Abstract
Identification of human CD1d-restricted T-cell receptor (TCR)-invariant natural killer T (iNKT) cells has been dependent on utilizing combinations of monoclonal antibodies or CD1d tetramers, which do not allow for the most specific analysis of this T-cell subpopulation. A novel monoclonal antibody (clone 6B11), specific for the invariant CDR3 loop of human canonical Vα24Jα18 TCR α chain, was developed and used to specifically characterize iNKT cells. In healthy individuals studied for up to 1 year, a wide but stable frequency of circulating iNKT cells (range: 0·01–0·92%) was observed, with no differences in frequency by gender. Four stable iNKT cell subsets were characterized in peripheral blood based on the expression of CD4 and CD8, with CD8+ iNKT cells being a phenotypic and functionally different subset from CD4+ and double negative iNKT cells; in particular, LAG-3 was preferentially expressed on CD8+ iNKT cells. In addition, a strong negative linear correlation between the frequency of total iNKT cells and percentage of the CD4+ subset was observed. In terms of their potential association with disease, patients at risk for type 1 diabetes had significantly expanded frequencies of double negative iNKT cells when compared to matched controls and first-degree relatives. Moreover, peripheral blood CD4+ iNKT cells were the highest producers of interleukin-4, while the production of interferon-γ and tumour necrosis factor-α was similar amongst all iNKT cell subsets. These differences in iNKT cell subsets suggest that in humans the relative ratio of iNKT cell subsets may influence susceptibility vs. resistance to immune-mediated diseases.
Keywords: invariant NKT cells, 6B11 clone, LAG-3, IFN-γ, IL-4
Introduction
CD1d-restricted T-cell receptor (TCR)-invariant natural killer T (iNKT) cells are a subset of CD3+ lymphocytes highly conserved through mammalian evolution and thought to subserve regulatory/effector functions in a wide variety of immune responses.1–4 Indeed, murine iNKT cells have been shown to play crucial roles in antitumour, autoimmune, and antimicrobial responses.5–7 These findings have encouraged further characterization of human iNKT cells and studies of their participation in diseases involving dysregulated immune functions. While murine iNKT cells have been subject to extensive investigation, less is known regarding human iNKT cells, particularly with respect to the influence of age, gender, racial background and environmental factors. Moreover, the contribution of these cells to human disease involving immune dysregulation remains controversial.8–11
The evaluation of human iNKT cells is particularly complicated because of their low frequency (i.e. ranging from 0·01% to 1%) amongst peripheral blood T cells and a lack of reagents specific for iNKT cells.2 As a result, human iNKT cells have largely been characterized using clones obtained following their expansion with α-galactosyl ceramide (α-GalCer) and long-term cell culture with autologous-irradiated mononuclear cells and cytokines (e.g. interleukin (IL)-2, IL-7);12,13 however, this process may change some of the surface markers and functional patterns originally expressed by unmanipulated, resting human iNKT cells. More recently, TCR-based strategies utilizing combinations of semiselective antibodies and CD1d tetramers have been developed.14,15 Despite these advances, difficult technical challenges remain when using these reagents in terms of defining specificity and sensitivity.15
In the TCR α chain of human iNKT cells, the Vα24 segment is joined with Jα18 in a germ-line configuration, resulting in an invariant CDR3 loop encoded by the mature TCR α chain.16,17 This α chain pairs with a restricted range of randomly rearranged Vβ chains, with Vβ11 being the most prominent in humans.18 Therefore, iNKT cells have classically been identified using monoclonal antibodies (mAbs) against the Vα24 and Vβ11 chains2,18. However, non-invariant and non-CD1d-restricted Vα24+ T cells can also pair with Vβ11, and contribute to the Vα24+/Vβ11+ subpopulation. This could lead to an overestimation of iNKT cell number, especially in individuals with decreased number of iNKT cells.2,19
In previous studies, iNKT cells have largely been evaluated using the coexpression of Vα24 and natural killer (NK) cell markers as CD161 or CD56.20–22 However, it is now clear that only a portion of iNKT cells express NK cell markers and this expression is associated with a late stage of cell differentiation.23 Furthermore, CD161 and other NK markers are expressed on activated CD8+ T lymphocytes. As 15–25% of peripheral blood T cells express CD161, this marker may include non-CD1d-restricted non-invariant Vα24+ T cells.24
The recent development of CD1d tetramers loaded with α-GalCer has significantly enhanced the specificity of iNKT cell evaluation.14,25,26 However, CD1d restriction is not limited to the iNKT cell population, because CD1d restriction has been found on T cells expressing CD161 with diverse repertoires.27,28 Using this approach, the identification is limited by the antigen loaded into the CD1d molecule, with background staining15 cross species variability and displacement of CD1d tetramers by combination with other monoclonal antibodies.28,29
The TCR of iNKT cells represents a unique case of a highly conserved antigen receptor rearrangement and a novel alternative for specific detection of human iNKT cells. Toward this end, we recently generated a mAb against the conserved CDR3 region of the canonical Vα24Jα18 TCR rearrangement (clone 6B11).19,30 The 6B11 mAb recognizes all T cells expressing the invariant TCR α chain, and can be used in combination with anti-Vα24, anti-Vβ11 or anti-CD3 with high specificity and sensitivity for iNKT cell detection.
In this study, we analysed the suitability of the mAb 6B11 for identification and quantification of iNKT cells in a large cohort of healthy individuals, an effort that involved both their phenotypic characterization as well the detection of activation-induced intracellular cytokines. In addition, as a model to determine the utility of the mAb 6B11 to study the frequency and phenotype of circulating iNKT cells in different human immune-mediated diseases, a cohort of type 1 diabetes patients, their relatives and a cohort of high-risk of diabetes individuals were evaluated.
Materials and methods
Study population
For characterization experiments, 90 healthy adults (40 males and 50 females; age range: 15–52 years), without clinical symptoms of acute or chronic immune-mediated diseases at the time of blood sampling, were evaluated in this study. iNKT cell frequency and subsets were delineated in a separate cohort of subjects with or at increased risk for type 1 diabetes (n = 183), participating in studies on the natural history of the disease at The University of Florida. The diagnosis of type 1 diabetes was according to American Diabetes Association (ADA) criteria, while the definition of ‘at risk’ was determined as previously described.31 Clear explanations of the objectives and implications of the results were given to each participant; subsequently, an institution-approved informed consent was signed. All study protocols were approved by the IRB of Rush University Medical Center or the University of Florida.
Antibodies and reagents
The following mAbs against human molecules were used: anti-CD3 fluorescein isothiocyanate (FITC), anti-CD3 peridin chlorphyll protein (PcP), anti-CD4 PcP, anti-CD4 allophycocyanin (APC), anti-CD8 FITC, anti-CD8 APC, anti-CD16 CyChrome, anti-CD25 APC, anti-CD27 FITC, anti-CD28 FITC, anti-CD38 APC anti-CD45RA FITC, anti-CD45RO APC, anti-CD56 CyChrome, anti-CD62L APC, anti-CD69 APC, anti-CD95 FITC, anti-CD154 APC, anti-CD161 APC, 6B11 phycoerythrin (PE; anti-invariant NKT cell TCR), anti-HLA-DR PcP, anti-interferon-γ (IFN-γ) APC, anti-IL-4 APC, anti-tumour necrosis factor-α (TNF-α) APC, and corresponding isotype control mAbs, all from Becton Dickinson-Pharmingen (San Jose, CA). FITC-labelled anti-Vβ11 and PE-labelled anti-Vα24 were from Coulter Immunotech (Marseille, France). PE-labelled human CD1d tetramers loaded with the α-GalCer analogue PB557 were from MHC Tetramer Core Facility (Emory University Vaccine Center, Atlanta, GA). Anti-LAG3 was obtained from Alexis/Axxora (San Diego, CA). FcγR blocking reagent was from Miltenyi Biotec (Bergisch Gladbach, Germany).
