. 2007;14:224–232. doi: 10.1007/s00534-006-1166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singh M, Maitra A. Precursor lesions of pancreatic cancer: molecular pathology and clinical implications. Pancreatology. 2007;7:9–19. doi: 10.1159/000101873. [DOI] [PubMed] [Google Scholar]
- 91.Rosty C, Christa L, Kuzdzal S, Baldwin WM, Zahurak ML, Carnot F, Chan DW, Canto M, Lillemoe KD, Cameron JL, Yeo CJ, Hruban RH, Goggins M. Identification of hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein I as a biomarker for pancreatic ductal adenocarcinoma by protein biochip technology. Cancer Res. 2002;62:1868–1875. [PubMed] [Google Scholar]
- 92.Koopmann J, Zhang Z, White N, Rosenzweig J, Fedarko N, Jagannath S, Canto MI, Yeo CJ, Chan DW, Goggins M. Serum diagnosis of pancreatic adenocarcinoma using surface-enhanced laser desorption and ionization mass spectrometry. Clin Cancer Res. 2004;10:860–868. doi: 10.1158/1078-0432.ccr-1167-3. [DOI] [PubMed] [Google Scholar]
- 93.Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 94.Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, Shi C, Danenberg K, Danenberg PV, Kuramochi H, Tanaka K, Singh S, Salimi-Moosavi H, Bouraoud N, Amador ML, Altiok S, Kulesza P, Yeo C, Messersmith W, Eshleman J, Hruban RH, Maitra A, Hidalgo M. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 95.Kelland LR. Of mice and men: values and liabilities of the athymic nude mouse model in anticancer drug development. Eur J Cancer. 2004;40:827–836. doi: 10.1016/j.ejca.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 96.Kerbel RS. Human tumor xenografts as predictive preclinical models for anticancer drug activity in humans: better than commonly perceived—but they can be improved. Cancer Biol Ther. 2003;2:S134–S139. [PubMed] [Google Scholar]
- 97.Loukopoulos P, Kanetaka K, Takamura M, Shibata T, Sakamoto M, Hirohashi S. Orthotopic transplantation models of pancreatic adenocarcinoma derived from cell lines and primary tumors and displaying varying metastatic activity. Pancreas. 2004;29:193–203. doi: 10.1097/00006676-200410000-00004. [DOI] [PubMed] [Google Scholar]
- 98.Ornitz DM, Hammer RE, Messing A, Palmiter RD, Brinster RL. Pancreatic neoplasia induced by SV40 T-antigen expression in acinar cells of transgenic mice. Science. 1987;238:188–193. doi: 10.1126/science.2821617. [DOI] [PubMed] [Google Scholar]
- 99.Sandgren EP, Luetteke NC, Palmiter RD, Brinster RL, Lee DC. Overexpression of TGF alpha in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell. 1990;61:1121–1135. doi: 10.1016/0092-8674(90)90075-p. [DOI] [PubMed] [Google Scholar]
- 100.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 101.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 102.Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bardeesy N, Sharpless NE. RAS unplugged: negative feedback and oncogene-induced senescence. Cancer Cell. 2006;10:451–453. doi: 10.1016/j.ccr.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 104.Grippo PJ, Nowlin PS, Demeure MJ, Longnecker DS, Sandgren EP. Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 2003;63:2016–2019. [PubMed] [Google Scholar]
- 105.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, Hanahan D, Redston MS, Chin L, Depinho RA. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 108.Wagner M, Greten FR, Weber CK, Koschnick S, Mattfeldt T, Deppert W, Kern H, Adler G, Schmid RM. A murine tumor progression model for pancreatic cancer recapitulating the genetic alterations of the human disease. Genes Dev. 2001;15:286–293. doi: 10.1101/gad.184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Garbe AI, Vermeer B, Gamrekelashvili J, von Wasielewski R, Greten FR, Westendorf AM, Buer J, Schmid RM, Manns MP, Korangy F, Greten TF. Genetically induced pancreatic adenocarcinoma is highly immunogenic and causes spontaneous tumor-specific immune responses. Cancer Res. 2006;66:508–516. doi: 10.1158/0008-5472.CAN-05-2383. [DOI] [PubMed] [Google Scholar]
- 110.Mukherjee P, Ginardi AR, Madsen CS, Sterner CJ, Adriance MC, Tevethia MJ, Gendler SJ. Mice with spontaneous pancreatic cancer naturally develop MUC-1-specific CTLs that eradicate tumors when adoptively transferred. J Immunol. 2000;165:3451–3460. doi: 10.4049/jimmunol.165.6.3451. [DOI] [PubMed] [Google Scholar]
- 111.McConnell EJ, Pathangey LB, Madsen CS, Gendler SJ, Mukherjee P. Dendritic cell-tumor cell fusion and staphylococcal enterotoxin B treatment in a pancreatic tumor model. J Surg Res. 2002;107:196–202. doi: 10.1006/jsre.2001.6497. [DOI] [PubMed] [Google Scholar]
- 112.Tevethia MJ, Bonneau RH, Griffith JW, Mylin L. A simian virus 40 large T-antigen segment containing amino acids 1 to 127 and expressed under the control of the rat elastase-1 promoter produces pancreatic acinar carcinomas in transgenic mice. J Virol. 1997;71:8157–8166. doi: 10.1128/jvi.71.11.8157-8166.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, DePinho RA. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, Grady WM, Deng CX, Hruban RH, Adsay NV, Tuveson DA, Hingorani SR. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–243. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 115.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, Wright CV, Moses HL. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 117.Pasca di Magliano M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Muller-Decker K, Furstenberger G, Annan N, Kucher D, Pohl-Arnold A, Steinbauer B, Esposito I, Chiblak S, Friess H, Schirmacher P, Berger I. Preinvasive duct-derived neoplasms in pancreas of keratin 5-promoter cyclooxygenase-2 transgenic mice. Gastroenterology. 2006;130:2165–2178. doi: 10.1053/j.gastro.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 119.Clerc P, Leung-Theung-Long S, Wang TC, Dockray GJ, Bouisson M, Delisle MB, Vaysse N, Pradayrol L, Fourmy D, Dufresne M. Expression of CCK2 receptors in the murine pancreas: proliferation, transdifferentiation of acinar cells, and neoplasia. Gastroenterology. 2002;122:428–437. doi: 10.1053/gast.2002.30984. [DOI] [PubMed] [Google Scholar]
- 120.Wang Z, Banerjee S, Li Y, Rahman KM, Zhang Y, Sarkar FH. Down-regulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res. 2006;66:2778–2784. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 121.Nawroth R, van Zante A, Cervantes S, McManus M, Hebrok M, Rosen SD. Extracellular sulfatases, elements of the wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS ONE. 2007;2:e392. doi: 10.1371/journal.pone.0000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zeng G, Germinaro M, Micsenyi A, Monga NK, Bell A, Sood A, Malhotra V, Sood N, Midda V, Monga DK, Kokkinakis DM, Monga SP. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8:279–289. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Laheru D, Jaffee EM. Immunotherapy for pancreatic cancer: science driving clinical progress. Nat Rev Cancer. 2005;5:459–467. doi: 10.1038/nrc1630. [DOI] [PubMed] [Google Scholar]
- 124.Jaffee EM, Hruban RH, Biedrzycki B, Laheru D, Schepers K, Sauter PR, Goemann M, Coleman J, Grochow L, Donehower RC, Lillemoe KD, O'Reilly S, Abrams RA, Pardoll DM, Cameron JL, Yeo CJ. Novel allogeneic granulocyte-macrophage colony-stimulating factor-secreting tumor vaccine for pancreatic cancer: a phase I trial of safety and immune activation. J Clin Oncol. 2001;19:145–156. doi: 10.1200/JCO.2001.19.1.145. [DOI] [PubMed] [Google Scholar]