Isolation and activation of mononuclear cells
Peripheral blood was collected in heparin containing tubes, with peripheral blood mononuclear cells (PBMC) obtained by density gradient centrifugation using Lymphoprep (Nycomed, Oslo, Norway). Viability of cells was determined by trypan blue exclusion. For stimulation experiments, PBMC (1 × 106/ml) were suspended in RPMI culture medium supplemented with 10% (v/v) of heat-inactivated fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. PBMC were cultured in 15 ml polypropylene tubes, stimulated with PMA (50 ng/ml, Sigma, St Louis, MO) plus ionomycin (500 ng/ml, Sigma) and incubated at 37°/5% CO2/6 hr. Brefeldin A solution 1× (Becton Dickinson) was added the last 4 hr of culture. Cells incubated in medium alone were used as background controls.
Flow cytometry
Phenotypic analysis of iNKT cells was performed by four-colour flow cytometry, both in whole blood and PBMC. To stain cell surface molecules in whole blood, 100 µl of anticoagulated blood was incubated with the corresponding specific fluorescent mAbs for 20 min/room temperature (RT) in the dark. The erythrocytes were lysed by incubating for 10 min with 2 ml of 1× fluoescence-activated cell sorting (FACS) Lysing Solution (Becton Dickinson). The cell suspension was centrifuged for 5 min at 300 g, the supernatant was discarded and remaining cells washed twice with 2 ml of cold phosphate-buffered saline (PBS) at 300 g/5 min. Finally, the cells were fixed with 250 µl of 2% formaldehyde.
For extracellular staining of isolated PBMC (fresh or phorbol 12-myristate 13-acetate (PMA)/ionomycin-stimulated), non-specific binding of mAbs was inhibited by addition 10 µl of blocking reagent (Miltenyi Biotec) per 1 × 106 cells, for 20 min/4°. Then, PBMC were washed with 2 ml PBS, suspended in cytometry buffer (PBS with 0·5% bovine serum albumin (BSA) and 0·1% NaN3) at 1 × 107/ml. The mAbs were then added to the cell suspension in the required combination and at manufacturer specifications. The suspension was incubated for 20 min/RT in the dark. For intracellular staining, PBMC were incubated with 500 µl of 1× Permeabilizing Solution 2 (Becton Dickinson) for 10 min/RT/dark. Cells were then washed with cytometry buffer and stained for intracellular antigens incubating 30 min at RT in the dark. Finally, stained cells were washed with cytometry buffer, fixed with 2% formaldehyde and stored at 4° until analysis.
CD1d tetramer staining was performed following a previously described protocol.14 Briefly, PBMC were first incubated with anti-CD3 FITC, anti-CD4 PcP and anti-CD8 APC, as described above. The cells were then washed with FACS buffer and incubated at 4°/60 min with PB557-loaded human CD1d tetramers (20 µl of 1 : 50 dilution); after, PBMC were washed twice with FACS buffer and fixed as stated above.
For all staining experiments, appropriate isotype-matched controls were included. Because of the low frequency of iNKT cells in PBMC, 5–10 × 105 lymphocyte gated cells were analysed. Flow cytometry was performed using the FACSCalibur instrument and analysed with CellQuest software (Becton Dickinson).
Statistical analysis
Results for iNKT cell percentages are presented as mean ± standard deviation (SD). Statistical comparisons among the different groups were performed using the anova non-parametric Kruskal–Wallis test, with a confidence level of 95%. A P < 0·05 value was considered significant. The degree of association between the percentage of total iNKT cells and the percentage of CD4+, CD8+, and double positive (DP) or DN iNKT cells was calculated using a Spearman's non-parametric correlation coefficient.
Results
The term ‘natural killer T cells (NKT)’ has widely been used to define a broad range of T-cell subsets. Currently, it is thought that iNKT cells define the major population of CD1d-restricted T cells that may or may not express NK markers.2,13,32 There are at least two populations of CD1d-restricted NKT cells, which can be distinguished based on their TCR repertoire. The best-studied subpopulations are iNKT cells, also known as type-I NKT cells.32 Recently, we developed a mAb against the CDR3 loop of the invariant a chain of iNKT cell TCR, clone 6B11, which was utilized in this study to specifically identify and characterize human iNKT cells.
Specific evaluation of iNKT cell frequency in healthy donors
The frequencies of iNKT cells in whole blood and PBMC using mAbs against the TCR chains Vα24 and Vβ11, PB557-loaded human CD1d tetramers, and the mAb 6B11 alone or in combination with anti-CD3, were determined. Figure 1(a) shows representative plots from the analysis of iNKT cell frequency in PBMC. A similar frequency of iNKT cells was observed using the mAb 6B11 alone or in combination with anti-CD3, as well as with CD1d tetramers or the combination anti-Vα24/anti-Vβ11 (Fig. 1b). However, we observed that the combination anti-Vα24 and anti-Vβ11 detected a higher percentage of iNKT cells (0·06 ± 0·05%) when compared to human CD1d tetramers with anti-CD3 (0·04 ± 0·04%) and 6B11 with anti-CD3 (0·04 ± 0·03%); these differences were not statistically significant.
Figure 1.
Frequency of iNKT cells and iNKT cell subsets in peripheral blood. (a) Representative profiles of iNKT cell frequencies detected in PBMC using anti-Vα24 and anti-Vβ11, human CD1d tetramers or 6B11 alone or in combination with anti-CD3. These representative plots show a similar frequency detection of iNKT cells in a region comprising the lymphocytes (R1, created from forward vs. side scatter). (b) Comparative analysis of the iNKT cell frequency in peripheral blood from healthy adult subjects (n = 10), showing that a similar percentage of iNKT cells in each individual is detected with these three combinations of reagents (mean ± standard deviation for the different combinations of reagents: 6B11 with anti-CD3 = 0·036 ± 0·030; anti-Vα24 and anti-Vβ11 = 0·055 ± 0·054; human CD1d tetramers with anti-CD3 = 0·040 ± 0·035). (c) Representative FACS used to quantitate CD4+, CD8+, CD4–/CD8– (DN), and CD4+/CD8+ (DP) iNKT cell subsets (in 6B11+/CD3+ cells in this example). The percentage of iNKT cells expressing CD4 or CD8 was determined in a region (R2) comprising the 6B11+/CD3+ cells. (d) Analysis of the frequencies of iNKT cell subsets using anti-Vα24 and anti-Vβ11, human CD1d tetramers or 6B11 in combination with anti-CD3, and the co-expression of CD4 and CD8 (n = 10). No differences in the frequency of iNKT cell subsets detected with the different combinations of reagents were observed; the results are presented as the mean ± standard deviation.
In mice, iNKT cells are distributed into CD4+ and DN subsets.5 In contrast, CD8αα has been identified on human iNKT cells, and at least four different subsets of iNKT cells exist based on the expression of CD4 and CD8 molecules.4,14,33 Using the same combinations of mAbs or tetramers, four unique iNKT cell subsets defined as CD4+/CD8– (CD4+), CD4–/CD8+ (CD8+), double negative (DN) and double positive (DP) were observed (representative plots in Fig. 1c). There were no significant differences in the percentage of each subset using different antibody combinations or tetramers for the purpose of defining iNKT cell populations (Fig. 1d).
Using mAbs 6B11 and anti-CD3, the frequency of peripheral blood iNKT cells was determined for 90 healthy adult volunteers (Fig. 2a). The frequency of iNKT cells varied widely ranging from 0·01% to 0·92% (mean = 0·17% ± 0·19%). No significant gender differences were noted; however, female subjects had a modest trend towards higher frequencies of circulating iNKT cells (Fig. 2b, c; mean = 0·21% ± 0·21% for females, vs. mean = 0·14% ± 0·13% for males; P = 0·0631). To evaluate whether the heterogeneity observed in peripheral blood iNKT cell frequency varied with time, the frequency of circulating iNKT cells was evaluated for a subset of 12 individuals every 2 months for at least 1 year. Interestingly, the pattern of iNKT cell frequency in peripheral blood was stable in these healthy donors for the period of evaluation (data not shown).
Figure 2.
Frequency of iNKT cells in a large cohort of healthy adult population. The percentage of peripheral blood iNKT cells was determined by flow cytometry analysis in 90 healthy adult individuals (50 female, 40 male), detecting at the lymphocyte's gate the cells positive for both 6B11 and CD3. There were no significant differences in the iNKT cell frequency among the total population (0·17%± 0·19%), female individuals (0·21%± 0·21%) and male subjects (0·14%± 0·13%). However, a heterogeneous distribution in iNKT cell frequency was observed in all the individuals, ranging from 0·01% to 0·92%(a). The same pattern of distribution was observed for female (b) and male (c) subgroups. Considering the mean value for iNKT cell frequency in all the study population (0·17%), the female subgroup had a trend to have a higher number of individuals with a higher frequency of iNKT cells (44%) than the male subgroup (25%).
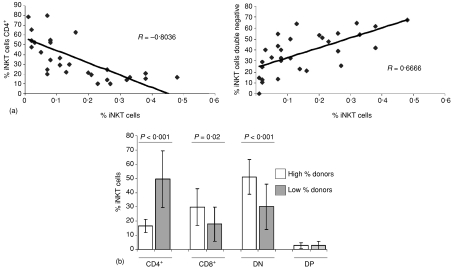
To confirm the finding previously published in a recent review34 we expanded the population of individuals evaluated to determine the frequency of the different iNKT cell subsets. In a randomly selected, sex balanced population of 30 normal donors (mean of iNKT cell frequency = 0·16% ± 0·13%), both the CD4+ and DN cells were the most frequent subsets and represented a similar percentage of iNKT cells (CD4+: mean = 37·7% ± 22·8%; DN: mean = 37·4% ± 17·7%), while CD8+ iNKT cells had an intermediate frequency (mean = 22·0% ± 13·4%) and DP cells were the least frequent iNKT subset (mean = 2·9% ± 2·4%). Notably, there was a direct linear and inverse correlation between the frequency of total iNKT cells and the percentage of CD4+ iNKT cells (R = −0·804, P < 0·0001; Fig. 3a, left panel), while a positive correlation between the frequency of iNKT cells and the percentage of DN iNKT cells (R = 0·666, P < 0·0001; Fig. 3b, right panel) was noted. Only a weak correlation between the frequency of iNKT cells and the percentage of CD8+ and DP iNKT cells was observed (R = 0·439, P = 0·0133 and R = −0·009, P = 0·9612, respectively).
Figure 3.
The percentage of CD4+ iNKT cells is inversely correlated to total iNKT cell frequency. The frequency of peripheral blood iNKT cells and their subsets were determined by flow cytometry in 30 healthy adults using mAbs 6B11 and anti-CD3, in combination with mAbs against CD4 and CD8 molecules. (a) The correlation analysis between the frequency of total iNKT cells and the percentage of iNKT cell subsets showed a negative correlation with the percentage of CD4+ iNKT cells (left panel) and a positive correlation with the percentage of DN iNKT cells (right panel). Based on this finding, these individuals were divided in two groups according to their mean frequency of iNKT cells (high-percentage donors, if iNKT cells were = 0·16%; and low-percentage donors, if iNKT cells were <0·16%). (b) Individuals with high frequency of iNKT cells had a significantly lower percentage of CD4+ iNKT cells and higher percentage of CD8+ and DN iNKT cells than subjects with low frequency of iNKT cells (P < 0·001, P = 0·02 and P < 0·001, respectively). The results are presented as the mean ± standard deviation of the percentage of iNKT cells expressing CD4 and/or CD8 molecules.
Further analysis of the data demonstrated that at least two groups of donors could be established according to the frequency of iNKT subsets and the total iNKT cells in peripheral blood, lower or higher than 0·16% of lymphocytes (i.e. the mean of the total iNKT cell frequency in this subgroup of 30 donors). Donors with total iNKT cells lower than 0·16% had a significantly higher percentage of CD4+ iNKT cells (P < 0·001), while those donors with a frequency of circulating iNKT cells greater than 0·16% demonstrated a significantly higher percentage of CD8+ and DN iNKT cells (P = 0·02 and P < 0·001, respectively; Fig. 3b). There was no significant difference in the percentage of DP iNKT cells between these two subgroups.
Given that CD4+ iNKT cells are thought to be immunoregulatory while the DN subset has a more pro-inflammatory and cytolytic phenotype, we hypothesized that it may be the CD4+ subset that is active in preventing autoimmunity while the DN could be pathogenic.1 To test whether this was the case, we used the mAb 6B11 to evaluate this question in patients at risk for or already having developed type 1 diabetes. Interestingly, there was a significant bias towards, and expansion of, DN iNKT cells in individuals at risk for type 1 diabetes (Table 1, P < 0·01).
Table 1.
Peripheral blood frequency of total iNKT cells and percentage of CD4+ and CD4 negative iNKT cells in a cohort of individuals from the DPT-1 trial
| Group | iNKT cell frequency* Mean (range) | CD4+ iNKT cells Mean | CD4– iNKT cells Mean |
|---|---|---|---|
| New onset1 DM (n = 37) | 0.085% (0·0–1·83) | 17·1% | 82·9% |
| Long-term2 DM (n = 49) | 0·11% (0·0–1·58) | 33·4% | 66·6% |
| Relatives3 (n = 17) | 0·07% (0·15–0·29) | 38·2% | 61·8% |
| High-risk4 individual (n = 33) | 0·21% (0·02–1·14) | 8·5% | 91·5% |
| Controls5 (n = 47) | 0·05% (0·02–0·54) | 47·4% | 52·6% |
DM: type 1 diabetes mellitus.
Total iNKT cell frequency defined as percentage of lymphocytes 6B11+/CD3+.
New onset: diabetes < 3 months of evolution.
Long-term: diabetes > 6 months of evolution.
First degree relative of a patient with type 1diabetes sharing at least one MHC allele.
High-risk subjects as defined and enrolled in the DPT-1 trial.
Controls were age and sex matched individuals.
Peripheral blood iNKT cells have a memory phenotype and express low levels of activation markers
The aforementioned experiments demonstrated that the mAb 6B11 was suitable in detecting iNKT cells in peripheral blood, as well as in subset analysis when used in combination with anti-CD4 and anti-CD8. Therefore, the 6B11 mAb was used to determine the expression of other phenotypic and functional molecules on resting peripheral blood iNKT cells (Fig. 4a and b). The majority of iNKT cells expressed the phenotypic T-cell markers CD27 (82·4% ± 8·4%) and CD28 (88·6% ± 6·7%). Regarding classical phenotypic NK-cell markers, a high percentage of iNKT cells expressed CD161 (84·9% ± 4·8%) while the frequency of iNKT cells expressing CD56 and CD16 was low (13·8% ± 7·2% and 3·4% ± 1·1%, respectively). As previously reported, a high percentage of peripheral blood iNKT cells were CD45R0+ (88·6% ± 6·4%), while a lower percentage were CD45RA+ (16·8% ± 7·3%). This memory phenotype corresponds predominantly to an effector type because just one-third of peripheral blood iNKT cells express CD62L (32·3% ± 12·8%), which suggests that most of the iNKT cells are programmed to migrate to peripheral tissues. Despite this memory phenotype, the expression of activation markers on peripheral blood iNKT cells was very low (CD25: 3·5% ± 2·8%; CD38: 2·9% ± 2·8%; CD95: 3·8% ± 2·6%; CD154: 0·28% ± 0·44%, and HLA-DR: 0·19% ± 0·02%), with a slightly higher expression of CD69 (10·6% ± 3·8%). There were no significant differences in the expression of these markers between female and male donors (data not shown).