- 125.van der Heijden MS, Brody JR, Dezentje DA, Gallmeier E, Cunningham SC, Swartz MJ, DeMarzo AM, Offerhaus GJ, Isacoff WH, Hruban RH, Kern SE. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res. 2005;11:7508–7515. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
- 126.Saur D, Seidler B, Schneider G, Algul H, Beck R, Senekowitsch-Schmidtke R, Schwaiger M, Schmid RM. CXCR4 expression increases liver and lung metastasis in a mouse model of pancreatic cancer. Gastroenterology. 2005;129:1237–1250. doi: 10.1053/j.gastro.2005.06.056. [DOI] [PubMed] [Google Scholar]
- 127.Guleng B, Tateishi K, Ohta M, Kanai F, Jazag A, Ijichi H, Tanaka Y, Washida M, Morikane K, Fukushima Y, Yamori T, Tsuruo T, Kawabe T, Miyagishi M, Taira K, Sata M, Omata M. Blockade of the stromal cell-derived factor-1/CXCR4 axis attenuates in vivo tumor growth by inhibiting angiogenesis in a vascular endothelial growth factor-independent manner. Cancer Res. 2005;65:5864–5871. doi: 10.1158/0008-5472.CAN-04-3833. [DOI] [PubMed] [Google Scholar]
- 128.Koschny R, Ganten TM, Sykora J, Haas TL, Sprick MR, Kolb A, Stremmel W, Walczak H. TRAIL/bortezomib cotreatment is potentially hepatotoxic but induces cancer-specific apoptosis within a therapeutic window. Hepatology. 2007;45:649–658. doi: 10.1002/hep.21555. [DOI] [PubMed] [Google Scholar]
- 129.Karikari CA, Roy I, Tryggestad E, Feldmann G, Pinilla C, Welsh K, Reed JC, Armour EP, Wong J, Herman J, Rakheja D, Maitra A. Targeting the apoptotic machinery in pancreatic cancers using small-molecule antagonists of the X-linked inhibitor of apoptosis protein. Mol Cancer Ther. 2007;6:957–966. doi: 10.1158/1535-7163.MCT-06-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Macdiarmid JA, Mugridge NB, Weiss JC, Phillips L, Burn AL, Paulin RP, Haasdyk JE, Dickson KA, Brahmbhatt VN, Pattison ST, James AC, Al Bakri G, Straw RC, Stillman B, Graham RM, Brahmbhatt H. Bacterially Derived 400 nm Particles for Encapsulation and Cancer Cell Targeting of Chemotherapeutics. Cancer Cell. 2007;11:431–445. doi: 10.1016/j.ccr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 131.Kunnumakkara AB, Guha S, Krishnan S, Diagaradjane P, Gelovani J, Aggarwal BB. Curcumin potentiates antitumor activity of gemcitabine in an orthotopic model of pancreatic cancer through suppression of proliferation, angiogenesis, and inhibition of nuclear factor-kappaB-regulated gene products. Cancer Res. 2007;67:3853–3861. doi: 10.1158/0008-5472.CAN-06-4257. [DOI] [PubMed] [Google Scholar]
- 132.Cruz-Correa M, Shoskes DA, Sanchez P, Zhao R, Hylind LM, Wexner SD, Giardiello FM. Combination treatment with curcumin and quercetin of adenomas in familial adenomatous polyposis. Clin Gastroenterol Hepatol. 2006;4:1035–1038. doi: 10.1016/j.cgh.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 133.Cheng AL, Hsu CH, Lin JK, Hsu MM, Ho YF, Shen TS, Ko JY, Lin JT, Lin BR, Ming-Shiang W, Yu HS, Jee SH, Chen GS, Chen TM, Chen CA, Lai MK, Pu YS, Pan MH, Wang YJ, Tsai CC, Hsieh CY. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001;21:2895–2900. [PubMed] [Google Scholar]
- 134.Lao CD, Ruffin MT, Normolle D, Heath DD, Murray SI, Bailey JM, Boggs ME, Crowell J, Rock CL, Brenner DE. Dose escalation of a curcuminoid formulation. BMC Complement Altern Med. 2006;6:10. doi: 10.1186/1472-6882-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bisht S, Feldmann G, Soni S, Ravi R, Karikar C, Maitra A, Maitra A. Polymeric nanoparticle-encapsulated curcumin (“nanocurcumin”): a novel strategy for human cancer therapy. J Nanobiotechnol. 2007;5:3. doi: 10.1186/1477-3155-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li L, Braiteh FS, Kurzrock R. Liposome-encapsulated curcumin: in vitro and in vivo effects on proliferation, apoptosis, signaling, and angiogenesis. Cancer. 2005;104:1322–1331. doi: 10.1002/cncr.21300. [DOI] [PubMed] [Google Scholar]