Figure 4.
Phenotypic and functional surface markers expressed on resting peripheral blood iNKT cells. (a) Representative dot plots showing the expression of surface markers (percentage of positive cells, and MFI in those positive cells) on resting peripheral blood iNKT cells (6B11+ cells). (b) The frequency of iNKT cells expressing each surface marker was evaluated by flow cytometry staining using the mAb 6B11 (n = 10, 5 females and 5 males). While the expression of NK markers (CD16, CD56 and CD161) was variable, more than 80% of peripheral blood iNKT cells expressed CD27 and CD28 and have an effector memory phenotype (CD45ROhigh/CD62Llow). However, the expression of activation markers (CD25, CD38, CD69, CD95, CD154, HLA-DR) was very low. (c) Differences in surface markers expression by subsets of resting peripheral blood iNKT cells. 6B11 in combination with specific mAbs against CD4, CD8 and the surface molecules noted were used to evaluate cells gated on lymphocytes by FSC/SSC and defined as CD4+, CD8+, DN, or DP as in Figure 1. Very few DP events were detected for statistical analysis. The molecule CD27 was expressed by a significantly lower percentage of CD4+ iNKT cells, while a higher percentage of them expressed CD28, in comparison with CD8+and DN iNKT cells. CD45RA was expressed by a significantly higher percentage of CD8+ iNKT cells; CD25 was expressed by a significantly higher percentage of CD4+ iNKT cells; CD95 was expressed by a significantly lower percentage of DN iNKT cells, and CD161 was expressed by a significantly higher percentage of CD8+ and DN than CD4+ iNKT cells. The results are presented as the mean ± standard deviation of the percentage of iNKT cells expressing each molecule.
The same 14 markers were evaluated on subsets of iNKT cells as defined by CD4 and CD8, and some important differences were found between them (Fig. 4c). CD27 was expressed on a significantly lower percentage of CD4+ iNKT cells (75·0% ± 8·1%) than on CD8+ iNKT cells (89·4% ± 8·7%; P < 0·01); there were no significant differences in the expression of this molecule between DN iNKT cells (82·1% ± 11·1%) and CD4+ or CD8+ iNKT cells. No significant difference was observed in the expression of CD28 on CD4+ (94·5% ± 5·1%), CD8+ (80·5% ± 16·2%) or DN (88·5% ± 5·2%) iNKT cells. A significantly higher percentage of CD8+ iNKT cells expressed CD45RA (28·8% ± 11·6%) in comparison with DN iNKT cells (13·6% ± 5·5%; P < 0·01) but not with CD4+ (19·3% ± 8·5%) iNKT cells. Despite the low expression of activation markers by the total peripheral blood iNKT cells, CD25 was expressed by a significantly higher percentage of CD4+ iNKT cells (7·3% ± 4·5%) than CD8+ or DN iNKT cells (1·9% ± 2·1%, P < 0·01 and 2·2% ± 2·0%, P < 0·05, respectively). CD95 was expressed by a significantly lower percentage of DN iNKT cells (0·5% ± 0·6%) than CD4+ or CD8+ cells (8·4% ± 4·4, P < 0·001 and 7·6% ± 9·4%, P < 0·01, respectively). CD161 was expressed on a significantly higher percentage of CD8+ (89·3% ± 7·4%) and DN (92·6% ± 4·7%) than CD4+ (69·0% ± 7·8%) iNKT cells (P < 0·01 and P < 0·001, respectively).
Recently, the CD4 homologue LAG-3 was reported to represent an important molecule for regulating the expansion of activated T cells, the generation of natural regulatory T cells (Treg), and differentiation of dendritic cells, a notable effector function of iNKT cells.35,36 In gene array experiments, LAG3 expression was observed preferentially on DN iNKT cell clones in vitro.37–39 Using the mAb 6B11 and four colour FACS analysis, the expression of LAG3 was determined for iNKT cell subsets (Fig. 5a). LAG3 was found to be expressed by a higher percentage of singly CD8+ (15%) iNKT cells, while its expression was low on DN (7%) and CD4+ (7%) iNKT cells (Fig. 5a); despite the observed LAG3 expression on DP (50%) iNKT cells, the number of these cells is small and significance of these population frequencies should be interpreted with cautions (Fig. 5a). Interestingly, LAG3 was selectively expressed on CD8+ iNKT cells from the subjects with the lowest frequencies of this subset, and was modestly negatively correlated with the frequency of singly CD8+ iNKT cells (R = −0·277; Fig. 5c, lower panel). The expression of CD8α by human iNKT cells is thought to be in part a reflection of activation.4,14,40 These results suggest that the mechanisms regulating LAG3 expression in the distinct iNKT cell subsets might be different.
Figure 5.
LAG-3 expression on resting peripheral blood iNKT cells. Expression of the CD4 homologue LAG3 is associated with co-expression of CD8. (a) Representative FACS profiles for LAG3 expression, determined on CD4+, CD8+, DP and DN iNKT cell subsets. (b) LAG3 expression on iNKT cell subsets: cumulative data for an additional cohort of 13 normal donors. The results are presented as the mean ± standard deviation of the percentage of iNKT cells expressing LAG3. (c) The expression of LAG3 is inversely related to the percentage of CD8+ iNKT cell frequency.
Cytokine production by activated peripheral blood iNKT cells
A role for iNKT cells in the regulation of immune responses has been suggested by their capacity to rapidly release large amounts of cytokines upon activation, and in those studies an analysis of the cytokine profile represented a key point for the functional characterization of iNKT cells. Previous ex vivo analysis using CD1d tetramers demonstrated that CD4+ CD1d-restricted NKT cells were responsible for the selective secretion of T helper 2 (Th2)-like cytokines.14,41 Nonetheless, iNKT cells have been classically defined as Th0-like cells that can simultaneously produce both Th1 and Th2 cytokines.13,42,43 However, most of these previous results were obtained with in vitro expanded iNKT cell clones or cell lines, components that may not necessarily reflect the potential production of cytokines by peripheral blood iNKT cells while, in addition, their subset distinctions were limited to a CD4+ versus CD4– comparison. The combination of intracellular staining for cytokines with surface staining for the canonical TCR (with the mAb 6B11), combined with additional phenotypic markers (e.g. CD4 and CD8), might allow for a more precise definition of the cytokines produced by peripheral blood iNKT cell subsets.
To induce the production of cytokines by iNKT cells, PBMC were stimulated with PMA and ionomycin, and the production of IFN-γ, IL-4 and TNF-α determined by intracellular cytokine staining (Fig. 6a). There was a strong induction of IFN-γ and TNF-α (62·9% ± 16·0% and 68·6% ± 6·6%, respectively), and an intermediate level of IL-4 up regulation (18·8% ± 9·5%) following PMA/ionomycin stimulation. As a control for activation, the expression of CD69 was noted to increase sixfold from 10·6 ± 3·8% to 61·8 ± 7·9% positivity in iNKT cells (data not shown).
Figure 6.
Cytokine profiles in subsets of iNKT cells activated ex vivo. PBMC from healthy individuals were suspended in complete culture media and incubated 6 hrs with PMA (50 ng/ml) and ionomycin (500 ng/ml); to block the secretion of the proteins synthesized and the down regulation of the TCR and CD4 molecules in response to activation, Brefeldin A (10 μg/ml) was added to the culture during the last 4 hrs of incubation. Cells were then stained for relevant cell surface while the production of cytokines was analyzed by intracellular staining. (a) Representative dot plots show the up-regulation of intracellular IFN-γ, IL-4 and TNF-α by peripheral blood iNKT cells (6B11+ cells) incubated with PMA/Ionomycin. (b) Activation-induced intracellular cytokine profiles for iNKT cell subsets. There were no significant differences in the up-regulation of IFN-γ and TNF-α expression among the different subsets of iNKT cells, while a significantly higher percentage of CD4+ iNKT cells expressed IL-4, in comparison to CD8+ and DN iNKT cells. The results are presented as the mean ± standard deviation of the absolute percentage of iNKT cells expressing each molecule, determined as: (% iNKT cells positive in activated samples) – (% iNKT cells positive in unstimulated samples).
The PMA/ionomycin-induced synthesis of cytokines was also evaluated for the subsets of iNKT cells noted earlier (Fig. 6b). There were no significant differences in the production of IFN-γ among CD4+, CD8+ and DN iNKT cells (73·5% ± 12·2%, 66·3% ± 11·7% and 62·5 ± 15·8, respectively). Similar results were seen when the production of TNF-α was evaluated (CD4+: 66·9% ± 11·6%; CD8+: 56·3% ± 8·3%; DN: 65·7% ± 6·8%). Statistically significant differences were, however, observed in the synthesis of IL-4, this cytokine being detected in a higher percentage of CD4+ iNKT cells (16·7% ± 7·0%) compared to CD8+ cells (6·7% ± 3·2%, P < 0·05) and DN cells (6·4% ± 2·5%, P < 0·05). As a control for activation in the different iNKT subsets, the expression of CD69 was evaluated and equivalent expression of CD69 on all iNKT cell subsets was observed (data not shown).
Discussion
A variety of methods have been used to identify iNKT cells. In the current study, we found no significant differences in the frequency of total iNKT cells and their subsets in peripheral blood comparing 6B11, human CD1d tetramers and anti-Vα24/anti-Vβ11 mAbs. It is possible that noninvariant or non-CD1d-restricted Vα24+ cells can also pair with Vβ11 and increase the detectable population of Vα24+/Vβ11+ cells. Thus, the population defined by the conventional Vα24/Vβ11 staining may include a proportion of non-iNKT cells. Staining with 6B11 (alone or in combination) is specific for iNKT cells as this mAb recognizes the CDR3 loop of the invariant α chain from CD1d-restricted iNKT cells.
CD1d tetramer staining alone invariably includes 0·01–0·05% non-Vα24 cells;41 even coexpression of Vα24 and Vβ11 does not completely overlap with CD1d-tetramer reactivity in humans, and could be complicated by competition between CD1d tetramers and anti-TCR mAbs (e.g. anti-Vα24).28,29 Concurrent preincubation with anti-Vα24 has been used to block tetramer binding to iNKT cells, and thus identify non-invariant CD1d-restricted populations.14,28,29 In other reports, the frequency of iNKT cells detected in PBMC using 6B11 was approximately 95% of the tetramer reactive CD1-restricted T cells.19 Additional studies on NKT cells in humans have focused on the Vα24+ population,11,44 the CD3+ CD56+ cells45 or the CD3+ CD161+ cells46 from peripheral blood or pathology tissues. As the accurate detection of iNKT cells is the first step in their analysis, we suggest that the best phenotypic definition of iNKT cells should be based on the invariant TCR α chain rearrangement (Vα24Jα18), specifically detected by the 6B11 mAb. We propose that 6B11, alone or in combination with anti-Vα24, anti-Vβ11 or anti-CD3 is an excellent tool for analysis of human iNKT cells.
Our results confirm previous reports regarding the heterogeneity of iNKT cell frequency in peripheral blood from healthy humans.11,22,41 However, in comparison with the study by Sandberg et al. we observed a greater range of iNKT cell frequency determined as the percentage of lymphocytes (0·01% to 0·92%), with no significant difference in that frequency between females and males.22 There was a modest trend towards increased frequency of iNKT cells in female subjects. The differences between these results and those of Sandberg et al. are likely the result of the use of anti-Vα24 and anti-CD161 in their study; when using these mAbs, there was a large population of non-iNKT cells that expressed Vα24 and CD161, while CD161 can be detected on up to 20% of circulating lymphocytes.2,24
The factors that regulate the size of the circulating iNKT cell pool in vivo are poorly understood. In previous studies, the frequency of peripheral blood iNKT cells was generally stable over time.11,16 In the cohort studied here, longitudinal analysis of iNKT cell frequency in peripheral blood from 12 healthy individuals was stable over at least a 12-month period. However, in one other report, it was found that an environmental factor (an unidentified respiratory infection) modified the frequency of circulating iNKT cells in a normal donor.22 Additionally, human iNKT cells are thought to fluctuate in the peripheral blood of patients with a variety of organ-specific and systemic autoimmune conditions and cancer.44,47–50
Human iNKT cells have long been known to segregate into CD4+ and CD4– subsets, with distinct phenotypic and functional characteristics;14 however, there are few descriptions evaluating subsets of iNKT cells that express CD8.14,51,52 Using different combinations of mAbs, four well-defined iNKT-cell subsets were noted based on TCR coreceptor expression. In contrast to previous studies22 where the CD4+ subset was predominant (59·5%), a similar frequency of CD4+ and DN iNKT cells was noted. In addition, as we defined in a preliminary report34 there was a direct inverse relationship between the percentage of CD4+ and total iNKT cell frequency. The different mAbs used for iNKT cell detection may partially explain these differences.
An expansion of the CD4– subset of peripheral blood iNKT cells was reported for healthy individuals when the frequency of total iNKT cells was above 0·1%.22 Our results show that when iNKT cells are higher than 0·16%, the DN cells were predominant while the CD4+ subset predominated in individuals with total iNKT cells below this percentage. Consistent with other studies, the frequency of iNKT cell subsets appears to remain stable in healthy individuals. This is in contrast to cancer and the limited, but controversial results in autoimmunity.10,47–49,53–55 The expansion of CD4– iNKT cells seems to be extra-thymic and driven by enhanced proliferation of CD4– relative to CD4+ iNKT cells.56,57 We recently found that Toll-like receptor-9-dependent signalling resulted in the preferential expansion and survival of CD4– iNKT cells.58 Given the association of type 1 diabetes development and recurrent enteroviral infections, we speculate that the expansion of DN iNKT cells seen in this study in patients at risk for type 1 diabetes might be driven by such a mechanism. This is of potential interest because DN iNKT cells are thought to be proinflammatory and effectors of tumour and viral surveillance1,14,59–61 and the CD4+ subset has been strongly associated with immunoregulatory function of these cells. Moreover, the selective loss of CD4+ iNKT cells in non-obese deiabetic mice resulted in accelerated diabetes.69 It should be noted that the findings on iNKT cell frequency in patients at risk have been controversial. Perhaps this is related to the reagents and methodologies used.10,37,42,55,57 However, further longitudinal population and effector studies will be needed to evaluate the relationship of this finding to type 1 diabetes.
Several other phenotypic and functional markers have been used to characterize iNKT cells. The studies are particularly extensive for mice while much less is known about human iNKT cells. iNKT cell subsets are distinct from each other in their expression of surface molecules and receptors, suggesting that they might be targeted to different tissues and meant to exert different immune functions.14,19,21,22,62 The expression of 15 surface markers on resting peripheral blood iNKT cells were evaluated to determine the physiological characteristics of these cells, avoiding methods that may alter their expression patterns (e.g. expansion with cytokines and CD1d-restricted molecules such as α-GalCer). As a whole population, circulating iNKT cells have a variable expression of NK markers, with a low percentage bearing CD16 and CD56, and CD161 being the most frequently expressed molecule. In contrast, the majority of iNKT cells have a memory-effector phenotype because a high percentage of them expressed CD45RO and CD28, and a low percentage expressed CD62L. The expression of activation markers (e.g. CD25, CD38, CD69, CD154 and HLA-DR) was low, suggesting that peripheral blood iNKT cells are physiologically programmed to migrate to inflamed peripheral tissues where they become activated. A similar phenotypic pattern was also described for peripheral blood Vα24+/Vβ11+ cells from healthy controls22 and patients chronically infected with hepatitis C virus;11,44 however, hepatic iNKT cells from those patients were highly activated (high expression of CD69), suggesting that the inflammatory environment in this organ provided the signals required to activate iNKT cells.
Examination of expression of these markers on iNKT subsets revealed group-specific differences. Interestingly, CD25 was mainly expressed on CD4+ iNKT cells and the 6B11+ CD4+ cells represent 6 ± 4% of the CD4+ CD25+ T cells in PBMC. Previous studies reported a similar tendency22 but in some studies the CD4+ CD25+ iNKT cells were found at higher percentages:41 10% to 80% versus 7·2% ± 4·5 in this report. These differences might be caused by the exposure to activation signals dependent on the incubation with PMA/ionomycin or αGalCer-loaded tetramers, or in differences is study populations. An important part of the CD4+ CD25+ T cell pool are Treg cells63 and considering the proposed immunoregulatory role of iNKT cells, it will be important to clarify the relationship of CD4+ CD25+ iNKT cells to Treg cells.
CD4+ and CD8+ iNKT expressed similar amounts of CD95 (8%), but expression was negligible on DN iNKT cells, suggesting different roles of these subsets in the control of immune reactions. Both DN and CD8+ iNKT cells expressed significantly more CD161 than CD4+ iNKT cells, consistent with previous reports for the DN subset.13,14,64 No differences in the expression of CD56 and CD62L by resting peripheral blood iNKT cell subsets were noted. Other investigators observed that CD56 was expressed by CD4– cells, while CD4+ iNKT cells were CD62L+ and CD4– iNKT cells were CD62L negative.65 These differences are likely to be caused, in part, by study subjects and reagents used.
iNKT cells were further characterized functionally using a combination of intracellular staining for cytokines with surface staining for the canonical TCR (6B11). The preferential production of IL-4 by CD4+ and similar production of proinflammatory cytokines by CD4+, DN, and now extended to CD8+ iNKT cells, was similar to previously reported studies.14,41 In in vitro studies of cell lines and clones, both CD4+ and DN iNKT cells displayed a Th0-like profile and were capable of simultaneously producing both Th1 and Th2 cytokines.13,43,66 The reasons for the discrepancies between ex vivo and in vitro studies remain unclear but are likely caused by changes induced by in vitro expansion of iNKT cell clones or cell lines.
Taken together, these results support the use of 6B11 for studies of iNKT cells, identifying at least four peripheral blood iNKT cell subsets based on classic T-cell coreceptor expression. Moreover, CD8+ iNKT cells are functionally very similar to DN iNKT cells.52 Considering that the frequency of these subsets is heterogeneous with an inverse relation between the CD4+ and DN subsets, that CD4+ iNKT cells display a Th0 phenotype while CD8+ and DN cells follow a Th1 pattern, and that the frequency of these last subsets is correlated with an inflammatory history, it seems reasonable to propose that the predominance of one or more of the iNKT-cell subsets may influence the susceptibility or resistant to immune-mediated diseases.42,48–50,67,68
Acknowledgments
This work was supported in part by 5PO1 AI055793 for L.A-H., C.M., A.M.K., A.L.L., S.B.W., and 2RO1 AI 45051 to S.B.W. This study was also supported by the Colombian Agency for the Development of Science and Technology (Colciencias for C.J.M., recipient of a doctoral scholarship) and by the Committee for the Development of Research (CODI) from the University of Antioquia.
Abbreviations
- α-GalCer
α-galactosylceramide
- DN
double negative
- DP
double positive
- iNKT
CD1d-restricted T-cell receptor invariant natural killer T cells
- mAbs
monoclonal antibodies
- NK
natural killer cells
- PBMC
peripheral blood mononuclear cells
- SD
standard deviation
- TCR
T-cell receptor
- Treg
natural regulatory T cells
References
- 1.Wilson SB, Delovitch TL. Janus-like role of regulatory iNKT cells in autoimmune disease and tumor immunity. Nat Rev Immunol. 2003;3:211–22. doi: 10.1038/nri1028. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey DI, Hammond KJ, Poulton LD, Smyth MJ, Baxter AG. NKT cells: facts, functions and fallacies. Immunol Today. 2000;21:573–83. doi: 10.1016/s0167-5699(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, Paul WE. Cultured NK1.1+ CD4+ T cells produce large amounts of IL-4 and IFN-γ upon activation by anti-CD3 or CD1. J Immunol. 1997;159:2240–9. [PubMed] [Google Scholar]
- 4.Porcelli SA, Modlin RL. The CD1 system. antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 5.Bendelac A, Rivera MN, Park HS, Roark JH. Mouse CD1-specific NK1 T cells: development, specifity and function. Annu Rev Immunol. 1997;15:535–62. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 6.Kawano T, Cui J, Koezuka Y, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–9. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 7.Brigl M, Brenner MB. CD1. antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–90. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 8.Godfrey DI, Kronenberg M. Going both ways: immune regulation via CD1d-dependent NKT cells. J Clin Invest. 2004;114:1379–88. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawano T, Nakayama T, Kamada N, et al. Antitumor cytotoxicity mediated by ligand-activated human V alpha24 NKT cells. Cancer Res. 1999;59:5102–5. [PubMed] [Google Scholar]
- 10.Lee PT, Putnam A, Benlagha K, Teyton L, Gottlieb PA, Bendelac A. Testing the NKT cell hypothesis of human IDDM pathogenesis. J Clin Invest. 2002;110:793–800. doi: 10.1172/JCI15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucas M, Gadola S, Meier U, et al. Frequency and phenotype of circulating Valpha24/Vbeta11 double-positive natural killer T cells during hepatitis C virus infection. J Virol. 2003;77:2251–7. doi: 10.1128/JVI.77.3.2251-2257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh N, Hong S, Scherer DC, Serizawa I, Burdin N, Kronenberg M, Koezuka Y, Van Kaer L. Cutting edge. activation of NK T cells by CD1d and alpha-galactosylceramide directs conventional T cells to the acquisition of a Th2 phenotype. J Immunol. 1999;163:2373–7. [PubMed] [Google Scholar]
- 13.Exley M, Garcia J, Balk SP, Porcelli S. Requirements for CD1d recognition by human invariant Valpha24+ CD4– CD8– T cells. J Exp Med. 1997;186:109–20. doi: 10.1084/jem.186.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by tetramer staining. J Exp Med. 2002;195:625–36. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metelitsa LS. Flow cytometry for natural killer T cells: multi-parameter methods for multifunctional cells. Clin Immunol. 2004;110:267–76. doi: 10.1016/j.clim.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Dellabona P, Casorati G, Friedli B, Angman L, Sallusto F, Tunnacliffe A, Roosneek E, Lanzavecchia A. In vivo persistence of expanded clones specific for bacterial antigens within the human T cell receptor alpha/beta CD4– 8– subset. J Exp Med. 1993;177:1763–71. doi: 10.1084/jem.177.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4– 8– alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant V alpha 24-J alpha Q/V beta 11 T cell receptor is expressed in all individuals by clonally expanded CD4– 8– T cells. J Exp Med. 1994;180:1171–6. doi: 10.1084/jem.180.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas SY, Hou R, Boyson JE, et al. CD1d-restricted NKT cells express a chemokine receptor profile indicative of Th1-type inflammatory homing cells. J Immunol. 2003;171:2571–80. doi: 10.4049/jimmunol.171.5.2571. [DOI] [PubMed] [Google Scholar]
- 20.Eberl G, Lees RK, Smiley ST, Taniguchi M, Grusby MJ, MacDonald HR. Tissue-specific segregation of CD1d-dependent and CD1d-independent NK T cells. J Immunol. 1999;162:6410–9. [PubMed] [Google Scholar]
- 21.Stenstrom M, Skold M, Ericsson A, Beaudoin L, Sidobre S, Kronenberg M, Lehuen A, Cardell S. Surface receptors identify mouse NK1.1+ T cell subsets distinguished by function and T cell receptor type. Eur J Immunol. 2004;34:56–65. doi: 10.1002/eji.200323963. [DOI] [PubMed] [Google Scholar]
- 22.Sandberg JK, Bhardwaj N, Nixon DF. Dominant effector memory characteristics, capacity for dynamic adaptive expansion, and sex bias in the innate Valpha24 NKT cell compartment. Eur J Immunol. 2003;33:588–96. doi: 10.1002/eji.200323707. [DOI] [PubMed] [Google Scholar]
- 23.Exley M, Porcelli S, Furman M, Garcia J, Balk S. CD161 (NKR-P1A) costimulation of CD1d-dependent activation of human T cells expressing invariant V alpha 24 J alpha Q T cell receptor alpha chains. J Exp Med. 1998;188:867–76. doi: 10.1084/jem.188.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanier LL, Chang C, Phillips JH. Human NKR-P1A. A disulfide linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol. 1994;153:2417–22. [PubMed] [Google Scholar]
- 25.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–54. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidobre S, Kronenberg M. CD1 tetramers. a powerful tool for the analysis of glycolipid-reactive T cells. J Immunol Meth. 2002;268:107–21. doi: 10.1016/s0022-1759(02)00204-1. [DOI] [PubMed] [Google Scholar]
- 27.Exley MA, Tahir SM, Cheng O, Shaulov A, Joyce R, Avigan D, Sackstein R, Balk SP. A major fraction of human bone marrow lymphocytes are Th2-like CD1d-reactive T cells that can suppress mixed lymphocyte responses. J Immunol. 2001;167:5531–4. doi: 10.4049/jimmunol.167.10.5531. [DOI] [PubMed] [Google Scholar]
- 28.Gadola SD, Dulphy N, Salio M, Cerundolo V. Valpha24-JalphaQ-independent, CD1d-restricted recognition of alpha-galactosylceramide by human CD4 (+) and CD8alphabeta (+) T lymphocytes. J Immunol. 2002;168:5514–20. doi: 10.4049/jimmunol.168.11.5514. [DOI] [PubMed] [Google Scholar]
- 29.Brigl M, van den Elzen P, Chen X, et al. Conserved and heterogeneous lipid antigen specificities of CD1d-restricted NKT cell receptors. J Immunol. 2006;176:3625–34. doi: 10.4049/jimmunol.176.6.3625. [DOI] [PubMed] [Google Scholar]
- 30.Boyson JE, Rybalov B, Koopman LA, et al. CD1d and invariant NKT cells at the human maternal–fetal interface. Proc Natl Acad Sci USA. 2002;99:13741–6. doi: 10.1073/pnas.162491699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schatz D, Cuthbertson D, Atkinson M, et al. Preservation of C-peptide secretion in subjects at high risk of developing type 1 diabetes mellitus – a new surrogate measure of non-progression? Pediatr Diabetes. 2004;5:72–9. doi: 10.1111/j.1399-543X.2004.00047.x. [DOI] [PubMed] [Google Scholar]
- 32.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol. 2004;4:231–7. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 33.Kronenberg M, Gapin L. The unconventional lifestyle of NKT cells. Nat Rev Immunol. 2002;2:557–68. doi: 10.1038/nri854. [DOI] [PubMed] [Google Scholar]
- 34.Bollyky PL, Wilson SB. CD1d-restricted T-cell subsets and dendritic cell function in autoimmunity. Immunol Cell Biol. 2004;82:307–14. doi: 10.1111/j.0818-9641.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- 35.Workman CJ, Vignali DA. The CD4-related molecule, LAG-3 (CD223), regulates the expansion of activated T cells. Eur J Immunol. 2003;33:970–9. doi: 10.1002/eji.200323382. [DOI] [PubMed] [Google Scholar]
- 36.Huang CT, Workman CJ, Flies D, et al. Role of LAG-3 in regulatory T cells. Immunity. 2004;21:503–13. doi: 10.1016/j.immuni.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Wilson SB, Kent SC, Horton HF, Hill AA, Bollyky PL, Hafler DA, Strominger JL, Byrne MC. Multiple differences in gene expression in regulatory Valpha 24Jalpha Q T cells from identical twins discordant for type I diabetes. Proc Natl Acad Sci USA. 2000;97:7411–6. doi: 10.1073/pnas.120161297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson SB, Byrne MC. Gene expression in NKT cells: defining a functionally distinct CD1d-restricted T cell subset. Curr Opin Immunol. 2001;13:555–61. doi: 10.1016/s0952-7915(00)00258-2. [DOI] [PubMed] [Google Scholar]
- 39.Strominger JL, Byrne MC, Wilson SB. Regulation of dendritic cell subsets by NKT cells. C R Biol. 2003;326:1045–8. doi: 10.1016/j.crvi.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 40.Kent SC, Hafler DA, Strominger JL, Wilson SB. Noncanonical Valpha24JalphaQ T cells with conservative alpha chain CDR3 region amino acid substitutions are restricted by CD1d. Hum Immunol. 1999;60:1080–9. doi: 10.1016/s0198-8859(99)00109-3. [DOI] [PubMed] [Google Scholar]
- 41.Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human Valpha24 natural killer T cells. J Exp Med. 2002;195:637–41. doi: 10.1084/jem.20011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson SB, Kent SC, Patton KT, et al. Extreme Th1 bias of invariant Valpha24JalphaQ T cells in type 1 diabetes. Nature. 1998;391:177–81. doi: 10.1038/34419. [DOI] [PubMed] [Google Scholar]
- 43.Hou R, Goloubeva O, Neuberg DS, Strominger JL, Wilson SB. Interleukin-12 and interleukin-2-induced invariant natural killer T-cell cytokine secretion and perforin expression independent of T-cell receptor activation. Immunology. 2003;110:30–7. doi: 10.1046/j.1365-2567.2003.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deignan T, Curry MP, Doherty DG, et al. Decrease in hepatic CD56 (+) T cells and V alpha 24 (+) natural killer T cells in chronic hepatitis C viral infection. J Hepatol. 2002;37:101–8. doi: 10.1016/s0168-8278(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 45.Norris S, Doherty DG, Collins C, McEntee G, Traynor O, Hegarty JE, O'Farrelly C. Natural T cells in the human liver. cytotoxic lymphocytes with dual T cell and natural killer cell phenotype and function are phenotypically heterogeneous and include Valpha24-JalphaQ and gamma-delta T cell receptor bearing cells. Hum Immunol. 1999;60:20–31. doi: 10.1016/s0198-8859(98)00098-6. [DOI] [PubMed] [Google Scholar]
- 46.Ishihara S, Nieda M, Kitayama J, et al. CD8+NKR-P1A+ T cells preferentially accumulate in human liver. Eur J Immunol. 1999;29:2406–13. doi: 10.1002/(SICI)1521-4141(199908)29:08<2406::AID-IMMU2406>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 47.van der Vliet HJ, von Blomberg BM, Nishi N, et al. Circulating V (alpha24+) Vbeta11+ NKT cell numbers are decreased in a wide variety of diseases that are characterized by autoreactive tissue damage. Clin Immunol. 2001;100:144–8. doi: 10.1006/clim.2001.5060. [DOI] [PubMed] [Google Scholar]
- 48.Sumida T, Sakamoto A, Murata H, et al. Selective reduction of T cells bearing invariant V alpha 24J alpha Q antigen receptor in patients with systemic sclerosis. J Exp Med. 1995;182:1163–8. doi: 10.1084/jem.182.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tahir SMA, Cheng O, Shaulov A, Koezuka Y, Bubley GB, Wilson SB, Balk SP, Exley MA. Loss of IFN gamma production by invariant NKT cells in advanced cancer. J Immunol. 2001;167:4046–50. doi: 10.4049/jimmunol.167.7.4046. [DOI] [PubMed] [Google Scholar]
- 50.Illes Z, Kondo T, Newcombe J, Oka N, Tabira T, Yamamura T. Differential expression of NK T cell V alpha 24J alpha Q invariant TCR chain in the lesions of multiple sclerosis and chronic inflammatory demyelinating polyneuropathy. J Immunol. 2000;164:4375–81. doi: 10.4049/jimmunol.164.8.4375. [DOI] [PubMed] [Google Scholar]
- 51.Kim CH, Butcher EC, Johnston B. Distinct subsets of human Valpha24-invariant NKT cells: cytokine responses and chemokine receptor expression. Trends Immunol. 2002;23:516–9. doi: 10.1016/s1471-4906(02)02323-2. [DOI] [PubMed] [Google Scholar]
- 52.Lin H, Nieda M, Rozenkov V, Nicol AJ. Analysis of the effect of different NKT cell subpopulations on the activation of CD4 and CD8 T cells, NK cells, and B cells. Exp Hematol. 2006;34:289–95. doi: 10.1016/j.exphem.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 53.van der Vliet HJ, Molling JW, Nishi N, et al. Polarization of Valpha24+ Vbeta11+ natural killer T cells of healthy volunteers and cancer patients using alpha-galactosylceramide-loaded and environmentally instructed dendritic cells. Cancer Res. 2003;63:4101–6. [PubMed] [Google Scholar]
- 54.van der Vliet HJ, von Blomberg BM, Hazenberg MD, et al. Selective decrease in circulating V alpha 24+V beta 11+ NKT cells during HIV type 1 infection. J Immunol. 2002;168:1490–5. doi: 10.4049/jimmunol.168.3.1490. [DOI] [PubMed] [Google Scholar]
- 55.Kukreja A, Cost G, Marker J, et al. Multiple immuno-regulatory defects in type-1 diabetes. J Clin Invest. 2002;109:131–40. doi: 10.1172/JCI13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baev DV, Peng XH, Song L, Barnhart JR, Crooks GM, Weinberg KI, Metelitsa LS. Distinct homeostatic requirements of CD4+ and CD4– subsets of Valpha24-invariant natural killer T cells in humans. Blood. 2004;104:4150–6. doi: 10.1182/blood-2004-04-1629. [DOI] [PubMed] [Google Scholar]
- 57.Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, Godfrey DI. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur J Immunol. 2005;35:1399–407. doi: 10.1002/eji.200425958. [DOI] [PubMed] [Google Scholar]
- 58.Montoya CJ, Jie HB, Al-Harthi L, et al. Activation of plasmacytoid dendritic cells with TLR9 agonists initiates invariant NKT cell-mediated cross-talk with myeloid dendritic cells. J Immunol. 2006;177:1028–39. doi: 10.4049/jimmunol.177.2.1028. [DOI] [PubMed] [Google Scholar]
- 59.Crowe NY, Coquet JM, Berzins SP, et al. Differential antitumor immunity mediated by NKT cell subsets in vivo. J Exp Med. 2005;202:1279–88. doi: 10.1084/jem.20050953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Exley MA, He Q, Cheng O, Wang RJ, Cheney CP, Balk SP, Koziel MJ. Cutting edge. Compartmentalization of Th1-like noninvariant CD1d-reactive T cells in hepatitis C virus-infected liver. J Immunol. 2002;168:1519–23. doi: 10.4049/jimmunol.168.4.1519. [DOI] [PubMed] [Google Scholar]
- 61.Levy O, Orange JS, Hibberd P, et al. Disseminated varicella infection due to the vaccine strain of varicella-zoster virus, in a patient with a novel deficiency in natural killer T cells. J Infect Dis. 2003;88:48–53. doi: 10.1086/378503. [DOI] [PubMed] [Google Scholar]
- 62.Hammond KJ, Pelikan SB, Crowe NY, et al. NKT cells are phenotypically and functionally diverse. Eur J Immunol. 1999;29:768–81. doi: 10.1002/(SICI)1521-4141(199911)29:11<3768::AID-IMMU3768>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 63.Scotto L, Naiyer AJ, Galluzzo S, et al. Overlap between molecular markers expressed by naturally occurring CD4+ CD25+ regulatory T cells and antigen specific CD4+ CD25+ and CD8+ CD28– T suppressor cells. Hum Immunol. 2004;65:297–306. doi: 10.1016/j.humimm.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 64.Exley M, Garcia J, Wilson SB, et al. CD1d structure and regulation on human thymocytes, peripheral blood T cells, B cells and monocytes. Immunology. 2000;100:7–47. doi: 10.1046/j.1365-2567.2000.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandberg JK, Fast NM, Palacios EH, et al. Selective loss of innate CD4+ Vα24 natural killer T cells in human immunodeficiency virus infection. J Virol. 2002;76:7528–34. doi: 10.1128/JVI.76.15.7528-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spada FM, Koezuka Y, Porcelli SA. CD1d-restricted recognition of synthetic glycolipid antigens by human natural killer T cells. J Exp Med. 1998;88:1529–34. doi: 10.1084/jem.188.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Araki M, Kondo T, Gumperz JE, Brenner MB, Miyake S, Yamamura T. Th2 bias of CD4+ NKT cells derived from multiple sclerosis in remission. Int Immunol. 2003;15:279–88. doi: 10.1093/intimm/dxg029. [DOI] [PubMed] [Google Scholar]
- 68.Akbari O, Faul JL, Hoyte EG, Berry GJ, Wahlstrom J, Kronenberg M, DeKruyff RH, Umetsu DT. CD4+ invariant T-cell-receptor+ natural killer T cells in bronchial asthma. N Engl J Med. 2006;354:1117–29. doi: 10.1056/NEJMoa053614. [DOI] [PubMed] [Google Scholar]
- 69.Chen YG, Chen J, Osborne MA, Chapman HD, Besra GS, Porcelli SA, Leiter EH, Wilson SB, Serreze DV. cd38 is required for the peripheral survival of immunotolerogenic CD4+ invariant NK T Cells in nonobese diabetic mice. J Immunol. 2006;177:2939–47. doi: 10.4049/jimmunol.177.5.2939. [DOI] [PubMed] [Google Scholar]